Abstract
Nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are important components of current combination therapies for human immunodeficiency virus type 1 (HIV-1) infection. However, their low genetic barriers against resistance development, cross-resistance, and serious side effects can compromise the benefits of the two current drugs in this class (efavirenz and nevirapine). In this study, we report a novel and potent NNRTI, VRX-480773, that inhibits viruses from efavirenz-resistant molecular clones and most NNRTI-resistant clinical HIV-1 isolates tested. In vitro mutation selection experiments revealed that longer times were required for viruses to develop resistance to VRX-480773 than to efavirenz. RT mutations selected by VRX-480773 after 3 months of cell culture in the presence of 1 nM VRX-480773 carried the Y181C mutation, resulting in a less-than-twofold increase in resistance to the compound. A virus containing the double mutation V106I-Y181C emerged after 4 months, causing a sixfold increase in resistance. Viruses containing additional mutations of D123G, F227L, and T369I emerged when the cultures were incubated with increasing concentrations of VRX-480773. Most of the resistant viruses selected by VRX-480773 are susceptible to efavirenz. Oral administration of VRX-480773 to dogs resulted in plasma concentrations that were significantly higher than those required for the inhibition of wild-type and mutant viruses. These results warrant further clinical development of VRX-480773 for the treatment of HIV infection in both NNRTI-naive and -experienced patients.
Standard HIV therapies consist of combinations of nucleoside reverse transcriptase (RT) inhibitors (NRTIs) and nonnucleoside reverse transcriptase inhibitors (NNRTIs) or protease inhibitors. Although they are generally effective and have resulted in reduced AIDS-related morbidity and mortality, none of them is curative. Treatment failures often occur when viruses that are resistant to one or more components of the regimens arise. Compared to the large numbers of drugs in the NRTI and protease inhibitor classes, the NNRTI class has only two drugs (efavirenz and nevirapine) in extensive use; delavirdine is rarely used because of its low efficacy and its three-times-per-day dosing requirement. There are two important issues that impact the continued use of efavirenz or nevirapine and call for newer drugs in this class: a low genetic barrier against resistance development and cross-resistance among approved NNRTIs (2).
In this report, we characterize a novel and potent nonnucleoside RT inhibitor of human immunodeficiency virus type 1 (HIV-1), designated VRX-480773, that resulted from lead optimization of a substituted triazole discovered from high-throughput screenings (6). It inhibits HIV-1 derived from the molecular clones carrying the RT mutations commonly observed in plasma samples of patients who failed efavirenz treatment. More importantly, VRX-480773 exhibits activity superior to those of efavirenz and nevirapine against a majority of clinical NNRTI-resistant HIV-1 isolates. In addition, VRX-480773 seems to impose a higher genetic barrier for resistance development than does efavirenz. A majority of the viruses selected by VRX-480773 can be inhibited by efavirenz, indicating that there is a low level of cross-resistance between these two NNRTIs. Pharmacokinetic analysis in dogs showed that it is orally bioavailable and reaches plasma concentrations above the 50% effective concentration (EC50) for both wild-type (wt) and mutant viruses. These data warrant further clinical development of VRX-480773 for its use in both naïve and NNRTI-experienced patients infected with HIV-1.
MATERIALS AND METHODS
Compounds.
VRX-387902 [N-(2-bromo-4-methylphenyl)-2-(5-methyl-4-phenyl-4H-1,2,4-triazol-3-ylthio)acetamide] was purchased from SPECS (Rijswijk, The Netherlands). VRX-480773 {2-[5-bromo-4-(4-cyclopropylnaphthalen-1-yl)-4H-1,2,4-triazol-3-ylthio]-N-(2-chloro-4-sulfamoylphenyl)acetamide} was synthesized at Valeant Research and Development. Efavirenz was obtained from the NIH AIDS Research and Reference Reagent Program. All test compounds were prepared from dimethyl sulfoxide (DMSO) stocks.
Cell lines, plasmids, and virus isolates.
HeLa-JC53 cells were obtained from David Kabat at the Oregon Health Sciences University (9). The HeLa-JC53-LTR-β-gal cell line was constructed by transduction of HeLa-JC53 cells with an HIV-1 vector carrying long terminal repeat (LTR)-β-galactosidase. The HeLa-JC53-LTR-Luci cell line was likewise constructed by transduction with an HIV-1 vector carrying LTR-luciferase.
The HIV-1 molecular clones pNL4-3 and pNL4-3.Luc.R−E− were obtained from the NIH AIDS Research and Reference Reagent Program (4). A panel of 20 HIV-1 isolates carrying NNRTI resistance mutations (Table 1) was constructed as described previously based on the molecular clone pNL4-3.Luc.R−E− (15).
TABLE 1.
Activities of VRX-480773 against HIV-1 derived from molecular clones carrying RT mutations found in viruses from patients experiencing efavirenz treatment failure
| RT mutationa | % of patientsb | EC50 ± SD (nM)c
|
Mean change (fold) in EC50d
|
||
|---|---|---|---|---|---|
| VRX-480773 | Efavirenz | VRX-480773 | Efavirenz | ||
| wt | 0.14 ± 0.032 | 0.17 ± 0.006 | |||
| K103N | 88.5 | 0.09 ± 0.027 | 2.73 ± 1.774* | 0.6 | 16.1 |
| K103N-P225H | 32.7 | 0.13 ± 0.000 | 34.88 ± 34.958* | 0.9 | 205.2 |
| K103N-V108I | 28.8 | 0.14 ± 0.006 | 8.63 ± 0.750* | 1.0 | 50.8 |
| K103N-K101Q | 16.3 | 0.15 ± 0.017 | 12.66 ± 4.328* | 1.1 | 74.5 |
| K103N-L100I | 10.6 | 0.05 ± 0.023 | 258.37 ± 63.998* | 0.4 | 1,519.8 |
| G190S | 10.6 | 0.23 ± 0.068 | 14.27 ± 9.963* | 1.6 | 83.9 |
| K103N-V108I-P225H | 9.6 | 0.15 ± 0.017 | 77.06 ± 23.958* | 1.1 | 458.3 |
| Y188L | 6.7 | 0.81 ± 0.026* | 24.43 ± 8.130* | 5.8 | 143.7 |
| K101E | 4.8 | 0.26 ± 0.025* | 0.74 ± 0.275 | 1.9 | 4.4 |
| K103N-F227L | 4.8 | 0.08 ± 0.009 | 2.26 ± 1.178* | 0.6 | 18.3 |
| V106I-Y188L | 4.8 | 2.27 ± 0.512* | 135.57 ± 100.645* | 16.2 | 797.5 |
| G190A | 3.8 | 0.19 ± 0.009 | 0.48 ± 0.238 | 1.4 | 2.8 |
| K103N-Y188L | 3.8 | 1.07 ± 0.138* | >2,500* | 7.6 | Max |
| K103N-G190A | 3.8 | 0.32 ± 0.062* | 25.14 ± 8.701* | 2.3 | 147.9 |
| K103N-K101Q-P225H | 3.8 | 0.11 ± 0.007 | 78.39 ± 49.039* | 0.8 | 461.1 |
| K103N-V108I-A98G | 3.8 | 0.24 ± 0.002 | 67.93 ± 18.038* | 1.7 | 395.6 |
| A98G | 2.9 | 0.3 ± 0.055* | 0.64 ± 0.123 | 2.1 | 3.8 |
| K103N-Y181C | 2.9 | 0.23 ± 0.025 | 4.24 ± 1.871* | 1.6 | 24.9 |
| V106I | 1.9 | 0.09 ± 0.000 | 0.27 ± 0.056 | 0.6 | 1.6 |
| Y181C | 1.9 | 0.23 ± 0.029 | 0.33 ± 0.092 | 1.6 | 1.9 |
RT mutations were built upon wt pNL4-3.Luc.R−E−.
