Abstract
Intergenic dyad sequences (IDS) are short repeated elements that have been described for several Haemophilus genomes and for only two other bacterial genera. We developed a repetitive-element sequence-based PCR using an IDS-specific primer as a typing method (IDS-PCR) for nonencapsulated Haemophilus strains and compared this technique with pulsed-field gel electrophoresis (PFGE) of DNA restricted with SmaI. IDS-PCR was rapid, easy to perform, and reproducible, with a high discriminatory capacity for nontypeable Haemophilus influenzae (NTHI) strains. The 69 NTHI strains tested generated 65 different banding patterns. Epidemiologically related strains gave similar or identical fingerprints, and all of the unrelated strains except two showed different patterns. These results were in agreement with those obtained by PFGE. For 20 genital strains usually identified as being biotype IV NTHI and belonging to a cryptic genospecies of Haemophilus with remarkable genetic homogeneity, four bands were significantly present and six bands were significantly absent from the fingerprints. The 20 strains were gathered in 11 closely related profiles, whereas PFGE provided no band when DNA was treated with SmaI. IDS-PCR improved the differentiation previously obtained within this species by ribotyping and multilocus enzyme electrophoresis. Our findings suggest that IDS-PCR is a rapid, reliable, and discriminatory method for typing NTHI strains and is currently the most efficient method for distinguishing strains within the cryptic genospecies of Haemophilus.
Nontypeable Haemophilus influenzae (NTHI) strains are human commensal bacteria that are responsible for a number of respiratory tract infections such as otitis media, bronchitis, and sinusitis. NTHI strains are also responsible for genital, mother-infant, and neonatal infections. A group of genital strains usually identified as biotype IV NTHI constitutes a cryptic genital genospecies that shows remarkable genetic homogeneity and forms a monophyletic unit with H. influenzae and Haemophilus haemolyticus (31, 32).
NTHI strains are extremely diverse, and many typing methods have been proposed for epidemiological studies and pathogenesis investigations. Biotyping and more-discriminatory phenotypic typing methods such as outer membrane protein analysis (2, 25, 26, 33) and lipooligosaccharide analysis (6, 13, 26) have gradually been replaced by genotypic techniques. Multilocus enzyme electrophoresis (MLEE) (27, 30, 31) is a discriminatory technique but is also very laborious. DNA restriction fragment length polymorphism (RFLP) analysis (4, 20, 42) has a high discriminatory capacity but gives overly complex banding patterns. Analysis of rRNA gene restriction patterns (ribotyping) (4, 16, 32, 46) is a discriminatory but time-consuming method. Analysis of pulsed-field gel electrophoresis (PFGE) patterns in which genomic DNA is digested with a rare cutting restriction enzyme (1, 36) is currently considered to be the “gold standard” for H. influenzae typing but is also a laborious and time-consuming method. In comparison to these labor-intensive methods, the analysis of randomly amplified polymorphic DNA (RAPD) profiles (17, 28) is rapid and can provide discriminatory results, particularly when several primer results are combined, but this method lacks reproducibility.
Several short (<200-bp) interspersed noncoding repetitive DNA sequences are widely distributed throughout prokaryotic genomes (21). Among these repeated elements, repetitive extragenic palindromic (REP) sequences and enterobacterial repetitive intergenic consensus (ERIC) sequences were initially identified in Escherichia coli and Salmonella enterica serovar Typhimurium but are also present in other enterobacterial strains and in numerous eubacterial species (21, 45). Enterobacterial strains have been typed by PCR using REP- and ERIC-specific primers (45). This typing method, called rep-PCR (repetitive-element sequence-based PCR), was rapidly developed and has been successfully applied to other groups of bacteria such as Staphylococcus aureus (41), Streptococcus pneumoniae (44), Mycobacterium tuberculosis (37), and Listeria monocytogenes (14).
The typing of NTHI strains using rep-PCR with REP- or ERIC-specific primers has given results consistent with those obtained by classical genotypic methods. rep-PCR assays are quick and inexpensive, are less laborious than ribotyping and PFGE, and generate results that are easier to interpret than those of RFLP and more reproducible than those of RAPD (10, 15, 29, 42). However, Pettigrew et al. recently reported that ERIC-PCR was less discriminatory than ribotyping and PFGE, and they recommended that ERIC-PCR be combined with other typing methods (29). These authors also pointed out the lack of true REP and ERIC sequences in the H. influenzae genome. The absence of such sequences was shown in the fully sequenced H. influenzae strain Rd (8, 29) and was confirmed by a full BLAST search of the sequenced regions of various Haemophilus strains in the EMBL and GenBank databases. Thus, low-stringency PCR conditions (up to 5 mM MgCl2 and annealing temperatures as low as 25°C) or degenerate primers were used in REP- and ERIC-PCR for H. influenzae typing. These conditions allowed nonspecific binding and decreased the reproducibility of the method.
rep-PCR using primers derived from genuine Haemophilus repeated sequences has not yet been performed for Haemophilus strains. Such primers would allow high-stringency PCR conditions, thus avoiding the drawbacks of REP- and ERIC-PCR. Mrázek and Karlin (24) have described a short noncoding repeated sequence that is widely distributed throughout the H. influenzae Rd genome and is often found in pairs as inverted repeats (18, 34). This 23-bp sequence, commonly called intergenic dyad sequence (IDS), has been found in several Haemophilus strains (3, 9, 22, 35, 43). A full and systematic BLAST search for IDSs in the combined EMBL and GenBank databases produced matches with sequences from only two other bacterial genera, Neisseria and Pseudomonas.
