Abstract
The chromosomal gene aph(3′)-IIb, encoding an aminoglycoside 3′-phosphotransferase in Pseudomonas aeruginosa, was cloned and overexpressed in Escherichia coli. The APH(3′)-IIb enzyme was purified as a monomer in a two-step procedure and was shown to phosphorylate its substrates at the C-3′-OH position, with kcat/Km values of 0.4 × 104 to 36 × 104 M−1 s−1.
Despite emerging bacterial resistance, aminoglycosides remain vital weapons against some multiresistant pathogens, such as Pseudomonas aeruginosa. The primary mechanism of resistance to aminoglycosides is the bacterial acquisition of enzymes that structurally modify these antibiotics. Among these enzymes, aminoglycoside 3′-phosphotransferases [APH(3′)s], which catalyze transfer of the γ-phosphoryl group of ATP to the 3′-hydroxyl of many drugs, are widely represented (5, 12). Ten years ago, the chromosomal gene aph(3′)-IIb was identified in P. aeruginosa (2), and it was believed that the “uniform resistance” of P. aeruginosa to kanamycin is greatly contributed to by the presence of this gene. aph(3′)-IIb is located in the pseudomonal genome in an operon downstream of the hpaA gene, which encodes an autoregulator involved in the metabolism of 4-hydroxyphenylacetic acid (4-HPA) (13). Interestingly, unlike most other aminoglycoside-modifying enzymes, which are constitutive, the hpaA-aph(3′)-IIb operon was found to be induced by 4-HPA. The present study describes the cloning, simple purification, and initial kinetic and biochemical characterization of the APH(3′)-IIb enzyme.
The aph(3′)-IIb gene (GenBank accession number X90856) was amplified from genomic DNA of P. aeruginosa ATCC 27538 by PCR (N-terminal primer, 5′-CGTCATATGGGTCCATGGATGATGCAGCCACCTCC-3′; C-terminal primer, 5′-CTGCTGCAGCACGGATCCTAGAAGAACTCGTCCAATAG-3′; the start codon is in bold). A 30-μl PCR mixture included 100 ng of chromosomal DNA, 10 ng/μl of each primer, 7.5% (vol/vol) dimethyl sulfoxide (DMSO), 0.2 mM deoxynucleoside triphosphates, ThermoPol buffer, and 2 units of Vent DNA polymerase (New England Biolabs). The PCR product was purified, digested with NcoI and BamHI, cloned into NcoI-BamHI-restricted plasmid pET11d (Novagen) to yield pET11d-aph(3′)-IIb, and sequenced.
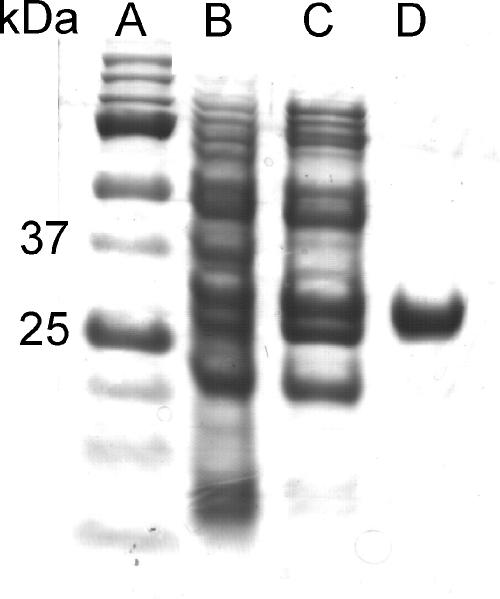
The procedure for purification of APH(3′)-IIb was adapted and optimized from protocols previously reported for other APH enzymes (7, 10). A 2-liter culture of Escherichia coli BL21(DE3) CodonPlus-RP (Stratagene) carrying the pET11d-aph(3′)-IIb plasmid, in Luria-Bertani medium containing 100 μg/ml ampicillin and 25 μg/ml kanamycin A, was grown with isopropyl-β-d-thiogalactopyranoside (2 mM) to a final turbidity of 2.5 (optical density at 600 nm). After harvesting, the cells were resuspended in 16 ml buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 200 mM NaCl, 0.1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride), disrupted (two passages through a French press), and centrifuged. The supernatant was diluted fourfold with buffer A1 (50 mM Tris-HCl [pH 8.0], 1 mM EDTA) and loaded onto a Q-Sepharose column (Pharmacia). The proteins were eluted with 1 column volume (CV) of a 0-to-20% linear gradient of buffer B1 (A1 plus 1 M NaCl), followed by 2 CVs of 20% B1, 6 CVs of a 20-to-50% B1 gradient, and 3 CVs of 50% B1. APH(3′) active fractions were pooled, diluted threefold with buffer A2 (50 mM Tris-HCl, pH 7.4), and loaded onto a neomycin-conjugated affinity column (prepared by coupling neomycin B with N-hydroxysuccinimide-activated Sepharose [HiTrap-HP, 5 ml; GE Healthcare], according to the manufacturer's protocol). The protein was eluted with 22 CVs of 0-to-100% buffer B2 (A2 plus 1.2 M KCl). Active fractions were pooled and concentrated to yield 18 mg of purified protein (Fig. 1; also Table S1 in the supplemental material). The Mr was determined using a Superdex 75 (10/30) gel filtration column (buffer A2 plus 100 mM KCl).