Appearance frequency among plasma HIV-1 samples from patients for whom combination therapies including efavirenz failed (1).
From two tests. *, EC50 was significantly higher than that against wild-type virus (P < 0.05).
Changes (n-fold) in EC50 relative to that against wt virus.
A panel of 94 clinical HIV-1 isolates carrying the most prevalent NNRTI-resistant mutations (Table 2) and 5 non-B wt clinical HIV-1 isolates (belonging to clades A, AC, C, D, and F) were generated at Monogram Biosciences Inc. (formerly ViroLogic, Inc., South San Francisco, CA) (12, 14). HIV-2(NIH-Z) was obtained from ABI Advanced Biotechnologies Inc. (Columbia, MD).
TABLE 2.
Activity of VRX-480773 against a panel of 94 clinical NNRTI-resistant HIV-1 isolates
| Isolate | NNRTI mutation(s) | % of NNRTI-resistant isolatesa | Change (fold) in EC50b
|
||
|---|---|---|---|---|---|
| VRX-480773 | Efavirenz | Nevirapine | |||
| 1 | 103N | 21.571 | 0.7 | 14.0 | 51.0 |
| 2 | 103N, 181C | 6.145 | 10.0 | >100 | >100 |
| 3 | 181C | 4.941 | 10.0 | 6.7 | >100 |
| 4 | 100I, 103N | 4.498 | 0.9 | >100 | 42.0 |
| 5 | 106I | 3.611 | 0.9 | 0.7 | 1.1 |
| 6 | 98G | 2.597 | 2.4 | 2.9 | 5.9 |
| 7 | 101Q | 2.534 | 2.2 | 2.5 | 2.8 |
| 8 | 103N, 225H | 2.407 | 1.6 | >100 | >100 |
| 9 | 103N, 225H | 2.407 | 1.0 | 89.0 | 93.0 |
| 10 | 188L | 2.312 | 11.0 | 86.0 | >100 |
| 11 | 103N, 108I | 2.249 | 1.3 | >100 | >100 |
| 12 | 179D | 1.932 | 3.3 | 5.7 | 4.2 |
| 13 | 190A | 1.457 | 13.0 | 18.0 | >100 |
| 14 | 181C, 190A | 1.457 | >100 | 14.0 | >100 |
| 15 | 103N, 190A | 1.425 | 76.0 | >100 | >100 |
| 16 | 101E | 1.362 | 11.0 | 16.0 | 42.0 |
| 17 | 108I | 1.362 | 1.5 | 1.7 | 2.3 |
| 18 | 103N, 181C, 190A | 1.172 | >100 | >100 | >100 |
| 19 | 101E, 190A | 1.109 | 83.0 | >100 | >100 |
| 20 | 103N, 238T | 1.077 | 0.6 | 31.0 | >100 |
| 21 | 98G, 103N | 1.014 | 4.4 | >100 | >100 |
| 22 | 101Q, 103N | 0.887 | 5.2 | >100 | >100 |
| 23 | 101P, 103N | 0.855 | 2.2 | >100 | >100 |
| 24 | 179E | 0.824 | 2.4 | 4.1 | 1.9 |
| 25 | 103N, 108I, 181C | 0.697 | 6.2 | 72.0 | >100 |
| 26 | 103N, 188L | 0.665 | 3.9 | >10.0 | >100 |
| 27 | 106A | 0.634 | 6.0 | 5.7 | >100 |
| 28 | 108I, 181C | 0.602 | 14.0 | 5.3 | >100 |
| 29 | 101E, 181C, 190A | 0.602 | >100 | >100 | >100 |
| 30 | 179A | 0.570 | 0.6 | 0.5 | 0.4 |
| 31 | 106A, F227L | 0.570 | >100 | >100 | >100 |
| 32 | 98G, 181C | 0.570 | 11.0 | 17.0 | >100 |
| 33 | 101Q, 181C, 190A | 0.570 | >100 | 43.0 | >100 |
| 34 | 190S | 0.507 | 3.9 | 97.0 | >100 |
| 35 | 101E, 190S | 0.475 | 16.0 | >100 | >100 |
| 36 | 190E | 0.443 | >100 | >100 | >100 |
| 37 | 106I, 188L | 0.443 | 62.0 | >100 | >100 |
| 38 | 103N, 106A | 0.412 | 2.6 | 26.0 | >100 |
| 39 | 179G | 0.348 | 0.3 | 0.2 | 0.1 |
| 40 | 227L | 0.348 | 0.47 | 0.80 | 2.10 |
| 41 | 103R, 179D, 188L | 0.348 | >100 | >100 | >100 |
| 42 | 101E, 181C | 0.317 | >100 | >100 | >100 |
| 43 | 103N, 106M | 0.317 | 12.0 | >100 | >100 |
| 44 | 103N, 106I | 0.317 | 1.8 | 28.0 | 122.0 |
| 45 | 103S, 190A | 0.285 | >100 | >100 | >100 |
| 46 | 181I | 0.285 | 5.4 | 1.4 | >100 |
| 47 | 179T | 0.285 | 1.3 | 0.8 | 0.5 |
| 48 | 101E, 181C, 190S | 0.285 | >100 | >100 | >100 |
| 49 | 101Q, 103R | 0.285 | 0.6 | 1.2 | 1.3 |
| 50 | 103R, 179D | 0.285 | 0.3 | 0.9 | 1.3 |
| 51 | 103N, 108I, 225H | 0.253 | 2.6 | >100 | >100 |
| 52 | 98G, 103N, 108I | 0.253 | 2.2 | >100 | >100 |
| 53 | 101P, 190A | 0.253 | 92.0 | >100 | >100 |
| 54 | 101R, 103N | 0.253 | 1.2 | 37.0 | >100 |
| 55 | 101Q, 190S | 0.253 | 2.4 | >100 | >100 |
| 56 | 98G, 190A | 0.253 | 16.0 | 64.0 | >100 |
| 57 | 106A, 190A | 0.253 | >100 | >100 | >100 |
| 58 | 101Q, 181C, 190S | 0.253 | >100 | >100 | >100 |
| 59 | 101E, 103N | 0.222 | 0.9 | 52.0 | >100 |
| 60 | 101Q, 181C | 0.222 | 8.9 | 4.4 | >100 |
| 61 | 236L | 0.222 | 0.9 | 0.2 | 0.5 |
| 62 | 101H, 190A | 0.222 | 32.0 | 77.0 | >100 |
| 63 | 98G, 188L | 0.222 | >100 | >100 | >100 |
| 64 | 98G, 103N, 181C | 0.190 | 4.2 | >100 | >100 |
| 65 | 101E, 108I, 181C, 190A | 0.190 | >100 | >100 | >100 |
| 66 | 103N, 108I, 238T | 0.190 | 2.5 | >100 | >100 |
| 67 | 98G, 100I, 103N | 0.190 | 1.4 | >100 | >100 |
| 68 | 100I, 103N, 108I | 0.190 | 1.9 | >100 | >100 |
| 69 | 103N, 188H | 0.190 | 3.5 | >100 | >100 |
| 70 | 188H | 0.190 | 1.6 | 8.7 | >100 |
| 71 | 98G, 101E, 181C, 190A | 0.190 | 65.0 | 71.0 | >100 |
| 72 | 103Q | 0.190 | 1.4 | 1.7 | 1.0 |
| 73 | 101H | 0.190 | 2.5 | 4.1 | 12.0 |
| 74 | 103N, 108I, 181C, 190A | 0.190 | >100 | >100 | >100 |
| 75 | 179D, 188L | 0.158 | >100 | >100 | >100 |