In the present study, PCR with an IDS-specific primer was used as a typing method for 90 Haemophilus strains. We evaluated the reproducibility of this technique, its discriminatory capacity, and its ability to cluster epidemiologically related strains. These findings were compared with those obtained by PFGE of DNA restricted with SmaI.
MATERIALS AND METHODS
Bacterial strains.
Ninety Haemophilus strains were included in this study (Table 1). All clinical isolates were capsule serotyped as previously described by Quentin et al. (31). The absence of capsular genes was confirmed by Southern blotting with capsular-type-specific probes prepared with the primers described by Falla et al. (7). All strains were biotyped with the API 20E system (bioMérieux sa, Marcy l'Etoile, France) as described by Holmes et al. (11). Isolates were tested for urease activity, indole production, and ornithine decarboxylase activity according to Kilian's classification (19). Fifty-four strains were isolated from patients with neonatal, mother-infant, and genital infections (genital strains). Twenty of these strains which were nonencapsulated and phenotypically identified as being biotype IV H. influenzae strains had previously been assigned to a cryptic genital genospecies of Haemophilus (32). Thirty-two strains were isolated from adults (14 strains) and children (18 strains) aged 1 month to 6 years, mostly with respiratory tract infections (28 strains). All strains were epidemiologically unrelated except for nine isolated from four distinct mother-infant infections. Four reference strains (H. influenzae Rd, H. haemolyticus CIP 103290, and NTHI strains CIP 102514 and CIP 5424) were also included in this study. All strains were stored at −80°C in Schaedler-vitamin K3 broth (bioMérieux sa) with 10% glycerol.
TABLE 1.
Characteristics of the Haemophilus isolates analyzed in this study
| Strain (other denomination) | Species | Biotype | Specimen | Country |
|---|---|---|---|---|
| Genital strains | ||||
| C1 (189) | Cryptic genital Haemophilus | Amniotic fluid | United States | |
| C2 (421) | Cryptic genital Haemophilus | Blood of neonate | United States | |
| C3 (422) | Cryptic genital Haemophilus | Blood of neonate | United States | |
| C4 (427) | Cryptic genital Haemophilus | Amniotic fluid | United States | |
| C5 (799) | Cryptic genital Haemophilus | Blood of neonate | United States | |
| C6 (847) | Cryptic genital Haemophilus | Scrotal abscess | United States | |
| C7 (911) | Cryptic genital Haemophilus | Cerebrospinal fluid of neonate | United States | |
| C8 (1595) | Cryptic genital Haemophilus | Blood of neonate | United States | |
| C9 (1610) | Cryptic genital Haemophilus | Blood of neonate | United States | |
| C10 (3N) | Cryptic genital Haemophilus | Gastric fluid of neonate | France | |
| C11 (10N) | Cryptic genital Haemophilus | Gastric fluid of neonate | France | |
| C12 (12N) | Cryptic genital Haemophilus | Gastric fluid of neonate | France | |
| C13 (15N) | Cryptic genital Haemophilus | Gastric fluid of neonate | France | |
| C14 (16N) | Cryptic genital Haemophilus | Gastric fluid of neonate | France | |
| C15 (10U) | Cryptic genital Haemophilus | Urethra (male) | France | |
| C16 (11PS) | Cryptic genital Haemophilus | Urethra (male) | France | |
| C17 (26E) | Cryptic genital Haemophilus | Uterus | France | |
| C18 (PIZ) | Cryptic genital Haemophilus | Endocervix | France | |
| C19 (2406) | Cryptic genital Haemophilus | Vagina | France | |
| C20 (TRA) | Cryptic genital Haemophilus | Endocervix | France | |
| G1 (NAJ) | H. influenzae | III | Intrauterine device | France |
| G2 (ROY) | H. influenzae | III | Vagina | France |
| G3 (BOU) | H. influenzae | III | Endocervix | France |
| G6 (GAR) | H. influenzae | II | Gastric fluid of neonate | France |
| G7 (GAR) | H. influenzae | II | Vagina | France |
| G9 (LOP) | H. influenzae | II | Bartholin's gland abscess | France |
| G11 (ELA) | H. influenzae | I | Intrauterine device | France |
| G12 (VIG) | H. influenzae | II | Intrauterine device | France |
| G13 (BER) | H. influenzae | II | Endocervix | France |
| G14 (ROB) | H. influenzae | II | Blood of neonate | France |
| G15 (SAU) | H. influenzae | V | Endocervix | France |
| G17 (PER) | H. influenzae | III | Intrauterine device | France |
| G18 (PEA) | H. influenzae | V | Endocervix | France |
| G19 (CAY) | H. influenzae | II | Vagina | France |
| G20 (MBA) | H. influenzae | II | Gastric fluid of neonate | France |
| G21 (GOU) | H. influenzae | I | Vagina | France |
| G22 (KUP) | H. influenzae | II | Gastric fluid of neonate | France |
| G23 (VER) | H. influenzae | II | Amniotic fluid | France |
| G24 (BAU) | H. influenzae | V | Endocervix | France |
| G25 (BAU) | H. influenzae | V | Placenta | France |
| G26 (BAU) | H. influenzae | V | Gastric fluid of neonate | France |
| G27 (SAY) | H. influenzae | III | Bartholin's gland abscess | France |
| G28 (DAC) | H. influenzae | II | Endocervix | France |
