Abstract
A class of antimicrobial peptides involved in host defense consists of sequences rich in Arg and Trp-R and -W. Analysis of the pharmacophore in these peptides revealed that chains as short as trimers of sequences such as WRW and RWR have antimicrobial activity (M. B. Strom, B. E. Haug, M. L. Skar, W. Stensen, T. Stiberg, and J. S. Svendsen, J. Med. Chem. 46:1567-1570, 2003). To evaluate the effect of chain length on antimicrobial activity, we synthesized a series of peptides containing simple sequence repeats, (RW)n-NH2 (where n equals 1, 2, 3, 4, or 5), and determined their antimicrobial and hemolytic activity. The antimicrobial activity of the peptides increases with chain length, as does the hemolysis of red blood cells. Within the experimental error, longer peptides (n equals 3, 4, or 5) show similar values for the ratio of hemolytic activity to antibacterial activity, or the hemolytic index. The (RW)3 represents the optimal chain length in terms of the efficacy of synthesis and selectivity as evaluated by the hemolytic index. Circular dichroism spectroscopy indicates that these short peptides appear to be unfolded in aqueous solution but acquire structure in the presence of phospholipids. Interaction of the peptides with model lipid vesicles was examined using tryptophan fluorescence. The (RW)n peptides preferentially interact with bilayers containing the negatively charged headgroup phosphatidylglycerol relative to those containing a zwitterionic headgroup, phosphatidylcholine.
Native and synthetic antimicrobial peptides (AMPs) have been considered as a potential alternative source of new antibiotics (4, 13, 59) for some time. As prospective antibiotics, AMPs show a broad spectrum of activities against gram-negative and gram-positive bacteria, including antibiotic-resistant bacterial strains and some fungi, viruses, and parasites (12). Unfortunately, due to relatively high inhibitory concentrations, sensitivity to salts, and cytotoxic effects (12), their utility is currently limited to topical applications, with the exception of injectable polymyxin (5). A further obstacle to the development of AMPs as therapeutics lies in the cost associated with manufacturing large quantities at competitive costs. Although some progress has been made in the recombinant expression of fusion proteins comprising multiple copies of AMPs (28), chemical peptide synthesis remains the preferred option for quality control purposes. Different approaches are being pursued in efforts to increase the effectiveness of AMPs, including alteration of sequences, inclusion of unnatural d-amino acids or beta-amino acids, cyclization of peptides, peptoid mimics, and synthesis of multivalent constructs of short peptides (8, 25, 26, 33, 35, 51). At present, short AMPs that minimize damage to host cells or tissues would appear to be the most promising candidates for large-scale production (47).
Known AMPs differ dramatically in size (from 12 to more than 50 amino acids), sequence, and structure and share only amphipathicity and positive charge (12, 59). This lack of sequence or structural homology makes it challenging to design potent synthetic antimicrobial peptides with the desired activities or to predict the activity of peptides in vivo (13). A comparison of AMP sequences reveals that two types of side chains are essential for antimicrobial activity. The cationic side chains arginine (R), lysine (K), and histidine (H) are thought to mediate peptide interactions with negatively charged membranes and/or cell walls of bacteria, including lipopolysaccharide (5). Bulky nonpolar side chains, such as proline (P), phenylalanine (F), and tryptophan (W), occur frequently in AMPs, presumably providing lipophilic anchors that ultimately induce membrane disruption (51). Certain AMPs are evidently helix forming, while others favor beta structure or neither of these structures. The role of conformation per se seems clear in helix-forming peptides but not universally (10, 15). How peptides as short as dimers achieve lytic activity relative to the longer membrane-spanning sequences also remains unclear (14). The effects of size, composition, and structure are not easy to deconvolute. One simplification of the problem is to investigate the effect of the length of peptides with simple repeated sequences (7) that contain balanced numbers of positively charged and hydrophobic side chains.
The side chains R and W appear in many AMPs that span a range of sizes and secondary structures. Lactoferricin B, indolicidin, and tritrpticin (2, 38, 40) are natural AMPs with sequences longer than 12 amino acids. PW2 and hexamers of the Ac-RRWWXX-NH2 sequence are active antibacterial peptides containing 6 to 12 amino acids derived from screening phage displays and synthetic combinatorial libraries, respectively (3, 52). Finally, antimicrobial tetrapeptides and modified dipeptides containing R and W have been shown to retain antibacterial activities (14, 47).