| 76 | 103N, 108I, 190A | 0.158 | 20.0 | >100 | >100 |
| 77 | 108I, 181C, 190A | 0.158 | >100 | 400 | >100 |
| 78 | 101Q, 103N, 225H | 0.158 | 3.9 | >100 | >100 |
| 79 | 98G, 103N, 181C, 190A | 0.158 | 82.0 | >100 | >100 |
| 80 | 103N, 190S | 0.158 | 18.0 | >100 | >100 |
| 81 | 106A, 181C | 0.158 | 16.0 | 8.2 | >100 |
| 82 | 98G, 108I, 181C | 0.158 | 15.0 | 47.0 | >100 |
| 83 | 238N | 0.158 | 0.7 | 0.8 | 1.9 |
| 84 | 238T | 0.127 | 2.2 | 3.0 | 100 |
| 85 | 103N, 238N | 0.127 | 2.3 | >100 | >100 |
| 86 | 103S | 0.127 | 2.7 | 8.6 | 39.0 |
| 87 | 230L | 0.127 | 11.0 | 16.0 | 46.0 |
| 88 | 101R, 103N, 181C, 190A | 0.127 | >100 | >100 | >100 |
| 89 | 101E, 103N, 190A | 0.127 | 50.0 | >100 | >100 |
| 90 | 101N, 103N | 0.127 | 0.6 | 26.0 | 32.0 |
| 91 | 181V | 0.127 | 20.0 | 4.5 | >100 |
| 92 | 106L, 188L | 0.127 | >100 | >100 | >100 |
| 93 | 179D, 181C | 0.127 | >100 | >100 | >100 |
| 94 | 100F | 0.127 | 1.8 | 1.6 | 2.4 |
| Total | 88.312 | ||||
Prevalance of mutation patterns was determined from sequence data in the Stanford HIV Resistance database (14).
Changes (n-fold) in EC50 relative to that against wt virus.
In vitro anti-HIV-1 activity assays.
The antiviral activity of VRX-480773 against NNRTI-resistant HIV-1 strains derived from both molecular clones and clinical isolates was evaluated. HeLa-JC53 cells were used for vesicular stomatitis virus G (VSV-G)-pseudotyped pNL4-3.Luc.R−E−-derived virus infections, while HeLa-JC53-LTR-Luci cells (expressing luciferase under the control of the HIV-1 LTR) were used for infections by HIV-2 (7, 9, 10, 13). Serial dilutions (1:4) of test compounds were performed in DMSO and then in culture medium (phenol red-free Dulbecco's modified Eagle's medium, 2% fetal bovine serum, and 1% penicillin-streptomycin) to yield a final DMSO concentration of 0.2% in the cell culture. Cells (15,000 cells in 85 μl of culture medium) were mixed with 5 μl of virus (multiplicity of infection of 0.03) and 10 μl of the serially diluted compounds in 96-well plates. All cultures were maintained at 37°C in 5% CO2 for 48 h. Following incubation, an equal volume (100 μl) of Bright-Glo luciferase reagent (Promega, Madison, WI) was added to each well, and chemiluminescence was read on an LJL Analyst (LJL BioSystems, Sunnyvale, CA). EC50 values (defined as the concentration of an inhibitor required to inhibit luciferase activity by 50%) were determined by nonlinear regression using XLfit4 or GraphPad Prism 4. One-way analysis of variance to compare mutant EC50 values to wild-type virus EC50 values was performed with GraphPad Prism4 after logarithmic transformation of the EC50 values.
The antiviral activity of VRX-480773 against a panel of 94 clinical HIV-1 isolates carrying NNRTI-resistant mutations and 5 wt non-B clinical HIV-1 isolates (belonging to clades A, AC, C, D, and F) was evaluated by Monogram Biosciences Inc. (South San Francisco, CA) (8).
In vitro cellular toxicity assay.
The cellular toxicity of VRX-480773 against a panel of tissue culture cell lines (HeLa, MT-2, SupT1, Panc 10.05, Hep3B, and ACHN) was determined by measuring cellular ATP levels in the presence of the compound. Cells (10,000 to 15,000 cells/well) were cultured with VRX-480773 in culture medium (phenol red-free Dulbecco's modified Eagle's medium or RPMI-1640 medium, 10% fetal bovine serum, and 1% penicillin-streptomycin) for 48 h at 37°C and 5% CO2, after which time an equal volume of CellTiter-Glo reagent (Promega, Madison, WI) was added to each well, and chemiluminescence was determined using an LJL Analyst system. The CC50 was defined as the concentration of inhibitor required to decrease cellular ATP levels by 50%.
In vitro RT assay.
HIV-1 RT was expressed in Escherichia coli and purified according to a procedure described previously by Boretto et al. (3). Expression plasmid p66RTB was a gift of B. Canard. Inhibition of HIV-1 RT was performed as described previously (15). Briefly, in vitro RT reactions were carried out for 1 h at 25°C in the presence of 16 μg/ml poly(rA)/oligo(dT)18, 2 μM TTP (labeled with 0.5 μCi of α-33P), 1 nM RT, and 0 to 100 μM inhibitor in a buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, and 60 μg/ml bovine serum albumin. Equal volumes of 20% trichloroacetic acid-1% sodium pyrophosphate were added, and radioactivity in the precipitated product was analyzed. The 50% inhibitory concentration was defined as the concentration of inhibitor required to inhibit RT activity by 50%.