| G29 (LEL) | H. influenzae | II | Blood of neonate | France |
| G30 (LEL) | H. influenzae | II | Placenta | France |
| G31 (PEG) | H. influenzae | III | Endocervix | France |
| G33 (BLI) | H. influenzae | III | Gastric fluid of neonate | France |
| G34 (IRL) | H. influenzae | II | Bartholin's gland abscess | France |
| G35 (HIN) | H. influenzae | I | Gastric fluid of neonate | France |
| G36 (PLA) | H. influenzae | I | Bartholin's gland abscess | France |
| G37 (HEN) | H. influenzae | III | Endocervix | France |
| G38 (VER) | H. influenzae | II | Anus of neonate | France |
| G39 (BEC) | H. influenzae | II | Endocervix | France |
| G40 (MER) | H. influenzae | II | Intrauterine device | France |
| Nongenital strains | ||||
| Isolates from children | ||||
| CH1 (PRO) | H. influenzae | II | Conjunctiva | France |
| CH3 (AUD) | H. influenzae | I | External ear | France |
| CH4 (FER) | H. influenzae | III | Trachea | France |
| CH5 (FOU) | H. influenzae | III | Middle ear | France |
| CH6 (GOB) | H. influenzae | II | Nasal swab | France |
| CH7 (RIB) | H. influenzae | I | External ear | France |
| CH8 (NGU) | H. influenzae | II | Mouth | France |
| CH9 (BOU) | H. influenzae | III | Nasal swab | France |
| CH10 (LOP) | H. influenzae | IV | External ear | France |
| CH11 (FAG) | H. influenzae | II | Urine | France/PICK> |
| CH12 (DEW) | H. influenzae | I | External ear | France |
| CH13 (MAN) | H. influenzae | III | Conjunctiva | France |
| CH14 (PIL) | H. influenzae | II | Middle ear | France |
| CH15 (MOU) | H. influenzae | II | Trachea | France |
| CH16 (ROB) | H. influenzae | III | Middle ear | France |
| CH17 (FOR) | H. influenzae | IV | Middle ear | France |
| CH18 (COR) | H. influenzae | II | Throat abscess | France |
| CH19 (COE) | H. influenzae | I | Cerebrospinal fluid | France |
| Isolates from adults | ||||
| A1 (REB) | H. influenzae | II | Sinus | France |
| A2 (HUE) | H. influenzae | III | Bronchial aspirate | France |
| A3 (MER) | H. influenzae | II | Bronchial aspirate | France |
| A4 (MEZ) | H. influenzae | II | Bronchoalveolar fluid | France |
| A5 (DUB) | H. influenzae | II | Protected distal brush | France |
| A6 (FER) | H. influenzae | II | Bronchial aspirate | France |
| A7 (COL) | H. influenzae | III | Protected distal brush | France |
| A8 (GUI) | H. influenzae | III | Bronchial aspirate | France |
| A10 (VOL) | H. influenzae | III | Pelvic abdominal abscess | France |
| A13 (BLA) | H. influenzae | II | Blood culture | France |
| A16 (GUI) | H. influenzae | I | Protected distal brush | France |
| A17 (LEB) | H. influenzae | II | Bronchial aspirate | France |
| A20 (MIC) | H. influenzae | II | Sinus | France |
| A21 (LAR) | H. influenzae | II | Bronchial aspirate | France |
| Reference strains | ||||
| CIP 5424 | H. influenzae | IV | Unknown | Unknown |
| CIP 102514 | H. influenzae | II | Unknown | United States |
| CIP 103290 | H. haemolyticus | Sputum | United Kingdom | |
| CIP 104746 (Rd) | H. influenzae | II | Unknown | Unknown |
Genomic DNA extraction.
The bacteria were grown for 24 h on chocolate agar plates (bioMérieux sa) at 37°C under 8% CO2-92% air. Bacterial cells were scraped from the plates and suspended in 1 ml of TEK buffer (40 mM Tris [pH 8]-2 mM EDTA). After addition of 150 μl of 1% lysozyme (Sigma, St. Louis, Mo.), the mixture was incubated for 30 min at 37°C with gentle agitation. Twenty-five microliters of 1% pronase (Calbiochem-Novabiochem Corporation, La Jolla, Calif.) and 150 μl of 10% sodium dodecyl sulfate (Bio-Rad Laboratories, Hercules, Calif.) were then added, and the mixture was incubated for 60 min at 60°C before undergoing two phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma) extractions. After centrifugation for 15 min at 18,800 × g, the DNA was precipitated in the presence of 30 mM NaCl in absolute ethanol for 15 min at −80°C. After centrifugation, the pellet was suspended in 100 μl of TE buffer (10 mM Tris [pH 8]-1 mM EDTA). DNA was quantified by photospectrometry.
PCR assay.
The IDS primer (5′ GTAGGGTGGGCGTAAGCCC 3′) (Eurogentec, Seraing, Belgium) was designed from the 23-bp repeated sequence of H. influenzae previously described by Read and Farley (34). The PCR mixture (20 μl) contained genomic DNA (20 ng), the IDS primer (4 μM), deoxynucleoside triphosphates (100 μM each) (Roche Diagnostics, Mannheim, Germany), Pfu Turbo DNA polymerase (1.25 U) (Stratagene, La Jolla, Calif.), 2 mM MgCl2, 20 mM Tris-HCl (pH 8.8), 10 mM (NH4)2SO4, 2 mM MgSO4, and 10 mM KCl (Stratagene). The PCR consisted of an initial 2.5-min hold at 94°C; 30 cycles, each consisting of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 2 min of elongation at 72°C; and a final 10-min elongation step (GeneAmp PCR System 9600; Perkin-Elmer, Norwalk, Conn.). The PCR products were subjected to electrophoresis on 1% agarose gels, and the resulting patterns were visualized by ethidium bromide staining and photographed.
Reproducibility.
