Abstract
The macrolide resistance plasmid pRSB111 was isolated from bacteria residing in the final effluents of a wastewater treatment plant. The 47-kb plasmid confers resistance to azithromycin, clarithromycin, erythromycin, roxithromycin, and tylosin when it is carried by Pseudomonas sp. strain B13 and is very similar to prototype IncP-1β plasmid pB3, which was previously isolated from an activated-sludge bacterial community of a wastewater treatment plant. The two plasmids differ in their accessory regions, located downstream of the conjugative transfer module gene traC. Nucleotide sequence analysis of the pRSB111 accessory region revealed that it contains a new macrolide resistance module composed of the genes mphR(E), mph(E), and mrx(E), which putatively encode a transcriptional regulator, a macrolide phosphotransferase, and a transmembrane transport protein, respectively. Analysis of the contributions of the individual genes of the macrolide resistance module revealed that mph(E) and mrx(E) are required for high-level macrolide resistance. The resistance genes are flanked by two insertion sequences, namely, ISPa15 and ISRSB111. Two truncated transposable elements, IS6100 and remnants of a Tn3-like transposon, were identified in the vicinity of ISRSB111. The accessory element of pRSB111 apparently replaced the Tn402-like element present on the sister plasmid, pB3, as suggested by the conservation of Tn402-specific terminal inverted repeats on pRSB111.
Considerable increases in the rates of antimicrobial resistance among clinical as well as environmental bacteria have been observed in recent years and are mainly due to the persisting selective pressure caused by the application of antimicrobial compounds and the dissemination of resistance determinants by means of horizontally mobile genetic elements (6, 7, 28).
Besides β-lactams, aminoglycosides, and fluoroquinolones, macrolide antibiotics, characterized by 14-, 15-, and 16-membered lactone ring systems, are among the most frequently administered antimicrobial drugs (2-4, 15, 30). They are effective against gram-positive bacteria as well as against some gram-negative bacteria (34, 37, 46) and are often used to treat community-acquired respiratory tract infections, skin and soft tissue infections, Legionnaires' disease, and venereal diseases (1, 8, 21, 24, 26, 36). Macrolides inhibit protein synthesis by binding to the 50S ribosomal subunit (15), and resistance to this class of antibiotics is mainly attributed to target site modifications (44), active efflux (38), or modification of the drug itself (13, 38). A number of different macrolide resistance genes have been identified in gram-positive bacteria as well as in gram-negative bacteria (33).
Wastewater treatment plants were identified as dissemination hot spots for resistance determinants. Antibiotic-resistant bacteria, resistance genes, and mobile genetic elements carrying resistance determinants are frequently isolated from bacterial communities residing in wastewater treatment plants (12, 27, 39, 40, 42, 43). Different erythromycin resistance plasmids obtained from among those antibiotic resistance plasmids from sewage bacteria were previously analyzed at the genomic level. For example, promiscuous, conjugative IncP-1β plasmid pB4 encodes a tripartite multidrug efflux system, consisting of an efflux permease, a periplasmic membrane fusion protein, and an outer membrane factor, which mediates high-level erythromycin and roxithromycin resistance (41). In contrast, the non-IncP plasmids pRSB101 and pRSB107, which were isolated from activated-sludge bacteria by a transformation-based approach, harbor the macrolide resistance determinant mph(A)-mrx-mphR(A) (39, 40). The mph(A) gene encodes a macrolide-2′-phosphotransferase I, and Mrx was predicted to be a hydrophobic transmembrane transport protein.
These findings motivated the search for mobile erythromycin resistance plasmids in the final effluents of a municipal wastewater treatment plant. Here we describe the new macrolide resistance determinant of the mobile, broad-host-range IncP-1β plasmid pRSB111 at the genomic level and its functional characterization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α mcr (19) containing the vector pBCSK (a derivative of pBluescript carrying a chloramphenicol resistance gene; Stratagene) with the cloned pRSB111 erythromycin resistance-conferring region was grown in Luria broth (LB) medium supplemented with erythromycin (300 μg/ml) at 37°C. Pseudomonas sp. strain B13 GFP1 (11, 14) containing the resistance plasmid pRSB111 was grown in LB medium supplemented with erythromycin (300 μg/ml) at 30°C.
Exogenous isolation of plasmids from bacteria of final effluents.