Selection and determination of VRX-480773 resistance mutations.
SupT1 cells (2 × 106 cells in 1 ml of RPMI 1640 containing 10% fetal bovine serum) were exposed to wt NL4.3 virus (multiplicity of infection of 0.05) for 3 h. The virus culture was subsequently maintained in 1 ml of growth medium containing 1 nM VRX-480773 or 2 nM efavirenz. Every 3 to 4 days, 100 μl of infected culture was transferred into 900 μl of medium containing fresh drug and 9 × 105 SupT1 cells. Virus replication was monitored microscopically by observing the formation of syncytia. At each virus breakthrough (massive syncytium formation), the concentration of inhibitor was doubled. Culture media and cell pellets from each breakthrough point were collected. Cellular DNA was purified with a Wizard genomic DNA isolation kit (Promega, Madison, WI). The protease and RT coding regions of proviruses were amplified using high-fidelity Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA). The entire protease and RT coding sequences of breakthrough viruses were determined by sequencing 10 to 15 individually recovered PCR clones at each point.
Construction of VRX-480773-resistant viruses.
The major patterns of HIV-1 RT mutations selected by VRX-480773 from the wt virus were engineered into the parental pNL4-3.Luc.R−E− molecular clone by exchanging a 1485-bp ApaI/AgeI RT fragment (nucleotides 2006 to 3490 in NL4.3) with recovered mutant PCR fragments. The resulting plasmids were sequenced throughout the insertion region. Virus stocks with the RT mutations were generated by transfection as previously described (15). The resulting viruses were tested for their susceptibilities to VRX-480773 and efavirenz.
Determination of replication capacities of VRX-480773-selected viruses.
The replication capacities (RCs) of viruses carrying VRX-480773-selected RT mutations relative to that of wt virus were studied in the absence of inhibitor in a single-round infection assay. Transfection-generated, VSV-G-pseudotyped viruses (normalized to 50 ng/ml p24) were used to infect 1.5 × 104 HeLa-JC53 cells. Infected cells were cultured at 37°C and 5% CO2 for 48 h. Following incubation, an equal volume of Bright-Glo luciferase reagent was added to each well, and chemiluminescence was read using an LJL Analyst system. The RC of the mutant viruses was defined as the percent luciferase activity relative to that of the wt virus control.
Determination of EC50 and EC90 values of VRX-480773 in 100% human serum.
The EC50 and EC90 values of VRX-480773 against wt and mutant Y188L viruses in 100% human serum were derived by fitting the increases in EC50 and EC90 values associated with increasing serum concentrations to a one-site binding model (hyperbola) and extrapolation to 100% serum using GraphPad Prism 4. HeLa-JC53 cells (1.5 × 104 cells in 35 μl of phenol red-free Dulbecco's modified Eagle's medium containing 2% fetal bovine serum and 1% penicillin-streptomycin) were mixed with 5 μl of VSV-G-pseudotyped wt or Y188L NL4-3.Luci.R−E− virus (multiplicity of infection of 0.03) and 10 μl of serially diluted VRX-480773. The cell-virus compound mixture was then mixed with heat-inactivated human serum (Bioreclamation Inc., Meadow, NY) to give final concentrations of human serum of 0, 5, 10, 20, 30, 40, and 50% in the culture medium in a final volume of 100 μl. Cultures were incubated at 37°C in 5% CO2 for 48 h. Following incubation, an equal volume of Bright-Glo luciferase reagent (Promega, Madison, WI) was added to each well, and chemiluminescence was read using an LJL Analyst system (LJL BioSystems, Sunnyvale, CA). The EC50 and EC90 values at different human serum concentrations were determined by nonlinear regression using GraphPad Prism 4.
Pharmacokinetics of VRX-480773 in dogs.
VRX-480773 was given to dogs intravenously at 10 mg/kg or orally at 20 mg/kg. Plasma samples were collected at predose and at 0.25, 1, 2, 3, 4, 6, 8, 12, and 24 h following administration. Bioanalytical methods have been validated for quantification of VRX-480773 in plasma containing tripotassium EDTA (K3EDTA) as an anticoagulant. [D6]VRX-480773 was used as internal reference standard. Plasma proteins were precipitated with acetonitrile. The extract was evaporated to dryness, reconstituted, and analyzed by high-performance liquid chromatography with tandem mass spectrometry. An API 4000 triple-quadruple mass spectrometer, operated in positive electrospray ion mode, was used to monitor the precursor→product ion transitions of m/z 592.0→386.0 and 598.0→392.0 for VRX-480773 and [D6]VRX-480773, respectively. The methods were linear over the concentration range of 1.00 to 1,000 ng/ml with a validated lower limit of quantitation of 1.00 ng/ml. Intra- and interbatch assay accuracy (percentage of nominal) and precision (percent coefficient of variation) were within ±15% for all levels and <±20% for the lower limit of quantitation, respectively.
RESULTS
Discovery of VRX-480773 as a novel and potent HIV-1 RT inhibitor.
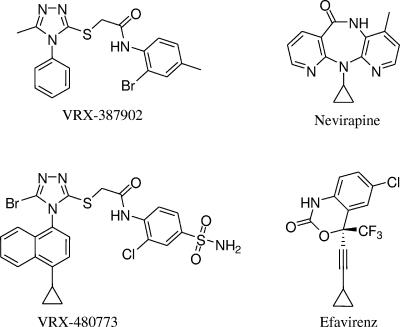
The small molecule VRX-387902 (Fig. 1) was originally discovered from a cell-based high-throughput screen for inhibitors of HIV-1 replication. Although VRX-387902 was a moderate inhibitor of wt HIV-1, with an in vitro EC50 value of 0.1 μM, it had reduced activity against an NNRTI-resistant virus containing the K103N-Y181C mutation (EC50 = 1.3 μM) (6). Lead optimization to increase potency against NNRTI-resistant mutants led to the synthesis of VRX-480773. VRX-480773 exhibited greatly increased in vitro activity, with wild-type EC50 values of 0.14 nM and K103N-Y181C mutant EC50 values of 0.23 nM (Table 1), and low cytotoxicity (CC50 of 3 to >100 μM). VRX-480773 was specific for HIV-1 inhibition, as it did not inhibit hepatitis B virus or hepatitis C virus replication, and, similar to other NNRTIs, it did not inhibit an HIV-2(NIH-Z) isolate (data not shown). However, VRX-480773 inhibited five non-B HIV-1 isolates (belonging to clades A, AC, C, D, and F) with an activity similar to that against wt clade B isolate NL4-3 (data not shown). In addition, enzymatic experiments demonstrated that VRX-480773 inhibits HIV-1 RT activity with wild-type 50% inhibitory concentration value of 4.0 nM, while it did not inhibit human DNA polymerases α and β at up to 45 μM (data not shown). Taken together, these results indicate that VRX-480773 is a potent and specific HIV-1 RT inhibitor.