FIG. 1.
SDS-PAGE of the purification procedure of APH(3′)-IIb. (A) Molecular size markers (bottom to top) of 10, 15, 20, 25, 37, 50, 75, and 100 kDa; (B) crude extract; (C) protein preparation after Q-Sepharose column elution; (D) purified APH(3′)-IIb after neomycin affinity column elution.
The enzymatic activity was monitored as previously described (3, 11). Km and kcat values were determined by nonlinear regression analysis using GraFit 5.0 (6). MICs were determined using the double-microdilution method according to the CLSI (formerly NCCLS) (8). The preparative phosphorylation of neomycin B was done with purified APH(3′)-IIb as previously described for kanamycin A (10).
During the cloning and expression of the APH(3′)-IIb protein, we encountered two problems which arose from the high GC content of the aph(3′)-IIb gene (2). The addition of DMSO to the PCR mixture was essential for successful amplification of the gene (1). Indeed, we were able to identify this gene in the standard P. aeruginosa strains ATCC 27853 and PAO1, as well as in six other clinical isolates, only after the addition of DMSO. Moreover, the APH(3′)-IIb enzyme contains five prolines encoded by the rare codon CCC; therefore, the overproduction of the protein in the CodonPlus-RP E. coli strain (which coexpresses the rare proline tRNA) allowed higher expression yields. In addition, the use of freshly transformed cells to inoculate the starter culture was crucial for overexpression of the protein, probably because of the unfavorable consumption of ATP by this enzyme in the bacterium, leading to selective pressure against maintenance of the plasmid (4).
Using anion-exchange and affinity columns, we obtained 18 mg of purified protein from a 2-liter culture. The purified APH(3′)-IIb protein is a monomer in solution with a Mr of about 31,000, in agreement with its calculated Mr (29,900). Unlike APH(3′)-Ia and APH(3′)-IIIa, which undergo spontaneous dimerization in the absence of dithiothreitol (7, 10), no dimerization was observed for APH(3′)-IIb. Based on the measured kcat/Km values (Table 1), amikacin is the poorest substrate for the enzyme. This decrease in specificity for amikacin is caused primarily by its poor ability to saturate the enzyme, as judged from its elevated Km value (440 μM) compared to the Km values of the other aminoglycosides (3.1 to 17.5 μM). Indeed, unlike other substrates tested, amikacin has a (S)-4-amino-2-hydroxy-butyryl substitution at the N-1 position, which is believed to be unfavorable for interactions with the enzyme active site. Lividomycin A, tobramycin, and gentamicin, which lack C-3′-OH, were not substrates for APH(3′)-IIb. The observed kinetic data are consistent with the antibacterial data (Table 2). Both P. aeruginosa and E. coli strains harboring APH(3′)-IIb are resistant to good substrates but are sensitive to those which were not substrates of the enzyme (tobramycin and gentamicin) or poor substrates (amikacin). The identical MICs of amikacin (12 μg/ml) against the two isogenic strains of E. coli indicate that the reason for the observed sensitivity of the E. coli strain harboring APH(3′)-IIb is the inferior activity of this enzyme towards amikacin, rather than reduced permeability of amikacin. In fact, comparison of its Km value (440 μM) with the MICs of 16 and 4 μM (12 and 3 μg/ml) against E. coli and P. aeruginosa, respectively, suggests that the bacteria are killed at far lower concentrations before the enzyme's full catalytic potential is reached. This observation with amikacin and the substrate profile of the purified APH(3′)-IIb are similar to those for the previously reported enzyme APH(3′)-IIa (9).