However, no obvious pattern of R and W emerges from such a survey. The role of RW-rich motifs in AMPs has been investigated using quantitative-structure-activity relationships (QSAR) (24, 45, 46, 53). Truncated peptide sequences from indolicidin and lactoferricin with conserved RW motifs retain antimicrobial activity even when their original secondary structure is lost (45, 53), suggesting that R and W content alone correlates with activity. QSAR analysis of the antimicrobial activities of R and W peptides suggests that charge and multiple W side chains are necessary, while W can be replaced by analogs with bulkier side chains (24, 46, 48). The results also suggest that, in short peptides, the order of amino acids is less important than the overall composition with respect to cationic and lipophilic residues (41, 46, 47).
Other studies suggest that the antimicrobial activity of peptides containing R is higher than those of peptides containing K (34, 41), while peptides containing W are more potent than those with either F or Y (8, 47, 49). The guanidinium group of R has a more dispersed positive charge than the single amine of K, possibly enhancing electrostatic interactions between peptides and the negatively charged bacterial membrane surface (39, 53). On the other hand, the bulkier W side chain may ensure more efficient interaction with membrane surfaces, allowing peptides to partition in the bilayer interface, in contrast with other nonpolar side chains such as F, P, or Y (46, 56). Recently, consideration of electrostatic effects, including dipole and quadrupole moments of R and W side chains, respectively, suggests that these properties may be involved in the ability to form hydrogen bonds once peptides associate with membranes (27, 53, 58).
While R and W play a role in many AMPs, the effects of chain length and composition on antimicrobial activity and selectivity have not been clearly distinguished. In this study, we synthesized a series of cationic peptides containing simple repeats, (RW)n-NH2 (where n equals 1, 2, 3, 4, or 5) (Table 1), and compared their antibacterial activities with their hemolytic activities. Since natural peptides, such as indolicidin (45), frequently have the C terminus amidated, removing a negative charge that appears to lower activity, we used N terminus-unprotected and C-terminal amide-protected peptides in this study. Selectivity was monitored by means of the hemolytic index (HI; also referred to as the membranolytic selectivity index), defined as the ratio of hemolytic to antibacterial activity (45, 51). Peptide conformation was investigated by circular dichroism (CD) spectroscopy. The interaction of peptides with two model lipids was examined using fluorescence spectroscopy.
TABLE 1.
Amino acid sequences and molecular weights of (RW)n-NH2 peptides
| Sequence | Formula | Molecular wt
|
|
|---|---|---|---|
| Calculated | Observed | ||
| RW-NH2 | C17H25N7O2 | 359.4 | 359.4 |
| (RW)2-NH2 | C34H47N13O4 | 701.8 | 701.8 |
| (RW)3-NH2 | C51H69N19O6 | 1,044.2 | 1,044.3 |
| (RW)4-NH2 | C68H91N25O8 | 1,386.6 | 1,386.7 |
| (RW)5-NH2 | C85H113N31O10 | 1,729.0 | 1,729.1 |
MATERIALS AND METHODS
Peptide design and synthesis.
The (RW)n-NH2 sequences were assembled on Rink amide resin from Nova Biochem (San Diego, CA) with a RAININ Instrument PS3 solid-phase synthesizer (Woburn, MA) using Fmoc (9-fluorenylmethoxycarbonyl) chemistry. Fmoc-Trp(butoxycarbonyl [Boc])/Arg(2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl [Pbf]), the coupling reagent HBTU [2-(1H-benzotriazol-1-yl) 1,1,3,3-tetramethyluroniumhexafluoro phosphate], and HOBT (N-hydroxybenzotriazol) were also purchased from Nova Biochem. Cleavage of the peptides from the resin was performed with 95% trifluoroacetic acid (TFA) in the presence of the scavenger, 2.5% triisopropylsilane (TIS), and 2.5% H2O. After precipitation with cold ether, samples were purified on a reverse-phase high-performance liquid chromatography C18 preparative column (2.2 by 25 cm, 300 Å; Grace Vydac Co., Hesperia, CA) with water and acetonitrile as eluants. Fractions containing product were pooled and lyophilized. The molecular weight of each peptide was confirmed by a Bruker matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (Billerica, MA), giving the observed molecular weights shown in Table 1.
Growth inhibition assays.