FIG. 1.
Chemical structures of VRX-387902, VRX-480773, nevirapine, and efavirenz.
Activity of VRX-480773 against efavirenz-resistant HIV-1 derived from molecular clones.
The anti-HIV-1 activity of VRX-480773 against the wt and a panel of 20 HIV-1 isolates (Table 1) that carry the HIV-1 RT gene mutations most frequently observed in patients who have failed efavirenz-containing regimens was evaluated (1). As shown in Table 1, VRX-480773 and efavirenz showed similar activities against wt virus. However, VRX-480773 was active against most (18 of 20) mutant isolates with EC50 values of less than 1.0 nM, while efavirenz was active against only five isolates at similar EC50 levels. More importantly, VRX-480773 was active against viruses that are most prevalent and highly resistant to efavirenz, such as those that carry RT mutations K103N, G190S, K103N-L100I, K103N-K101Q, K103N-V108I, and K103N-P225H. This result suggests that VRX-480773 may be used in both NNRTI-naïve patients and patients who are experiencing efavirenz treatment failure.
Activity of VRX-480773 against clinical HIV-1 isolates carrying NNRTI resistance mutations.
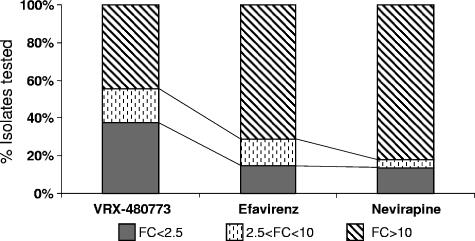
The Stanford HIV Resistance database contains 3,157 HIV-1 sequences associated with NNRTI resistance (12, 14). These sequences can be clustered into 218 different patterns of amino acid changes in the RT gene, with the K103N mutation being the most prevalent one (681 sequences or 21.6% of the total). The activity of VRX-480773 against a panel of 94 clinical isolates containing the most prevalent NNRTI resistance mutation patterns was tested (Table 2). These 94 mutant viruses represent 88.3% of the total NNRTI resistance isolates (prevalence) in the Stanford HIV Resistance database. As summarized in Fig. 2, there are significantly more isolates in this panel that are susceptible (<2.5-fold change) to VRX-480773 than to the current NNRTIs efavirenz or nevirapine (37% versus 15% or 14%, respectively). This represents 49% of the total NNRTI-resistant isolates present in the Stanford database that can be effectively inhibited (<2.5-fold change) by VRX-480773, compared to only 10.6% and 8.6% for efavirenz and nevirapine, respectively. Conversely, significantly fewer isolates showed high resistance (>10-fold change) to VRX-480773 than to efavirenz or nevirapine (45% versus 71% or 82%, respectively, representing 20%, 65%, and 72.5% of the total NNRTI-resistant isolates in the Stanford database, respectively) (Fig. 2). Table 3 further breaks down the 94 isolates into those with less-than- or greater-than-10-fold changes in resistance against VRX-480773 and efavirenz. Among the 67 isolates with which efavirenz has a greater-than-10-fold reduction in potency, 28 isolates are susceptible to VRX-480773 (with less than a 10-fold change in potency). Although there were 39 mutants with a greater-than-10-fold change in resistance against both VRX-480773 and efavirenz, viruses with these mutation patterns represent less than 20% of the total NNRTI-resistant isolates in the Stanford database (Table 3). The susceptibility or resistance of viruses containing the remaining 125 mutation patterns (11.7% of the isolates) is not known and requires further testing.
FIG. 2.
Profiles of activity of VRX-480773, efavirenz, and nevirapine against 94 clinical NNRTI-resistant HIV-1 isolates. FC, change (n-fold) in EC50 relative to EC50 against wt virus.
TABLE 3.
Cross-resistance of 94 clinical HIV-1 isolates between efavirenz and VRX-480773
| Efavirenz fold change in EC50 value | No. (%) of isolates with VRX-480773 fold change in EC50 value ofa:
|
|
|---|---|---|
| <10 | >10 | |
| <10 | 24 (22.4) | 3 (0.9) |
| >10 | 28 (45.9) | 39 (19.1) |
Numbers in parentheses correspond to the percentage of total isolates in the Stanford database.
Mutation selection and durability of VRX-480773 suppression.
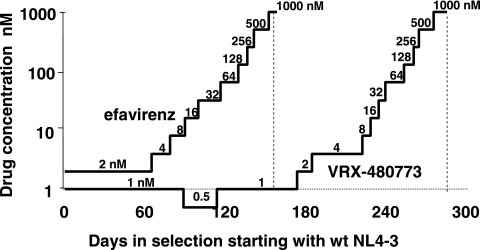
Wild-type virus strain NL4-3 was cultured in the presence of increasing concentrations of VRX-480773 or efavirenz to determine the times required for viral breakthrough and resistance pathways. This experiment also allowed us to compare the genetic barriers imposed by VRX-480773 and efavirenz against resistance development in cell culture conditions. As shown in Fig. 3, VRX-480773 exhibited a more durable pressure on the virus than efavirenz. It took 180 days for mutant viruses to emerge and break through the initial selection of VRX-480773 at 1 nM. In contrast, mutant virus emerged in 60 days under the selection of 2 nM efavirenz (Fig. 3). Note that the virus replicated so slowly in the presence of VRX-480773 that we had to drop the initial concentration of VRX-480773 from 1 nM to 0.5 nM after 90 days of selection. This 0.5 nM concentration was kept for 30 days to allow the recovery of virus replication (Fig. 3).
FIG. 3.
Time courses of resistance mutation selection for VRX-480773 and efavirenz. Wild-type NL4.3 was used to infect SupT1 cells (2 × 106 cells in 1 ml of RPMI 1640 medium containing 10% fetal bovine serum) in the initial presence of 1 nM VRX-480773 or 2 nM efavirenz. Subsequently, every 3 to 4 days, 100 μl of infected culture was transferred into 900 μl of medium containing fresh drug and 9 × 105 SupT1 cells. Virus replication was monitored microscopically by observing the formation of syncytia. At each virus breakthrough (massive syncytium formation), the concentration of the inhibitor was doubled.
To determine the mutation pathway to VRX-480773 resistance from wild-type virus, the sequences of 10 to 15 proviral DNA clones at each breakthrough concentration point were examined. Table 4 lists the individually cloned sequences. Although it needs to be further confirmed, the virus seemed to evolve in the presence of increasing concentrations of VRX-480773 through the following pathway: wt to Y181C to V106I-Y181C to V106I-Y181C-F227L to V106I-D123G-Y181C-F227L or V106I-Y181C-F227L-T369I to V106I-D123G-Y181C-F227L-T369I and, finally, to V106I-D123G-V179D-Y181C-F227L-T369I (Table 4). Notably, the D123G and T369I mutations are two mutations that have not previously been reported to be associated with NNRTI resistance.
TABLE 4.