A sample (1 liter of water) from the final effluents of the wastewater treatment plant Bielefeld-Heepen (Germany) was taken in April 2003. The bacteria in the sample were collected by centrifugation. Exogenous isolation of plasmids from the bacteria in the final effluents was done in three parallel mating experiments with Pseudomonas sp. strain B13 GFP1 as the recipient strain, as described previously (12). Pseudomonas sp. strain B13 transconjugants which potentially received a plasmid from endogenous bacteria of the final effluents were selected on LB medium containing gentamicin (30 μg/ml), kanamycin (50 μg/ml), and erythromycin (300 μg/ml).
Determination of resistance patterns.
Antimicrobial susceptibility testing was done according to the standards outlined by the Clinical and Laboratory Standards Institute (5). Antimicrobial disk diffusion assays were done by using the following disks purchased from Oxoid GmbH (Wesel, Germany): azithromycin (AZM15), clarithromycin (CLR15), clindamycin (DA10), erythromycin (E30), and lincomycin (MY15). The antibiotics roxithromycin (ROX15) and tylosin (TY30) were purchased from Mast Diagnostica GmbH (Reinfeld, Germany). MIC tests were performed as described by Dröge et al. (12).
Transfer activity of plasmid pRSB111.
Transfer tests with plasmid pRSB111 were done as described previously (12). Ralstonia eutropha GFP3 (a β-proteobacterium) (43), Pseudomonas sp. strain B13 GFP1 (a γ-proteobacterium) (11, 14), and E. coli CV60 (a γ-proteobacterium) (40) were used as the recipients in mating experiments. Selection of transconjugants was done on LB medium containing kanamycin (50 μg/ml) and erythromycin (300 μg/ml). All recipients used here are resistant to kanamycin, and plasmid pRSB111 confers erythromycin resistance. The transfer frequencies for each recipient were calculated.
Standard DNA techniques.
The plasmid content of the transconjugants was checked by Eckhardt analysis (23), and plasmid DNA was isolated from Pseudomonas sp. strain B13 GFP1(pRSB111) by using the Nucleobond kit PC100 and AX100 columns (Macherey-Nagel, Düren, Germany), according to the protocol supplied by the manufacturer. Recombinant pBCSK derivatives were isolated by using a QIAprep spin miniprep kit (QIAGEN, Hilden, Germany), according to the manufacturers' instructions. Restriction fragments were extracted from agarose gels by use of the Sephaglas BandPrep kit (Amersham Pharmacia Biotech, Freiburg, Germany). Restriction enzyme digestion, agarose gel electrophoresis, DNA cloning, and transformation of E. coli DH5α were carried out as described by Sambrook et al. (35). The IncP-1-specific PCR with trfA-specific primers was carried out as described by Götz et al. (18). The mph(A)-specific PCR was done as described by Szczepanowski et al. (40).
Sequencing of pRSB111 8-kb NotI restriction fragment and annotation.
Purified plasmid pRSB111 DNA was restricted with the restriction endonuclease NotI. An 8-kb NotI restriction fragment was subcloned into the vector pBCSK. Subfragments of the NotI fragment were generated by using the enzymes BamHI, EcoRI, PstI, and SphI and were cloned into pZErO-2 (Invitrogen, Karlsruhe, Germany), pGEM-T Easy (Promega Corporation, Madison, WI), and pUC19 (45). Sequencing of recombinant plasmids was achieved by using standard and walking primers and ET-Dye Terminator mix chemistry (Applera; Applied Biosystems). The extension products were separated on an ABI 377 sequencer (IIT Biotech GmbH, Bielefeld, Germany). Gap closure and polishing of the sequence were done by applying a primer walking strategy with appropriate restriction fragment clones. Computer-assisted assembly and sequence quality control were carried out with the CONSED/AUTOFINISH software tool (16, 17). Annotation of the finished sequence was done by using the GenDB (version 2.2) annotation tool (29).
Amplification and cloning of individual genes encoded in pRSB111 macrolide resistance-conferring region.
Individual genes of the pRSB111 macrolide resistance-conferring region were amplified by PCR. The primer pairs used and the corresponding amplicon sizes are listed in Table 1. The amplicons were restricted with BamHI and cloned into the BamHI-digested pUC18 vector (45). The identities of the cloned fragments and their orientations within the vector were verified by digestion with different restriction enzymes. All genes were cloned in the sense orientation with respect to the lac promoter of pUC18. The original ribosomal binding sites of each gene were retained. PCRs were done under standard conditions with the proofreading polymerase Pfu (Fermentas, St. Leon-Rot, Germany).