To check for reproducibility, IDS-PCR was performed on five separate occasions on 10 Haemophilus strains (6 genital strains, 3 respiratory tract strains, and strain Rd). Both the GeneAmp PCR System 9600 and the GeneAmp PCR System 2400 (Perkin-Elmer) were used.
PFGE.
PFGE was performed as described by Moor et al. (23). Briefly, bacteria grown on chocolate agar plates (bioMérieux) were suspended in TE buffer, mixed with an equal volume of low-melting-temperature agarose (FMC BioProducts, Rockland, Maine), and placed in a slot former (Bio-Rad Laboratories). The resulting inserts were placed in 2 ml of lysis solution (0.5 M EDTA-1% Sarkosyl) and after the addition of 10 μl of proteinase K (20 mg/ml) were incubated for 48 h at 37°C. After washes, the DNA was digested overnight with SmaI (Roche Diagnostics). Restriction fragments were subjected to electrophoresis in a 1% agarose gel (FMC BioProducts) in TBE (4.5 mM Tris-HCl, 4.5 mM borate, 0.125 mM EDTA [pH 7.7]) with a contour-clamped homogeneous electric field (CHEF DRIII; Bio-Rad Laboratories). Pulse times were ramped from 6 to 8 s for 7 h, followed by a second ramp of 1 to 38 s for 17 h at 6 V/cm. Gels were stained with ethidium bromide, and PFGE patterns were detected by UV transillumination.
Analysis of patterns.
Photographs of each gel obtained by IDS-PCR and by PFGE of DNA restricted with SmaI were digitized with a video camera connected to a microcomputer (Bio-Profil; Vilber-Lourmat, Marne la Vallée, France). Numerical analysis was carried out with the Taxotron package (Taxolab; Institut Pasteur, Paris, France), which includes the RestrictoScan and RestrictoTyper programs. All of the bands on each gel were taken into account, even the faint ones. The band migration distances for each lane were determined in pixel units with RestrictoScan. The molecular size of each fragment was calculated from the migration distances by using cubic spline algorithms with RestrictoTyper. A 1-kb DNA ladder (Invitrogen Life Technologies, Cergy Pontoise, France) and a Lambda ladder (Bio-Rad Laboratories) were used as molecular weight standards for size determination.
Statistical analysis and discrimination index.
To evaluate differences in the presence or absence of DNA fragments in the banding patterns obtained by IDS-PCR with the different groups of strains, the chi-square test or the two-tailed Fisher’s exact test was performed using EpiInfo software, version 6.04. P values of <0.05 were considered to be statistically significant.
The single numerical discrimination index (D) was calculated by the method of Hunter and Gaston to evaluate the discriminatory power of the IDS-PCR and PFGE typing methods (12). The probability of two unrelated strains being placed in different typing groups was calculated by the equation
 |
where N is the total number of strains in the sample population, S is the total number of types described, and nj is the number of strains belonging to the jth type.
RESULTS
General characteristics and diversity of the IDS-PCR patterns of the Haemophilus strains.
PCR with IDS primers produced banding patterns with 2 to 10 fragments ranging from 0.3 to 3.8 kb. The distinct patterns obtained with 18 Haemophilus strains are shown in Fig. 1. Among the 69 H. influenzae strains studied, 64 non-epidemiologically-related strains gave 63 different patterns. D was greater than 0.99. Close or identical patterns were obtained for epidemiologically related strains isolated from patients with neonatal and mother-infant infections (Fig. 2). Two pairs of strains (G6-G7 and G29-G30), each comprising one strain isolated from the mother and one strain isolated from her child, gave identical patterns. One pair (G23-G38) and one group of three strains (G24-G25-G26), isolated from a mother and her child, gave closely related banding patterns. Only two non-epidemiologically-related strains (G22 and G23) gave identical patterns. The 20 non-epidemiologically-related genital strains belonging to the cryptic genospecies of Haemophilus showed 11 closely related patterns (D = 0.91) (Fig. 3). Thirteen of these 20 strains were grouped into four banding patterns including 5, 3, 3, and 2 strains, respectively.
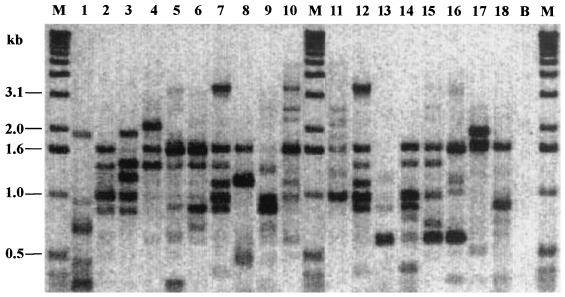
FIG. 1.
Banding patterns obtained by IDS-PCR for 18 Haemophilus strains. Lane 1, cryptic genital strain C2; lanes 2 to 8, genital strains G1 and G35 to G40; lanes 9 to 18, nongenital strains A3 to A7 and CH3 to CH7; lane B, reagent blank; lanes M, size markers.
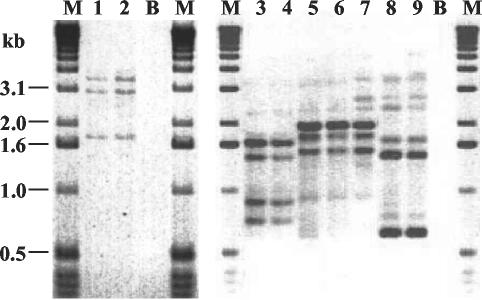
FIG. 2.