TABLE 1.
Kinetic parameters for phosphorylation of aminoglycosides by APH(3')-IIb
| Compound | Km (μM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|
| Kanamycin A | 3.1 ± 0.4 | 1.1 ± 0.1 | (3.6 ± 0.6) × 105 |
| Neomycin B | 17.5 ± 0.9 | 2.1 ± 0.1 | (1.2 ± 0.1) × 105 |
| Neamine | 10.3 ± 0.5 | 1.7 ± 0.1 | (1.7 ± 0.1) × 105 |
| Ribostamycin | 10.0 ± 0.5 | 1.9 ± 0.1 | (1.9 ± 0.1) × 105 |
| Amikacin | 440 ± 20 | 1.6 ± 0.1 | (4.0 ± 0.3) × 103 |
TABLE 2.
MICs of several aminoglycosides against P. aeruginosa, the E. coli background strain, and its engineered varianta
| Compound | MIC (μg/ml)
|
||
|---|---|---|---|
| P. aeruginosa | E. coli | E. coli R | |
| Kanamycin A | 256 | 12 | 768 |
| Neomycin B | 128 | 12 | 96 |
| Neamine | 512 | 96 | 1,536 |
| Ribostamycin | 384 | 12 | 768 |
| Amikacin | 3 | 12 | 12 |
| Tobramycin | 1 | 6 | 6 |
| Gentamicin | 2 | 6 | 6 |
P. aeruginosa ATCC 27853, E. coli BL21(DE3) CodonPlus-RP, and E. coli BL21(DE3) CodonPlus-RP/pET11d-aph(3′)-IIb expressing the APH(3')-IIb enzyme (E. coli R).
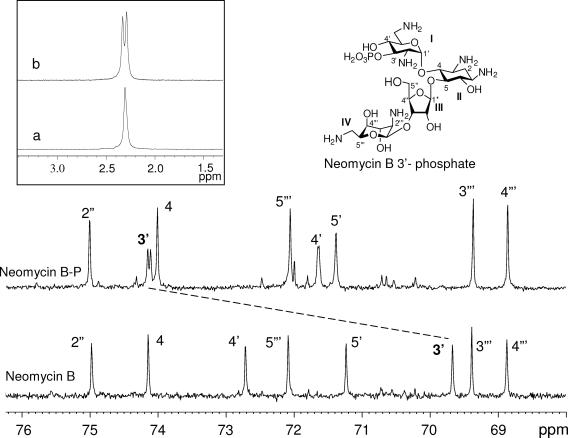
Monitoring the time courses of APH(3′)-IIb-catalyzed phosphorylations by proton-coupled 31P nuclear magnetic resonance (31P-NMR) [50 mM Tris-HCl buffer (pD 7.1), 40 mM KCl, 10 mM MgCl2, 132 mM ATP, 150 μl purified APH(3′)-IIb (0.3-U/ml stock solution), and 44 mM aminoglycoside in a total volume of 0.65 ml D2O] (3) revealed a single phosphate resonance (doublet, with a typical three-bond P—O—C—H coupling constant [J] of ≈8 Hz [inset in Fig. 2 ], indicative of phosphate attached to a carbon bearing only one hydrogen) for all substrates but no phosphorylation product for lividomycin, indicating that APH(3′)-IIb is highly specific for C-3′-OH. As such, APH(3′)-IIb differs from other APH(3′) enzymes, which perform multiple phosphorylations on neomycin B and C-5" phosphorylation on lividomycin (3, 11). For example, proton-coupled 31P-NMR experiments with APH(3′)-IIIa revealed two phosphorylation sites for neomycin B and multiple phosphorylations on other synthetic derivatives (3).
FIG. 2.
Partial 13C proton-decoupled NMR spectra of neomycin B and phosphorylated neomycin B (neomycin B-P) at 125.77 MHz. The dashed line shows the deshielding of the neomycin B C-3′ carbon (singlet) to its phosphorylated product (doublet). The inset shows proton-decoupled (a) and proton-coupled (b) 31P-NMR spectra of neomycin B-P at 202.46 MHz. All the spectra were recorded on a Bruker Avance-500 spectrometer.