TABLE 1.
Primer pairs used to amplify individual genes of the pRSB111 macrolide resistance region and expected amplicon sizes
| Gene(s) | Primer sequencea
|
Size (bp) | |
|---|---|---|---|
| Left | Right | ||
| mph(E) | 5′-GAGAGGATCCCAGCTGGACCTGATCTTTCC | 5′-GAGAGGATCCCGGCAAGTACACCCAGGAG | 1,042 |
| mrx(E) | 5′-GAGAGGATCCAGCGCAGAGCATCTGGAG | 5′-GAGAGGATCCCTTATGCCGAGGGAATTGAG | 1,266 |
| mph(E)-mrx(E) | 5′-GAGAGGATCCCAGCTGGACCTGATCTTTCC | 5′-GAGAGGATCCCTTATGCCGAGGGAATTGAG | 2,212 |
| mphR(E)-mph(E)-mrx(E) | 5′-GAGAGGATCCTGATATTCCGTGCAGTCGTC | 5′-GAGAGGATCCCTTATGCCGAGGGAATTGAG | 2,876 |
The underscored letters represent primer extensions, including the BamHI restriction site, used for subcloning of the amplicons into vector pUC18.
Nucleotide sequence accession number.
The nucleotide sequence of the pRSB111 macrolide resistance gene region was submitted to the GenBank database and is available under accession number AM260957.
RESULTS AND DISCUSSION
Isolation of self-transmissible erythromycin resistance plasmids from bacteria of final effluents of a wastewater treatment plant.
The objective of this work was to isolate self-transmissible erythromycin resistance plasmids from bacteria of the final effluents of a wastewater treatment plant to determine whether bacteria carrying mobile genetic elements for erythromycin resistance are released into the environment. For this purpose we applied the exogenous plasmid isolation method and Pseudomonas sp. strain B13 GFP1 as the recipient (12). Transconjugants which potentially received a plasmid from endogenous bacteria of the final effluents were selected on medium containing erythromycin. Twenty-five transconjugants possessed a plasmid of the same size that conferred resistance to erythromycin. Isolation of three of these plasmids and subsequent restriction analyses revealed that all plasmids had identical restriction patterns. One of these plasmids, designated pRSB111, was chosen for further analysis.
Plasmid pRSB111 is closely related to prototype IncP-1β resistance plasmid pB3.
Restriction of plasmid pRSB111 with the restriction enzyme NotI elucidated that its restriction pattern is very similar to that of prototype IncP-1β plasmid pB3, which was previously isolated from an activated-sludge bacterial community of a wastewater treatment plant (12, 22). The NotI patterns of the two plasmids differed by only one restriction fragment. Plasmid pRSB111 possesses an 8-kb fragment that is missing in the pB3 pattern. Instead, pB3 contains a 17-kb NotI fragment that is known to contain the accessory region of the plasmid. It could therefore be assumed that the 8-kb NotI fragment of pRSB111 represents the resistance gene region of this plasmid. To confirm that pRSB111 is a member of the IncP-1β plasmid family, part of its replication initiation gene, trfA, was amplified by using IncP-1 trfA-specific primers (18). Sequencing of the amplicon obtained showed that the trfA nucleotide sequence is nearly identical (99.6%) to the corresponding sequence of plasmid pB3 (22).
Plasmid pRSB111 apparently does not harbor the mph(A) erythromycin resistance gene since it could not be detected on the plasmid genome by PCR. The mph(A) gene was previously identified on many other plasmids originating from activated-sludge bacteria (40). Therefore, we decided to subclone the pRSB111 8-kb NotI fragment containing the new erythromycin resistance determinant for complete nucleotide sequencing.
The accessory region on plasmid pRSB111 contains a new macrolide resistance operon.