Banding patterns obtained by IDS-PCR. Lanes 1 and 2, identical profiles of strains G6 and G7; lanes 3 and 4, identical profiles of strains G22 and G23; lanes 5 to 7, closely related profiles of strains G24, G25, and G26; lanes 8 and 9, identical profiles of strains G29 and G30; lanes B, reagent blank; lanes M; size markers.
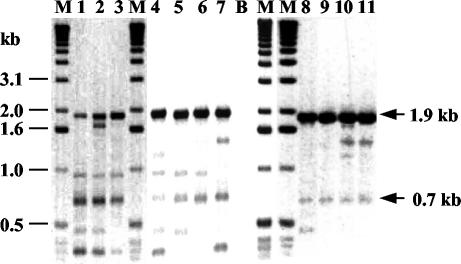
FIG. 3.
Banding patterns obtained by IDS-PCR with the cryptic genital strains C2, C3, C7, C8, and C10 (lane 1), C4, C6, and C14 (lane 2), C19 (lane 3), C5 (lane 4), C9, C11, and C12 (lane 5), C13 and C20 (lane 6), C17 (lane 7), C1 (lane 8), C15 (lane 9), C16 (lane 10), and C18 (lane 11). Lane B, reagent blank; lanes M, size markers. The sizes of the two fragments common to the 20 cryptic genital strains are indicated on the right.
Eleven DNA fragments appeared to be significantly associated with one or several groups of strains as determined by the chi-square test or the two-tailed Fisher’s exact test (P < 0.01) (Table 2). An intensely staining 1.9-kb DNA fragment and a 0.7-kb fragment were present in all of the 20 cryptic genital strains (Fig. 3) and were found in only 18 and 21 of the 70 other Haemophilus (69 H. influenzae and one H. haemolyticus) strains, respectively (P ≤ 0.0000001). Two other fragments of approximately 0.9 and 0.5 kb were present in 15 and 13 of the 20 cryptic genital strains, respectively, and were found in only 27 (P < 0.01) and 7 (P < 0.000001) of the 70 other Haemophilus strains, respectively. Five fragments of 1.6, 0.6, 3.3, 1.0, and 2.6 kb were absent from all of the 20 cryptic genital strains and were found in 44 (P < 0.00001), 34 (P < 0.001), 27 (P < 0.01), 27 (P < 0.01), and 21 (P < 0.01) of the 70 other Haemophilus strains, respectively. Two other fragments of approximately 2.2 and 1.4 kb were significantly associated with the noncryptic genital strains. These bands were present in 19 and 21 of the 34 noncryptic genital strains, respectively, and were found in only 9 (P < 0.001) and 14 (P < 0.01) of the 56 other strains, respectively. The 2.2-kb fragment was absent from all of the 20 cryptic genital strains.
TABLE 2.
Frequency of specific IDS-PCR amplification products among 90 Haemophilus strains
| Fragment size (kb) | No. of strains exhibiting the fragment
|
||
|---|---|---|---|
| Cryptic genital strains (n = 20) | Other genital strains (n = 34) | Nongenital strains (n = 36) | |
| Fragments significantly present in cryptic genital strains | |||
| 1.9 | 20 (P < 0.0000001)a | 11 | 7 |
| 0.9 | 15 (P = 0.0086)a | 14 | 13 |
| 0.7 | 20 (P = 0.0000001)a | 11 | 10 |
| 0.5 | 13 (P = 0.0000019)b | 3 | 4 |
| Fragments significantly absent in cryptic genital strains | |||
| 3.3 | 0 (P = 0.0023)a | 13 | 14 |
| 2.6 | 0 (P = 0.0027)b | 14 | 7 |
| 1.6 | 0 (P = 0.0000025)a | 23 | 21 |
| 1.0 | 0 (P = 0.0023)a | 11 | 16 |
| 0.6 | 0 (P = 0.00022)a | 13 | 21 |
| Fragments significantly present in noncryptic genital strains | |||
| 2.2 | 0 | 19 (P = 0.00019)a | 9 |
| 1.4 | 3 | 21 (P = 0.0011)a | 11 |
P values were calculated by the chi-square test.
P values were calculated by the two-tailed Fisher’s exact test.
Reproducibility of the IDS-PCR method.
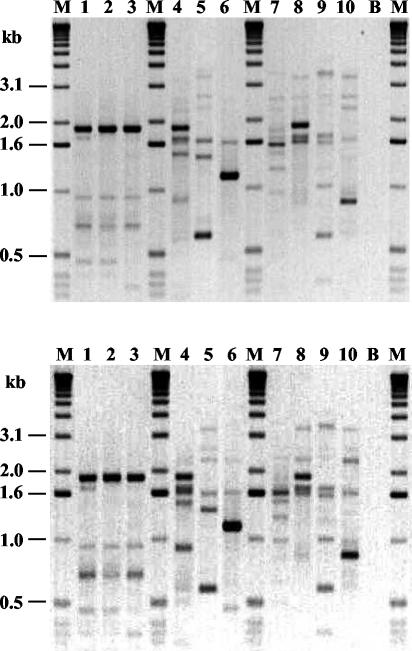
To assess whether this technique was reproducible, IDS-PCR was performed on five separate occasions with 10 Haemophilus strains. Three genital strains belonging to the cryptic genospecies of Haemophilus that showed closely related patterns and seven strains that showed distinct patterns (three noncryptic genital strains, three strains isolated from respiratory tracts, and strain Rd) were selected. The banding patterns obtained on two different days are shown in Fig. 4. The profiles observed were very similar for each strain. The only differences that could be observed were in the staining intensity of some DNA fragments or the absence of weakly staining fragments.
FIG. 4.