VRX-480773-selected RT mutations starting from wt HIV-1(NL-4.3)
| Concn (nM) | Clone | Known NNRTI resistance mutation(s) | Novel mutation(s) | Other mutation(s) |
|---|---|---|---|---|
| 2 | 181 | E240G, V559I | ||
| 183 | E240G, T131A | |||
| 185 | G18D, A371T | |||
| 186 | Y181C | V241I, A437T | ||
| 187 | P176L | |||
| 188 | K220E, V245A, A376V, Q509R, S515L | |||
| 189 | V106I | D123G | D471N | |
| 231 | V118I, Q207R, V254I | |||
| 233 | G15R, T470A | |||
| 235 | G555R | |||
| 237 | V10I, E300G, A327T | |||
| 238 | Y181C | A129V, K259R, V518I | ||
| 240 | F116S | |||
| 243 | E240G, A446V | |||
| 3 | 280 | Y181C | ||
| 283 | Y181C | L303P, E430K, I556T | ||
| 285 | Y181C | C162Y | ||
| 287 | I37N | |||
| 288 | Y181C | R172K, E204G, E328G | ||
| 290 | Y181C | A445V | ||
| 292 | V8I, K82R, V189I, Q407R | |||
| 293 | D121N, H221R | |||
| 296 | Y181C | E29K, V559I | ||
| 297 | D123G | E194K, P225S, V276A, A304T, K395R | ||
| 298 | Y181C | |||
| 300 | V8I, Q197R, D250G | |||
| 301 | Y181C | V108A | ||
| 32 | 205 | Y181C | T409A | |
| 213 | V179A | E204G | ||
| 214 | E204G | |||
| 251 | Y181C | |||
| 252 | Y181C | V276I, E291G, V317A | ||
| 253 | F214L | |||
| 254 | Y181C | |||
| 255 | V261M, E291G, P313S, E430G | |||
| 256 | Y181C | K43R, E404G, P433L, A446V | ||
| 259 | V106I, Y181C | D123G | ||
| 260 | Y181C | D123G | K43R | |
| 261 | V106I, Y181C | Q242R, D471N | ||
| 262 | V106I, Y181C | |||
| 64 | 151 | V106I, Y181C, F227L | T369I | D471N |
| 153 | V106I, Y181C, F227L | T369I | K103R, CI62R, K424R, S519N | |
| 155 | V106I, Y181C | D123G | ||
| 156 | V106I, Y181C, F227L | D123G | ||
| 157 | V106I, Y181C, F227L | D123G, T369I | ||
| 158 | V106I, Y181C, F227L | T369I | T296A, I326M | |
| 159 | V106I, Y181C, F227L | T369I | E204G, A267T | |
| 160 | V106I, Y181C | D123G, T369I | E204G | |
| 161 | V106I, Y181C, F227L | T369I | A408T, Q524R | |
| 162 | V106I, Y181C, F227L | D123G, T369I | ||
| 163 | V106I, Y181C, F227L | D123G, T369I | M184V | |
| 128 | 61 | V106I, Y181C, F227L | D123G, T369I | |
| 62 | V106I, Y181C, F227L | D123G, T369I | F61S | |
| 63 | V106I, Y181C, F227L | D123G, T369I | W71R | |
| 65 | V106I, Y181C, F227L | T369I | T351A | |
| 66 | V106I, Y181C, F227L | |||
| 67 | V106I, Y181C | P4T, G99E, K353R, M357I, P412S | ||
| 68 | V106I, Y181C, F227L | D123G, T369I | P4T, H198Q | |
| 69 | V106I, Y181C, F227L | D123G, T369I | V189A | |
| 70 | V106I, Y181C, F227L | T369I | S513N | |
| 71 | V106I, Y181C, F227L | T369I | G40R | |
| 500 | 91 | V106I, Y181C, F227L | D123G, T369I | |
| 92 | V106I, Y181C, F227L | D123G, T369I | H221Y | |
| 93 | V106I, Y181C, F227L | D123G, T369I | Y188H, V467I | |
| 95 | V106I, Y181C, F227L | D123G, T369I | V118A, V467I | |
| 96 | V106I, Y181C, F227L | D123G, T369I | I257V, D488G, L551S | |
| 98 | V106I, Y181C | D123G | P236L | |
| 99 | V106I, Y181C, F227L | T369I | ||
| 100 | V106I, Y181C, F227L | T369I | ||
| 101 | V106I, Y181C, F227L | T369I | ||
| 102 | V106I, Y181C, F227L | D123G, T369I | P236L, I329T | |
| 103 | V106I, Y181C, F227L | T369I | ||
| 1,000 | 3 | V106I, V179D, Y181C, F227L | T369I | |
| 5 | V106I, Y181C, F227L | D123G, T369I | V314I, G384R | |
| 8 | V106I, Y181C, F227L | T369I | D460N | |
| 11 | V106I, V179D, Y181C, F227L | D123G, T369I | D17N | |
| 13 | V106I, Y181C, F227L | T369I | I495T | |
| 14 | V106I, Y181C, F227L | T369I | ||
| 15 | V106I, V179D, Y181C, F227L | D123G, T369I | ||
| 18 | V106I, Y181C, F227L | D123G, T369I | ||
| 23 | V106I, Y181C, F227L | T369I | ||
| 28 | V106I, V179D, Y181C, F227L | D123G, T369I | ||
| 31 | V106I, Y181C, F227L | D123G, T369I | K431R |
Resistance conferred by VRX-480773-selected mutations.
In order to determine the levels of resistance to VRX-480773 and to efavirenz conferred by VRX-480773-selected mutations, these mutations, individually or in combination, were engineered back into the wt NL4-3 molecular clone by site-directed mutagenesis. The resulting viruses were tested for their resistance to VRX-480773 and efavirenz and for their replication capacity in the absence of an inhibitor (Table 5). The single mutations V106I, D123G, Y181C, F227L, and T369I did not confer much resistance (<2-fold change) to VRX-480773. Multiple mutations were required before significant shifts in susceptibility were observed. Importantly, all mutant viruses remained susceptible to efavirenz (≤8-fold change). These data suggest a reduced level of cross-resistance between VRX-480773 and efavirenz.
TABLE 5.