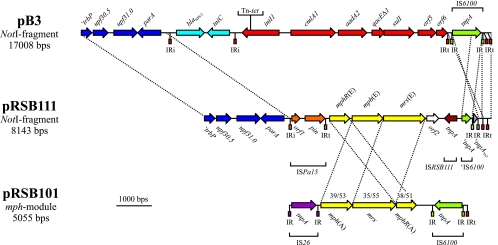
The complete nucleotide sequence of the 8-kb NotI fragment subcloned from pRSB111 was determined. The left-hand part of the sequenced fragment contains the IncP-1-specific genes ′trbP (mating pair formation), upf30.5 (unknown function), upf31.0 (restriction protection), and parA (multimer resolution). A 2,430-bp region spanning ′trbP to parA is 100% identical to the corresponding segment on pB3, demonstrating that both plasmids are very closely related (Fig. 1). Plasmid pRSB111 contains a copy of insertion sequence (IS) ISPa15 inserted 166 bp upstream of parA. The element is bordered by inverted repeat motifs that are nearly identical to the IRi and IRt inverted repeat (IR) ends of class 1 integron-containing Tn402 transposons. The parA upstream region seems to represent a hot spot for the insertion of mobile elements related to Tn402 since plasmid pB3 contains an insertion of a so-called cryptic integron 253 bp upstream of parA (22). ISPa15 elements were previously found on the conjugative IncU tetracycline resistance plasmid pFBAOT6 of Aeromonas punctata isolated from hospital effluent (32) and the clinical Pseudomonas aeruginosa IncP-6 plasmid Rms149 (20), which could mean that the original pRSB111 host bacterium, A. punctata, and P. aeruginosa share a common pool of mobile genetic elements. The occurrence of ISPa15 on plasmids of different incompatibility groups isolated from different hosts in different countries (Aeromonas punctata harboring pFBAOT6 was isolated in the United Kingdom, whereas Pseudomonas aeruginosa carrying Rms149 originates from Frankfurt, Germany) indicates the wide dissemination of this element and/or of cognate plasmids.
FIG. 1.
Comparison of the accessory resistance-conferring regions of plasmids pB3 (22), pRSB111, and pRSB101 (40). Coding regions are shown as arrows indicating the direction of transcription. Genes ′trbP, upf30.5, upf31.0, and parA represent IncP-1β-specific backbone module genes (blue). Plasmid pRSB111 contains the macrolide resistance-conferring module mphR(E)-mph(E)-mrx(E) (yellow) flanked by insertion sequences (ISPa15 and ISRSB111). Two truncated transposable elements (IS6100 and Tn3-like elements) were identified in the vicinity of ISRSB111. Plasmid pB3 contains a class 1 integron (red) composed of the integrase gene intI1, the resistance gene cassettes cmlA1 and aadA2, and the 3′ conserved module genes qacEΔ1, sul1, orf5, and orf6. The pB3 integron is flanked by a copy of IS6100 (green) downstream of orf6. Upstream of parA pB3 carries an insertion of a so-called cryptic integron (light blue), consisting of the β-lactamase gene blaNPS-2 and the resolvase gene tniC. Dashed lines between plasmids pB3 and pRSB111 indicate regions showing 100% nucleotide sequence identity. The macrolide resistance-conferring region of plasmid pRSB101 is flanked by the insertion sequences IS26 (lilac) and IS6100. Homologous regions of plasmids pRSB111 and pRSB101 are marked by dashed lines, and the numbers above the pRSB101 macrolide resistance-conferring genes indicate the identities and similarities of the encoded gene products to the corresponding proteins of pRSB111. Terminal IRs of transposable elements are symbolized by rectangles.
The insertion sequence ISPa15 flanks a macrolide resistance-conferring region (whose role in conferring macrolide resistance was experimentally demonstrated [see below]) consisting of three genes, namely, mphR(E), mph(E), and mrx(E). Putative promoters are located upstream of mphR(E) and mph(E). The deduced gene product of mphR(E) is a putative transcriptional regulator of the TetR/AcrR family (COG1309, pfam00440) and shows 40% identity to MphR(A), which is encoded by the multiresistance plasmid pRSB101 (40). The second gene product of this region, Mph(E), is a putative macrolide phosphotransferase (COG3173, pfam01636) and shares 40% sequence identity with Mph(A) encoded on pRSB101. The mrx(E) gene encodes a putative transmembrane transport protein (pfam05877) that is 34% identical to the hydrophobic protein Mrx of plasmid pRSB101. Mrx(E) also shows limited similarity to permeases of the major facilitator superfamily. The mph(E) and mrx(E) genes are probably translationally coupled since the mph(E) stop codon overlaps with the predicted mrx(E) start codon. Remarkably, the arrangement of genes in the pRSB111 macrolide determinant mphR(E)-mph(E)-mrx(E) differs from the arrangement of the mph(A)-mrx-mphR(A) operon present on the multiresistance plasmids pRSB101 and pRSB107 (39) (Fig. 1). In summary, plasmid pRSB111 contains a new macrolide resistance determinant (indexed by an “E”) that is only distantly related to the well-known mph(A)-mrx-mphR(A) macrolide resistance operon.