Reproducibility of IDS-PCR. Shown are banding patterns obtained on two different days with 10 Haemophilus strains. Lanes 1 to 3, cryptic genital strains C4, C11, and C19; lanes 4 to 6, genital strains G24, G30, and G40; lane 7, strain Rd; lanes 8 to 10, nongenital strains A8, CH8, and CH9; lanes B, reagent blank; lanes M, size markers.
PFGE analysis.
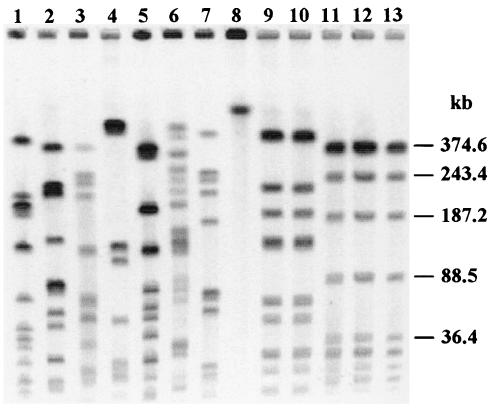
Eighty-seven Haemophilus strains, including 66 NTHI strains, the 20 genital strains belonging to the cryptic genospecies of Haemophilus, and the H. haemolyticus strain, were subjected to PFGE. Fingerprints were analyzed and compared with the Taxotron package and were then evaluated using the criteria developed by Tenover et al. (40). The banding patterns obtained by PFGE with 13 Haemophilus strains are shown in Fig. 5. The 66 NTHI strains gave patterns consisting of 4 to 15 fragments ranging from 27.5 to 482.8 kb. For these strains, D was greater than 0.99. Fifty-nine different restriction patterns were identified for 61 non-epidemiologically-related strains. Epidemiologically related strains gave identical patterns (G24-G25-G26 and G29-G30) (Fig. 5) or patterns that differed by one to three bands (G23-G38 and G6-G7). Only two pairs of non-epidemiologically-related strains that gave distinct patterns with IDS-PCR gave identical patterns with PFGE (G12-G40 and E1-A5). The two non-epidemiologically-related strains that exhibited identical patterns with IDS-PCR, G22 and G23, gave patterns that differed by five bands with PFGE and thus are possibly related according to Tenover's criteria (40). For the 20 genital strains belonging to the cryptic genospecies of Haemophilus, PFGE of DNA restricted with SmaI was performed several times and no DNA restriction products were observed. As shown in Fig. 5, lane 8, the DNA remained at the top of the gel. The H. haemolyticus strain gave a pattern that was distinct from those of all the other strains.
FIG. 5.
Banding patterns obtained by PFGE of DNA restricted with SmaI for 13 Haemophilus strains. Lanes 1 to 3, genital strains G1, G35, and G39; lanes 4 to 7, nongenital strains A4, A7, CH3, and CH6; lane 8, cryptic genital strain C7; lanes 9 and 10, identical profiles of strains G29 and G30; lanes 11 to 13, identical profiles of strains G24, G25, and G26.
DISCUSSION
None of the genotypic methods that have been described for typing of Haemophilus strains appear both to be simple and rapid and to provide reproducible and highly discriminatory results. PFGE, which is considered the gold standard, is laborious and time-consuming, and RAPD, which is a rapid and discriminatory technique, lacks reproducibility (15, 17, 36). PCR-based methods such as rep-PCR, which is based on repetitive REP and ERIC sequences, have been developed to offer the advantages of simple, rapid, reproducible, and discriminatory techniques. Nevertheless, for Haemophilus typing, REP- and ERIC-PCR are always carried out under low-stringency conditions to allow primer binding. Thus, an annealing temperature between 25 and 40°C (10, 15, 28, 38, 42), 5 mM MgCl2 (29), or degenerate primers (15) are used. These conditions, which are probably used to compensate for the absence of true REP and ERIC sequences in the Haemophilus genome, result in a lack of reproducibility (10, 29).
The aims of our study were to develop a rep-PCR method based on a genuine Haemophilus-specific repeated sequence and to validate this technique for typing of Haemophilus strains. The uptake signal sequence (USS) is a repeated element present in 1,465 copies on the H. influenzae Rd genome (39). rep-PCR using USS-specific primers generated overly complex banding patterns (data not shown) due to the large number of USS copies. This sequence was therefore not selected for rep-PCR. IDS is another repeated element that is present in 14 pairs throughout the H. influenzae Rd genome (24, 34). IDS also flanks the hif genes, which encode the LKP fimbriae and are absent from strain Rd (3, 35, 43). Thus, IDS-PCR could theoretically generate about 10 DNA fragments with sizes up to 5 kb that would make the analysis possible. We studied 90 strains, including 20 genital Haemophilus strains that exhibit a high degree of genetic homogeneity and are differentiated with difficulty by methods such as MLEE and ribotyping (31, 32). We used a nondegenerate primer that is specific for IDS and high-stringency annealing conditions.
This method was rapid (6 h for amplification and electrophoresis) and easy to perform. It did not require specific and expensive equipment. The banding patterns obtained were simple to read and to compare visually or with the help of a computer program. They were composed of as many as 10 bands with most of the fragments ranging from 150 bp to 2 kb, as expected from the distribution of IDS throughout the H. influenzae Rd genome. When IDS-PCR was repeated on 10 Haemophilus strains on different days and with two thermocyclers, the resulting patterns were essentially the same, indicating that this technique also had good reproducibility. IDS-PCR is thus more reproducible than RAPD. Its reproducibility is similar to that of ribotyping, for which differences in the staining intensity and the absence of weakly staining DNA fragments can also be observed (32).