To confirm our results, as an example, the product of phosphorylation of neomycin B by the APH(3′)-IIb enzyme was isolated, and the modification was confirmed to occur solely at the 3′-OH (Fig. 2; see also Table S2 in the supplemental material). Relatively large deshielding of the C-3′ proton and the C-3′ carbon resonances in the phosphorylated product (4.43 and 74.11 ppm) from the corresponding resonances in neomycin B (3.97 and 69.67 ppm) are indicative that the phosphate group is linked at the C-3′ position. Furthermore, additional splitting observed for both the C-3′ proton (J = 8.2 Hz) and the C-3′ carbon (J = 4.6 Hz) in the phosphorylated product is consistent with three-bond (P—O—C—H) and two-bond (P—O—C) couplings of these nuclei with phosphorus (11). More biochemical and structural studies of the APH(3′)-IIb enzyme are under way.
Supplementary Material
Acknowledgments
This research was supported by the Israel Science Foundation, founded by the Israel Academy of Sciences and Humanities (grant no. 766/04), and in part by the Technion V.P.R. Fund. D. Shallom-Shezifi is supported in part at the Technion by a Neaman Fellowship.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 6 November 2006.
REFERENCES
- 1.Chakrabarti, R., and C. E. Schutt. 2001. The enhancement of PCR amplification by low molecular-weight sulfones. Gene 274:293-298. [DOI] [PubMed] [Google Scholar]
- 2.Hachler, H., P. Santanam, and F. H. Kayser. 1996. Sequence and characterization of a novel chromosomal aminoglycoside phosphotransferase gene, aph(3′)-IIb, in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:1254-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hainrichson, M., V. Pokrovskaya, D. Shallom-Shezifi, M. Fridman, V. Belakhov, D. Shachar, S. Yaron, and T. Baasov. 2005. Branched aminoglycosides: biochemical studies and antibacterial activity of neomycin B derivatives. Bioorg. Med. Chem. 13:5797-5807. [DOI] [PubMed] [Google Scholar]
- 4.Kim, C., J. Y. Cha, H. Yan, S. B. Vakulenko, and S. Mobashery. 2006. Hydrolysis of ATP by aminoglycoside 3′-phosphotransferases: an unexpected cost to bacteria for harboring an antibiotic resistance enzyme. J. Biol. Chem. 281:6964-6969. [DOI] [PubMed] [Google Scholar]
- 5.Kim, C., and S. Mobashery. 2005. Phosphoryl transfer by aminoglycoside 3′-phosphotransferases and manifestation of antibiotic resistance. Bioorg. Chem. 33:149-158. [DOI] [PubMed] [Google Scholar]
- 6.Leatherbarrow, R. J. 2001. GraFit 5. Erithacus Software Ltd., Horley, United Kingdom.
- 7.McKay, G. A., P. R. Thompson, and G. D. Wright. 1994. Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification, and substrate specificity. Biochemistry 33:6936-6944. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 1994. Performance standards for antimicrobial susceptibility testing. Fifth information supplement: approved standard M100-S5. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 9.Siregar, J. J., S. A. Lerner, and S. Mobashery. 1994. Purification and characterization of aminoglycoside 3′-phosphotransferase type IIa and kinetic comparison with a new mutant enzyme. Antimicrob. Agents Chemother. 38:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siregar, J. J., K. Miroshnikov, and S. Mobashery. 1995. Purification, characterization, and investigation of the mechanism of aminoglycoside 3′-phosphotransferase type Ia. Biochemistry 34:12681-12688. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, P. R., D. W. Hughes, and G. D. Wright. 1996. Regiospecificity of aminoglycoside phosphotransferase from Enterococci and Staphylococci (APH(3′)-IIIa). Biochemistry 35:8686-8695. [DOI] [PubMed] [Google Scholar]
- 12.Wright, G. D., and P. R. Thompson. 1999. Aminoglycoside phosphotransferases: proteins, structure, and mechanism. Front. Biosci. 4:D9-D21. [DOI] [PubMed] [Google Scholar]
- 13.Zeng, L., and S. Jin. 2003. aph(3′)-IIb, a gene encoding an aminoglycoside-modifying enzyme, is under the positive control of surrogate regulator HpaA. Antimicrob. Agents Chemother. 47:3867-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.