On the right-hand side the macrolide resistance-conferring region is bordered by the insertion sequence ISRSB111, which encodes a transposase (COG3293) that is 89% identical to a putative transposase from Paracoccus denitrificans (GenBank accession no. ZP_00629048). Inverted repeats could not be identified in the vicinity of the ISRSB111 tnpA gene. Thus, it appears that the pRSB111 macrolide resistance-conferring region is flanked by IS elements (ISPa15 and ISRSB111) on each side, which resembles the arrangement of the macrolide resistance operon mph(A)-mrx-mphR(A), which is flanked by IS26 and IS6100 (Fig. 1). IS elements therefore could have played an important role in the acquisition of the erythromycin resistance determinants. ISRSB111 inserted into a copy of IS6100, thereby deleting the 5′ part of the corresponding transposase gene tnpAIS6100. On the right-hand side, IS6100 is terminated by the IS6100-specific inverted repeat motif (Fig. 1). Insertion of IS6100 truncated another transposable element that is related (96% identity) to a Tn3 family transposon present on pFBAOT6 from Aeromonas punctata (32). The Tn3-like transposon transposed into the terminal end (IRt end) of a Tn402-like element that had probably been present on the pRSB111 ancestor plasmid. The remaining part of the NotI fragment sequenced (251 bp), including two IRt motifs, is completely identical to the corresponding region of pB3, again supporting the possibility that pRSB111 and pB3 derived from a common ancestor plasmid that already contained two interleaved, nested Tn402-like elements. Starting from a common ancestor, evolution of the accessory module on pRSB111 might have involved subsequent insertions of two transposable elements (namely, a Tn3-like transposon and IS6100), the incorporation of a module containing the macrolide resistance-conferring region, and the deletion of most of the Tn402 derivative. In contrast, the accessory module on pB3 contains a Tn402-based class 1 integron consisting of the integrase gene intI1, a resistance gene cassette region, and an integron-specific 3′ conserved segment (Fig. 1). Insertion of additional resistance-conferring modules into the integron platform seems to occur very frequently, thus facilitating extension of antibiotic resistance-conferring regions (22, 25, 31, 39, 40).
The pRSB111 resistance-conferring region confers resistance to the macrolides azithromycin, clarithromycin, erythromycin, roxithromycin, and tylosin.
Plasmid pRSB111 contains a so far unknown macrolide resistance-conferring region. Therefore, we analyzed the mediated resistance profile using the macrolides azithromycin, clarithromycin, erythromycin, roxithromycin, and tylosin, as well as the lincosamines clindamycin and lincomycin, in a disk diffusion assay. The pRSB111-encoded macrolide resistance-conferring region confers resistance to the macrolide antibiotics tested but not to the lincosamines tested (data not shown).
Since the pRSB111 macrolide resistance-conferring region shows weak similarity to the corresponding region on the multiresistance plasmid pRSB101, we performed MIC tests using both plasmids to investigate possible differences in their resistance profiles. MIC tests were done with erythromycin, roxithromycin, and tylosin (Table 2). Both plasmids conferred high-level erythromycin and roxithromycin resistance to the host bacterium, Pseudomonas sp. strain B13 GFP1, while only pRSB111 was able to decrease the strain's susceptibility to tylosin.
TABLE 2.