Replication capacity and cross-resistance of VRX-480773-selected viruses
| VRX-480773-selected mutationa | % wt RCb | Mean change (fold) in EC50 ± SDc
|
|
|---|---|---|---|
| VRX-480773 | Efavirenz | ||
| wt | 100 | ||
| V106I | 54.56 ± 5.02 | 1.0 ± 0.11 | 0.7 ± 0.07 |
| D123G | 89.5 ± 19.09 | 1.0 ± 0.02 | 1.0 ± 0.03 |
| Y181C | 87.8 ± 10.96 | 1.9 ± 0.37 | 1.3 ± 0.03 |
| F227L | 114.5 ± 2.12 | 0.7 ± 0.09 | 0.5 ± 0.07 |
| T369I | 30.5 ± 3.46 | 1.3 ± 0.08 | 2.1 ± 0.07 |
| V106I-Y181C | 43.8 ± 10.25 | 5.4 ± 0.72* | 3.3 ± 1.05* |
| V106I-Y181C-F227L | 0.635 ± 0.09 | 18.7 ± 3.95* | 2.5 ± 0.90 |
| V106I-Y181C-F227L-T369I | 0.2 ± 0.00 | 111.9 ± 22.28* | 8.0 ± 1.72* |
| V106I-D123G-Y181C-F227L | 64.0 ± 14.14 | 9.1 ± 0.08* | 1.5 ± 0.18 |
| V106I-D123G-Y181C-F227L-T369I | 11.3 ± 2.40 | 31.2 ± 13.96* | 2.9 ± 0.09 |
Molecular clones were built in the reporter vector pNL4-3.Luc.R−E−, and EC50 values were determined by using Hela-JC-53 cells.
Virus replication capacity was determined in the absence of inhibitor.
Mean change (n-fold) in EC50 values ± standard deviations were from two tests. *, EC50 was statistically significantly higher than that against wild-type virus (P < 0.05).
Prediction of EC50 and EC90 values of VRX-480773 in 100% human serum.
The EC50 and EC90 values of VRX-480773 against wt and mutant Y188L NL4-3 viruses determined with 0 to 50% human serum in cell culture were used to project its EC50 and EC90 values in 100% human serum (Table 6 and data not shown). Using this method, the EC50 and EC90 of VRX-480773 in 100% human serum are predicted to be 23 nM and 68.5 nM, respectively, for wt virus. For Y188L mutant virus, the predicted EC50 and EC90 values in 100% human serum were 89 nM and 840.2 nM, respectively. Notably, the EC50 values of efavirenz were similarly affected by increasing amounts of human serum in culture medium (Table 6), indicating that the potency of these two inhibitors is similarly affected by binding to serum proteins. The EC50 values predicted for efavirenz in 100% human serum are 24 nM and 2,524 nM for wt and Y188L mutant viruses, respectively.
TABLE 6.
EC50 values of VRX-480773 and efavirenz under different concentrations of human serum
| Strain | Treatment | Mean EC50 (nM) ± SD under human serum concn in culture mediuma (%) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 20 | 30 | 40 | 50 | 100b | ||
| WT NL4-3 | VRX-480773 | 0.6 ± 0.21 | 2.4 ± 0.44 | 3.9 ± 0.92 | 7.1 ± 3.82 | 8.6 ± 3.10 | 9.5 ± 2.58 | 13.0 ± 1.98 | 23 ± 3.0 |
| Efavirenz | 0.8 ± 0.04 | 2.1 ± 0.76 | 3.5 ± 1.21 | 5.6 ± 1.38 | 8.5 ± 3.82 | 10.3 ± 3.80 | 12.8 ± 3.58 | 24.2 ± 0.07 | |
| Y188L NL4-3 | VRX-480773 | 3.7 ± 0.15 | 13.7 ± 2.60 | 20.7 ± 1.45 | 43.3 ± 1.95 | 45.9 ± 7.88 | 54.0 ± 2.28 | 79.9 ± 1.51 | 89.1 ± 0.07 |
| Efavirenz | 85.9 ± 1.56 | 298.5 ± 6.77 | 416.6 ± 41.55 | 814.9 ± 160.71 | 1,016.1 ± 107.10 | 1,382.1 ± 240.47 | 1,608.4 ± 22.34 | 2,524 ± 0.07 | |
Culture medium contained basal 2% fetal bovine serum.
EC50 values in 100% human serum were derived by fitting the increases in EC50 values associated with increasing serum concentrations to a one-site binding model (hyperbola) and extrapolation to 100% serum using GraphPad Prism 4.
Pharmacokinetics of VRX-480773 in dogs.
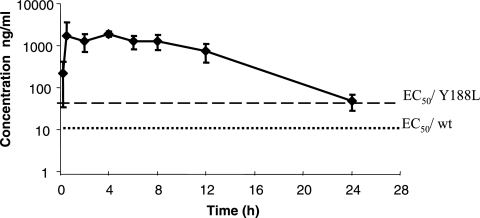
Pharmacokinetic studies demonstrated that VRX-480773 was rapidly absorbed in dogs. Following a single oral administration of 20 mg/kg, an average time to maximum concentration of drug in serum of 5.3 h and a maximum concentration of drug in serum of 1,980 ng/ml were observed (Table 7). The bioavailability was estimated to be 47% for VRX-480773. The plasma concentration-versus-time profile of VRX-480773 in dogs is presented in Fig. 4. At 12 h postadministration, a plasma concentration of VRX-480773 of 742 ng/ml (1,251 nM) was observed in dogs (Table 7). Based on the predicted EC50 values of VRX-480773 of 23 nM for wt virus and 89 nM for Y188L virus, the inhibitory quotient (IQ) (IQ = concentration of drug in plasma at 12 h/predicted EC50 in 100% human serum) for VRX-480773 given 20 mg/kg twice daily was calculated to be 54 for wt virus and 14 for Y188L virus (Table 7).
TABLE 7.
Pharmacokinetic parameters of VRX-480773 in dogs following single intravenous or oral administrations
| Parameter | Dosing
|
|
|---|---|---|
| Intravenous | Oral | |
| Dose (mg/kg) | 10 | 20 |
| No. of animalsa | 3 | 3 |
| Tmax (h)b | 0.28 (0.083-0.50) | 5.3 (4.0-8.0) |
| Cmax (ng/ml)c | 4,420 ± 465 | 1,980 ± 246 |
| T1/2 (h)d | 2.89 ± 0.490 | 3.36 ± 0.558 |
| AUClast (ng · h/ml)e | 20,900 ± 224 | 19,600 ± 4,480 |
| C12 h (ng/ml)f | 283 ± 73 | 742 ± 343 |
| IQwtg | 20.7 | 54.4 |
| IQY188Lh | 5.4 | 14.1 |
| Bioavailability (%)i | 47.1 | |
n = 3 for each time point with multiple groups due to limit of sample volume.
Tmax, time at which dosed drug reached maximum concentration in plasma.
Cmax, maximum concentration of dosed drug in plasma.
T1/2, elimination half-time of dosed drug in plasma.
AUClast, total area under concentration-time curve.
C12 h, drug concentration in plasma at 12 h postadministration.
IQwt = C12 h (nM)/23 nM (EC50 against wt virus predicted in 100% human serum).
IQY188L = C12 h (nM)/89 nM (EC50 against Y188L virus predicted in 100% human serum).
Bioavailability (percent) = (oral AUC0-∞/intravenous AUC0-∞) × 100.
FIG. 4.
Plasma concentration-versus-time profile of VRX-480773 following oral administration in dogs. EC50/wt and EC50/Y188L are the predicted EC50 values of VRX-480773 in 100% human serum for wt and Y188L HIV-1, respectively.