IDS-PCR showed a very high discriminatory capacity within the H. influenzae species. The 69 NTHI strains generated 65 different fingerprints (D > 0.99). Very similar results were obtained by PFGE of DNA restricted with SmaI, which is considered the gold standard for H. influenzae typing. Sixty-six NTHI strains gave 62 different patterns (D > 0.99). These results are in agreement with those obtained by different techniques, showing that NTHI strains are extremely diverse (10, 15, 17, 29, 31, 32, 36). Among the 66 NTHI clinical isolates tested by IDS-PCR, only the epidemiologically related strains (nine strains) and two non-epidemiologically-related strains gave identical or closely related patterns. Similar results were obtained by PFGE. IDS-PCR is consequently a discriminatory method for typing of NTHI strains, as is PFGE, and is considerably less labor-intensive, costly, and time-consuming than PFGE.
IDS-PCR grouped the 20 genital strains belonging to the cryptic genospecies of Haemophilus into only 11 closely related fingerprints. Four fragments appeared to be significantly associated with the cryptic genital strains, and six fragments were significantly absent. In this group of strains only, identical banding patterns were obtained for non-epidemiologically-related strains, clustering as many as five strains. The particular homogeneity of this group of strains obtained by this typing method is in agreement with their remarkable genetic homogeneity previously shown and contrasts with the diversity observed in the H. influenzae species in general (31, 32). Ribotyping grouped 18 genital strains into seven profiles, two of which included a total of 11 strains (32). MLEE grouped 25 strains into seven electrophoretic types, one of which included 18 strains (31). The discrimination index was thus greater for IDS-PCR within this group (D = 0.91) than for ribotyping (D = 0.81) or MLEE (D = 0.49).
By PFGE, SmaI did not restrict the DNA from the 20 cryptic genital strains. Neither did ApaI, another endonuclease that is sometimes used to type Haemophilus strains by PFGE (23). This could be explained by an inhibition of DNA restriction caused by a methylation of cytosines at the restriction site (5). This is another unusual characteristic of this particular group of genital strains in addition to those previously shown (31, 32).
In conclusion, this study was the first practical application of a rep-PCR assay using a primer designed from the Haemophilus-specific repeated sequence IDS as a typing method for nonencapsulated Haemophilus strains. Our results suggest that IDS-PCR is a reliable and discriminatory method for typing of NTHI strains, as is PFGE, and is considerably more rapid and less labor-intensive than PFGE. In addition, IDS-PCR is currently the most efficient method for distinguishing strains within the cryptic genospecies of Haemophilus.
REFERENCES
- 1.Aparicio, P., F. Roman, and J. Campos. 1996. Epidemiological characterization of Haemophilus influenzae using molecular markers. Enferm. Infecc. Microbiol. Clin. 14:227-232. [PubMed] [Google Scholar]
- 2.Barenkamp, S. J., R. S. Munson, Jr., and D. M. Granoff. 1982. Outer membrane protein and biotype analysis of pathogenic nontypeable Haemophilus influenzae. Infect. Immun. 36:535-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruant, G., N. Gousset, R. Quentin, and A. Rosenau. 2002. Fimbrial ghf gene cluster of genital strains of Haemophilus spp. Infect. Immun. 70:5438-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruce, K. D., and J. Z. Jordens. 1991. Characterization of noncapsulate Haemophilus influenzae by whole-cell polypeptide profiles, restriction endonuclease analysis, and rRNA gene restriction patterns. J. Clin. Microbiol. 29:291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butkus, V., L. Petrauskiene, Z. Maneliene, S. Klimasauskas, V. Laucys, and A. Janulaitis. 1987. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 15:7091-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campagnari, A. A., M. R. Gupta, K. C. Dudas, T. F. Murphy, and M. A. Apicella. 1987. Antigenic diversity of lipooligosaccharides of nontypeable Haemophilus influenzae. Infect. Immun. 55:882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falla, T. J., D. W. M. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 9.Geluk, F., P. P. Eijk, S. M. van Ham, H. M. Jansen, and L. van Alphen. 1998. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect. Immun. 66:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-De-Leon, P., J. I. Santos, J. Caballero, D. Gomez, L. E. Espinosa, I. Moreno, D. Piñero, and A. Cravioto. 2000. Genomic variability of Haemophilus influenzae isolated from Mexican children determined by using enterobacterial repetitive intergenic consensus sequences and PCR. J. Clin. Microbiol. 38:2504-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes, R. L., L. M. De Franco, and M. Otto. 1982. Novel method of biotyping Haemophilus influenzae that uses API 20E. J. Clin. Microbiol. 15:1150-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inzana, T. J. 1983. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J. Infect. Dis. 148:492-499. [DOI] [PubMed] [Google Scholar]
- 14.Jeršek, B., P. Gilot, M. Gubina, N. Klun, J. Mehle, E. Tcherneva, N. Rijpens, and L. Herman. 1999. Typing of Listeria monocytogenes strains by repetitive-element sequence-based PCR. J. Clin. Microbiol. 37:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordens, J. Z. 1998. Characterization of non-capsulate Haemophilus influenzae by repetitive extragenic palindromic (REP)-PCR. J. Med. Microbiol. 47:1031-1034. [DOI] [PubMed] [Google Scholar]
- 16.Jordens, J. Z., and N. I. Leaves. 1997. Source of variation detected in ribotyping patterns of Haemophilus influenzae: comparison of traditional ribotyping, PCR-ribotyping and rDNA restriction analysis. J. Med. Microbiol. 46:763-772. [DOI] [PubMed] [Google Scholar]