MICs mediated by macrolide resistance plasmid pRSB111 compared to those conferred by multiresistance plasmid pRSB101
| Antibiotic | MIC (μg/ml)
|
||
|---|---|---|---|
| Pseudomonas sp. strain B13 GFP1 | Pseudomonas sp. strain B13 GFP1(pRSB101) | Pseudomonas sp. strain B13 GFP1(pRSB111) | |
| Erythromycin | 20 | 2,500 | 2,500 |
| Roxithromycin | 25 | 1,200 | 1,400 |
| Tylosin | 150 | 150 | 400 |
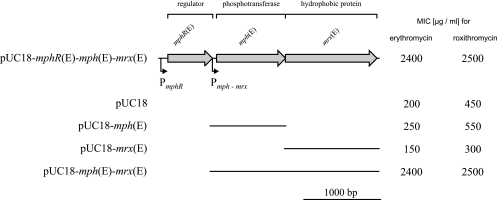
To test the extent to which individual genes of the macrolide resistance-conferring region contribute to the observed resistance phenotype, mphR(E), mph(E), and mrx(E) were amplified by PCR and subcloned into vector pUC18 (Fig. 2). It appeared that mph(E) or mrx(E) alone did not significantly affect susceptibility to erythromycin and roxithromycin, while the presence of both genes was necessary to confer high-level resistance to the two macrolides. Due to the high intrinsic insensitivity of E. coli DH5α, tylosin MICs could not be determined for the constructs.
FIG. 2.
Contributions of selected genes encoded in the pRSB111 macrolide resistance-conferring region to the erythromycin and roxithromycin resistance phenotype. The coding regions of mphR(E), mph(E), and mrx(E) are indicated by arrows. Promoter structures upstream of mphR(E) and mph(E) are marked by little arrows. The bars below the genetic map represent fragments of the macrolide resistance-conferring region that were amplified by PCR and cloned into vector pUC18. The MICs for E. coli strain DH5α carrying the different recombinant plasmids and pUC18 (control) are given on the right.
In summary, the macrolide resistance-conferring regions of pRSB101 and pRSB111 considerably differ at the nucleotide sequence level and can be distinguished with respect to their resistance profiles.
Plasmid pRSB111 is self-transmissible and has a broad host range.
Restriction analysis of the plasmids pB3 and pRSB111 showed that their backbones, including the conjugative transfer (tra) and mating pair formation (trb) modules, are conserved. For plasmid pB3 it is known that it transfers at high frequencies between different classes of proteobacteria (22). To examine the transfer properties of plasmid pRSB111, mating experiments with different recipient strains were done. Plasmid pRSB111 transfers at high frequencies from E. coli DH5α to E. coli CV60 (1.05 × 10−1) and Pseudomonas sp. strain B13 GFP1 (6.1 × 10−2) and transfers at a lower frequency to Ralstonia eutropha GFP3 (1.4 × 10−6). Different studies have shown that plasmid transfer greatly depends on the plasmid donor and recipient strains (9, 10, 22). The transfer frequencies of pRSB111 are comparable to those of other IncP-1β resistance-conferring plasmids isolated from wastewater treatment plant bacteria (12, 22). Consequently, pRSB111 was also classified as a highly mobile, self-transmissible broad-host-range plasmid.
Acknowledgments
We thank the Bioinformatics Resource Facility at the Center for Biotechnology (Bielefeld University) for bioinformatic support. We also thank Michael Stiens for critically reading the manuscript.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Bell, S. C., S. L. Senini, and J. G. McCormack. 2005. Macrolides in cystic fibrosis. Chron. Respir. Dis. 2:85-98. [DOI] [PubMed] [Google Scholar]
- 2.Blondeau, J. M. 2002. The evolution and role of macrolides in infectious diseases. Expert Opin. Pharmacother. 3:1131-1151. [DOI] [PubMed] [Google Scholar]
- 3.Blondeau, J. M., E. DeCarolis, K. L. Metzler, and G. T. Hansen. 2002. The macrolides. Expert Opin. Investig. Drugs 11:189-215. [DOI] [PubMed] [Google Scholar]
- 4.Chu, D. T. 1999. Recent developments in macrolides and ketolides. Curr. Opin. Microbiol. 2:467-474. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. 15th informational Supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 7.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 8.Debremaeker, D., D. Visky, H. K. Chepkwony, A. Van Schepdael, E. Roets, and J. Hoogmartens. 2003. Analysis of unknown compounds in azithromycin bulk samples with liquid chromatography coupled to ion trap mass spectrometry. Rapid Commun. Mass. Spectrom. 17:342-350. [DOI] [PubMed] [Google Scholar]