DISCUSSION
In this report, we have described VRX-480773 as being a novel nonnucleoside RT inhibitor of HIV-1. VRX-480773 was as potent as efavirenz against wt virus but was more potent than efavirenz against HIV-1 carrying the RT mutations most commonly observed in plasma samples from patients experiencing efavirenz treatment failure. More importantly, VRX-480773 was superior to efavirenz and nevirapine against a majority of NNRTI-resistant HIV-1 clinical isolates. In addition, the longer time for mutant viruses to break through suppression by VRX-480773 suggests that it has a higher genetic barrier for resistance development than efavirenz. Most of the resistant viruses selected by VRX-480773 can be inhibited by efavirenz, indicating a low level of cross-resistance between these two NNRTIs. Finally, levels of VRX-480773 that were higher than its EC50 values for inhibiting wt and mutant Y188L viruses were observed in dog plasma following oral administration. These data support the further clinical development of VRX-480773 for its use in both naïve and NNRTI-experienced patients infected by HIV-1.
The more durable suppression of virus replication exerted by VRX-480773 than that by efavirenz seems to stem from the more flexible nature of VRX-480773. In contrast to the rigid structures of efavirenz and nevirapine, VRX-480773 has multiple linkers connecting the “ring systems,” which allows them to rotate within the hydrophobic pocket of the HIV-1 RT to accommodate amino acid changes, similar to another NNRTI, TMC-125, which has been described as being able to bind the RT enzyme in more than one conformationally distinct mode (5). This improved molecular flexibility seems to enable the inhibitor to adapt to conformational changes due to mutations. Thus, single amino acid changes selected by VRX-480773, such as Y181C, V106I, or F227L, do not cause significant resistance to VRX-480773. The V106I-Y181C double mutation caused merely a 5.4-fold increase in resistance to VRX-480773 (Table 5).
During the in vitro selection for viruses that were resistant to VRX-480773, Y181C seemed to be the first single mutation selected; the other four mutations (V106I, D123G, F227L, and T369I) appeared in association with either this mutation or other unreported mutations in the RT (Table 4). Analysis of the replication and resistance patterns of the reconstituted viruses carrying these mutations suggests that V106I, F227L, and T369I appear to contribute directly to virus resistance, since adding V106I toY181C increased the resistance from 1.9- to 5.4-fold and decreased the RC from 88% to 44% (Table 5). Similarly, adding F227L to V106I-Y181C resulted in an increase in resistance from 5.4- to 18.7-fold, while dramatically decreasing the RC from 44% to 0.6% of that of wt virus. Adding T369I to V106I-Y181C-F227L further increased the resistance from 18.7- to 112-fold and further dropped the virus RC from 0.6% to 0.2%. Although T369I is clearly associated with increased resistance to VRX-480773, its mechanism is unclear, since this residue is located far from the NNRTI-binding pocket (11).
In contrast to the other mutations, the D123G mutation seems to play a compensatory role that increases the virus RC. Adding the D123G mutation to V106I-Y181C-F227L dramatically increased the virus RC from 0.6% to 64%, while the virus resistance decreased from 18.7- to 9-fold (Table 5). Similarly, adding the D123G mutation to V106I-Y181C-F227L-T369I increased the virus RC from 0.2% to 11.3%, while decreasing the virus resistance from 112- to 31-fold. Although the Y181C mutation alone occurs frequently in the Stanford Drug Resistance database (156 sequences, or 4.9% of the total NNRTI-resistant sequences), the prevalence of the Y181C mutation in combination with the other mutations is rather low. There are only two sequences containing the V106I-Y181C combination and none containing V106I-Y181C-F227L (12, 14).
Although, like efavirenz, VRX-480773 is highly protein bound, the VRX-480773 level in dogs seems to be high enough at 12 h to effectively inhibit both wt and mutant Y188L viruses (with predicted IQ values of 54 and 14, respectively) (Table 7). VRX-480773 is currently under phase I clinical studies to determine its pharmacokinetics and safety profile in humans.
Acknowledgments
We thank Yolanda Lie and Neil Parkin at Monogram Biosciences Inc. for some of the phenotypic tests, Dan Bellows, Heli Walker, and Xiaofeng Guo for their technical supports, and Bruno Canard for the kind gift of plasmid p66RTB.
Footnotes
Published ahead of print on 20 November 2006.
REFERENCES
- 1.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boone, L. R. 2006. Next-generation HIV-1 non-nucleoside reverse transcriptase inhibitors. Curr. Opin. Investig. Drugs 7:128-135. [PubMed] [Google Scholar]
- 3.Boretto, J., S. Longhi, J. M. Navarro, B. Selmi, J. Sire, and B. Canard. 2001. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 292:139-147. [DOI] [PubMed] [Google Scholar]
- 4.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 5.De Corte, B. L. 2005. From 4,5,6,7-tetrahydro-5-methylimidazo[4,5,1-jk](1,4)benzodiazepin-2(1H)-one (TIBO) to etravirine (TMC125): fifteen years of research on non-nucleoside inhibitors of HIV-1 reverse transcriptase. J. Med. Chem. 48:1689-1696. [DOI] [PubMed] [Google Scholar]
- 6.De La Rosa, M., H.-W. Kim, E. Gunic, C. Jenket, U. Boyle, Y.-H. Koh, I. Korboukh, M. Allan, W. Zhang, H. Chen, W. Xu, S. Nilar, N. Yao, R. Hamatake, S. Lang, Z. Hong, Z. Zhang, and J. L. Girardet. 2006. Tri-substituted triazoles as potent non-nucleoside inhibitors of the HIV reverse transcriptase. Bioorg. Med. Chem. Lett. 16:4444-4449. [DOI] [PubMed] [Google Scholar]
- 7.Harrington, R., L. Wu, H. Pullen, and M. Emerman. 2000. Direct detection of infectious HIV-1 in blood using a centrifugation-indicator cell assay. J. Virol. Methods 88:111-115. [DOI] [PubMed] [Google Scholar]
- 8.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popik, W., T. M. Alce, and W. C. Au. 2002. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4+ T cells. J. Virol. 76:4709-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren, J., J. Milton, K. L. Weaver, S. A. Short, D. I. Stuart, and D. K. Stammers. 2000. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 8:1089-1094. [DOI] [PubMed] [Google Scholar]
- 12.Rhee, S. Y., T. Liu, J. Ravela, M. J. Gonzales, and R. W. Shafer. 2004. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob. Agents Chemother. 48:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 14.Shafer, R. W. 19 May 2005, posting date. NNRTI mutation pattern & susceptibility. http://hivdb.stanford.edu/pages/phenoSummary/Pheno.NNRTI.Simple.html.
- 15.Zhang, Z., M. Walker, W. Xu, J. H. Shim, J. L. Girardet, R. K. Hamatake, and Z. Hong. 2006. Novel nonnucleoside inhibitors that select nucleoside inhibitor resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 50:2772-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]