- 17.Jordens, J. Z., N. I. Leaves, E. C. Anderson, and M. P. Slack. 1993. Polymerase chain reaction-based strain characterization of noncapsulate Haemophilus influenzae. J. Clin. Microbiol. 31:2981-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlin, S., J. Mrázek, and A. M. Campbell. 1996. Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res. 24:4263-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 93:9-62. [DOI] [PubMed] [Google Scholar]
- 20.Loos, B. G., J. M. Bernstein, D. M. Dryja, T. F. Murphy, and D. P. Dickinson. 1989. Determination of the epidemiology and transmission of nontypeable Haemophilus influenzae in children with otitis media by comparison of total genomic DNA restriction fingerprints. Infect. Immun. 57:2751-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 180:4693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moor, P. E., P. C. Collignon, and G. L. Gilbert. 1999. Pulsed-field gel electrophoresis used to investigate genetic diversity of Haemophilus influenzae type b isolates in Australia shows differences between aboriginal and non-aboriginal isolates. J. Clin. Microbiol. 37:1524-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mrázek, J., and S. Karlin. 1996. A new significant recurrent dyad pairing in Haemophilus influenzae. Trends Biochem. Sci. 21:201-202. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, T. F., K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, T. F., J. M. Bernstein, D. M. Dryja, A. A. Campagnari, and M. A. Apicella. 1987. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to nontypable Haemophilus influenzae: pathogenic and epidemiological observations. J. Infect. Dis. 156:723-731. [DOI] [PubMed] [Google Scholar]
- 27.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypeable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peerbooms, P. G. H., M. N. Engelen, D. A. J. Stokman, B. H. B. van Benthem, M.-L. van Weert, S. M. Bruisten, A. van Belkum, and R. A. Coutinho. 2002. Nasopharyngeal carriage of potential bacterial pathogens related to day care attendance, with special reference to the molecular epidemiology of Haemophilus influenzae. J. Clin. Microbiol. 40:2832-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettigrew, M. M., B. Foxman, Z. Ecevit, C. F. Marrs, and J. Gilsdorf. 2002. Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 40:660-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porras, O., D. A. Caugant, T. Lagergard, and C. Svanborg-Eden. 1986. Application of multilocus enzyme gel electrophoresis to Haemophilus influenzae. Infect. Immun. 53:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quentin, R., A. Goudeau, R. J. Wallace, Jr., A. L. Smith, R. K. Selander, and J. M. Musser. 1990. Urogenital, maternal and neonatal isolates of Haemophilus influenzae: identification of unusually virulent serologically non-typable clone families and evidence for a new Haemophilus species. J. Gen. Microbiol. 136:1203-1209. [DOI] [PubMed] [Google Scholar]
- 32.Quentin, R., C. Martin, J. M. Musser, N. Pasquier-Picard, and A. Goudeau. 1993. Genetic characterization of a cryptic genospecies of Haemophilus causing urogenital and neonatal infections. J. Clin. Microbiol. 31:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quentin, R., J. M. Musser, M. Mellouet, P. Y. Sizaret, R. K. Selander, and A. Goudeau. 1989. Typing of urogenital, maternal, and neonatal isolates of Haemophilus influenzae and Haemophilus parainfluenzae in correlation with clinical source of isolation and evidence for a genital specificity of H. influenzae biotype IV. J. Clin. Microbiol. 27:2286-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read, T. D., and M. M. Farley. 1997. Conserved extragenic DNA elements in Haemophilus influenzae. Mol. Microbiol. 23:627-628. [DOI] [PubMed] [Google Scholar]
- 35.Read, T. D., S. W. Satola, and M. M. Farley. 2000. Nucleotide sequence analysis of hypervariable junctions of Haemophilus influenzae pilus gene clusters. Infect. Immun. 68:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito, M., A. Umeda, and S.-I. Yoshida. 1999. Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 37:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sechi, L. A., S. Zanetti, I. Dupré, G. Delogu, and G. Fadda. 1998. Enterobacterial repetitive intergenic consensus sequences as molecular targets for typing of Mycobacterium tuberculosis strains. J. Clin. Microbiol. 36:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma, A., R. Kaur, N. K. Ganguly, P. D. Singh, and A. Chakraborti. 2002. Subtype distribution of Haemophilus influenzae isolates from North India. J. Med. Microbiol. 51:399-404. [DOI] [PubMed] [Google Scholar]
- 39.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Belkum, A., R. Bax, P. Peerbooms, W. H. Goessens, N. van Leeuwen, and W. G. Quint. 1993. Comparison of phage typing and DNA fingerprinting by polymerase chain reaction for discrimination of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 31:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Belkum, A., B. Duim, A. Regelink, L. Moller, W. Quint, and L. van Alphen. 1994. Genomic DNA fingerprinting of clinical Haemophilus influenzae isolates by polymerase chain reaction amplification: comparison with major outer-membrane protein and restriction fragment length polymorphism analysis. J. Med. Microbiol. 41:63-68. [DOI] [PubMed] [Google Scholar]
- 43.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. M. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 44.Versalovic, J., V. Kapur, E. O. Mason, Jr., U. Shah, T. Koeuth, J. R. Lupski, and J. M. Musser. 1993. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J. Infect. Dis. 167:850-856. [DOI] [PubMed] [Google Scholar]
- 45.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, C.-C., L. K. Siu, M.-K. Chen, Y. L. Yu, F. M. Lin, M. Ho, and M.-L. Chu. 2001. Use of automated riboprinter and pulsed-field gel electrophoresis for epidemiological studies of invasive Haemophilus influenzae in Taiwan. J. Med. Microbiol. 50:277-283. [DOI] [PubMed] [Google Scholar]