- 9.De Gelder, L., F. P. Vandecasteele, C. J. Brown, L. J. Forney, and E. M. Top. 2005. Plasmid donor affects host range of promiscuous IncP-1β plasmid pB10 in an activated-sludge microbial community. Appl. Environ. Microbiol. 71:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionisio, F., I. Matic, M. Radman, O. R. Rodrigues, and F. Taddei. 2002. Plasmids spread very fast in heterogeneous bacterial communities. Genetics 162:1525-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn, E., M. Hellwig, W. Reineke, and H. J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 12.Dröge, M., A. Pühler, and W. Selbitschka. 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263:471-482. [DOI] [PubMed] [Google Scholar]
- 13.Fluit, A. C., M. R. Visser, and F. J. Schmitz. 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 14:836-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillard, M., T. Vallaeys, F. J. Vorhölter, M. Minoia, C. Werlen, V. Sentchilo, A. Pühler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaynor, M., and A. S. Mankin. 2003. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top Med. Chem. 3:949-961. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11:614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Götz, A., R. Pukall, E. Smit, E. Tietze, R. Prager, H. Tschäpe, J. D. van Elsas, and K. Smalla. 1996. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halpern, M. T., J. K. Schmier, L. M. Snyder, C. Asche, P. W. Sarocco, B. Lavin, R. Nieman, and L. A. Mandell. 2005. Meta-analysis of bacterial resistance to macrolides. J. Antimicrob. Chemother. 55:748-757. [DOI] [PubMed] [Google Scholar]
- 22.Heuer, H., R. Szczepanowski, S. Schneiker, A. Pühler, E. M. Top, and A. Schlüter. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591-3599. [DOI] [PubMed] [Google Scholar]
- 23.Hynes, M. F., R. Simon, and A. Pühler. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate Patc58. Plasmid 13:99-105. [DOI] [PubMed] [Google Scholar]
- 24.Jain, R., and L. H. Danziger. 2004. The macrolide antibiotics: a pharmacokinetic and pharmacodynamic overview. Curr. Pharm. Des. 10:3045-3053. [DOI] [PubMed] [Google Scholar]
- 25.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mabe, S., J. Eller, and W. S. Champney. 2004. Structure-activity relationships for three macrolide antibiotics in Haemophilus influenzae. Curr. Microbiol. 49:248-254. [DOI] [PubMed] [Google Scholar]
- 27.Mach, P. A., and D. J. Grimes. 1982. R-plasmid transfer in a wastewater treatment plant. Appl. Environ. Microbiol. 44:1395-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nwosu, V. C. 2001. Antibiotic resistance with particular reference to soil microorganisms. Res. Microbiol. 152:421-430. [DOI] [PubMed] [Google Scholar]
- 31.Partridge, S. R., and R. M. Hall. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185:6371-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes, G., J. Parkhill, C. Bird, K. Ambrose, M. C. Jones, G. Huys, J. Swings, and R. W. Pickup. 2004. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70:7497-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts, M. C. 2004. Distribution of macrolide, lincosamide, streptogramin, ketolide and oxazolidinone (MLSKO) resistance genes in gram-negative bacteria. Curr. Drug Targets Infect. Disord. 4:207-215. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Shinkai, M., and B. K. Rubin. 2005. Macrolides and airway inflammation in children. Paediatr. Respir. Rev. 6:227-235. [DOI] [PubMed] [Google Scholar]
- 37.Stratton, C. W. 1998. Macrolides, lincosamides, and streptogramins: new agents and new roles. Antimicrob. Infect. Dis. Newsl. 17:89-92. [Google Scholar]
- 38.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Pühler, and A. Schlüter. 2005. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]
- 40.Szczepanowski, R., I. Krahn, B. Linke, A. Goesmann, A. Pühler, and A. Schlüter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 41.Tauch, A., A. Schlüter, N. Bischoff, A. Goesmann, F. Meyer, and A. Pühler. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene blaNPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Genet. Genomics 268:570-584. [DOI] [PubMed] [Google Scholar]
- 42.Tennstedt, T., R. Szczepanowski, S. Braun, A. Pühler, and A. Schlüter. 2003. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 45:239-252. [DOI] [PubMed] [Google Scholar]
- 43.Tennstedt, T., R. Szczepanowski, I. Krahn, A. Pühler, and A. Schlüter. 2005. Sequence of the 68,869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53:218-238. [DOI] [PubMed] [Google Scholar]
- 44.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 46.Zhanel, G. G., M. Dueck, D. J. Hoban, L. M. Vercaigne, J. M. Embil, A. S. Gin, and J. A. Karlowsky. 2001. Review of macrolides and ketolides: focus on respiratory tract infections. Drugs 61:443-498. [DOI] [PubMed] [Google Scholar]