The antimicrobial activity of each peptide was tested by following standard broth microdilution protocols recommended by the National Committee for Clinical Laboratory Standard (30). Ampicillin- and streptomycin-resistant Escherichia coli (D31) and the multidrug-resistant Staphylococcus aureus strain ATCC BAA-44 were obtained from the E. coli Genetic Resource Center (Yale University, New Haven, CT) and the American Type Culture Collection (Rockville, MD), respectively. Bacteria were grown overnight in Mueller-Hinton broth at 37°C. Then, cultures were diluted in Mueller-Hinton broth to a final concentration range of 2 × 104 to 2 × 105 CFU/ml. Bacterial inocula were incubated at 37°C in phosphate-buffered saline (PBS) buffer (pH 7.2) with various volumes of twofold dilutions of peptide stock. The 18-hour absorbance data were used to calculate the percentage of inhibition for each sample by comparison with the absorbance of cultures without peptides. Bacterial growth was measured by turbidity as the optical density at 600 nm (OD600), using a Genesys 5 Spectrophotometer (Rochester, NY). All assays were carried out in triplicate. The concentration of peptide that resulted in 50% inhibition of growth was recorded as the IC50.
Minimal bactericidal concentrations.
The inhibitory concentrations of each peptide were determined through macrodilution antimicrobial testing. One hundred microliters of the initial inoculum of 5 × 105 CFU/ml were plated on Mueller-Hinton agar as the positive control, and 100 μl of the post-18-h inhibitory concentration test sample was plated on Mueller-Hinton agar to determine the minimal bactericidal concentrations (MBCs). The MBC50 is the lowest concentration of peptide that kills 50% of the strains.
Hemolysis assays.
The hemolytic activity of model peptides was assessed on fresh sheep erythrocytes (Fitzgerald Inc., Concord, MA). Peptide concentrations yielding 50% hemolysis were used as the hemolytic dose (HD50), determined from dose-response curves (32). The red blood cell suspension was incubated in PBS buffer (pH 7.2) with various volumes of peptide stocks at 37°C for 30 min and then spun down at 3,000 rpm for 10 min. The resulting supernatant was diluted by a factor of 40 in distilled water. The absorbance levels of the supernatants at λ was 540 nm (OD540) were measured with a UV spectrophotometer. Zero-hemolysis and 100%-hemolysis controls were obtained by incubating the cells with buffer and 1% Triton-X, respectively. The HI was defined as HD50/IC50.
Preparation of liposomes.
Small unilamellar lipid vesicles (SUV) for fluorescence spectroscopy were prepared as described by Morrissey (29). Briefly, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids in chloroform were purchased from Avanti Polar Lipids (Alabaster, AK). Following chloroform evaporation, the POPG and POPC lipids were resuspended in 20 mM sodium phosphate buffer (pH 7.4) with 100 mM NaCl. The suspensions were vigorously vortexed, and the samples were subsequently sonicated in an ultrasonic cleaner, Branson B-220 (Danbury, CT), until the solutions clarified.
CD spectra.
Far-UV CD spectra were recorded on an Aviv 202 CD spectrometer (Lakewood, NJ) using 0.1-cm-path-length Hellma CD cuvettes (Forest Hills, NY). The instrument was calibrated with (+)-10-camphorsulfonic acid standard purchased from Sigma-Aldrich Co. (St. Louis, MO). Spectra were recorded with 40 μM peptide in 20 mM phosphate buffer (pH 7.4, 100 mM NaCl) and 2 mM POPG or POPC SUV suspensions at 25°C. The concentration of the peptides was calibrated by the UV absorbance of tryptophan residue at 280 nm. All CD spectra shown have had the corresponding peptide-free solvent baselines subtracted. The results are expressed in terms of molar residue CD.
Peptide binding to lipids measured by tryptophan fluorescence.
The tryptophan fluorescence spectra of the peptides were measured using an F-2500 fluorescence spectrophotometer from Hitachi High-Technologies America, Inc. (Chicago, IL). Tryptophan residues were excited at a wavelength of 295 nm, and emission spectra were scanned from 300 to 450 nm using a scanning speed of 10 nm/s. Spectra were baseline corrected by subtracting blank spectra of the corresponding solutions without peptide. Experiments were carried out in 20 mM phosphate buffer (pH 7.4) with 100 mM NaCl. The concentration of peptides in all experiments was 10 μM, calibrated by UV absorbance of tryptophan residue at 280 nm. Measurements were made in triplicate for each peptide in buffer and in the presence of 500 μM each of POPG or POPC vesicles.
Fluorescence quenching experiments were conducted using acrylamide as a quencher. The concentrations of acrylamide ranged from 0.01 to 0.40 M, and the intensity of the spectra was recorded. The peak maxima (F) were then compared to those recorded in the absence of quenching reagents (F0). The effects of the quenching reagent on peptide fluorescence intensities were compared by means of the quenching constant (KSV) as determined by the Stern-Vollmer equation F0/F = 1 + KSV(Q), where Q is the concentration of quencher.
RESULTS
Comparison of antimicrobial and hemolytic activities of the peptides.
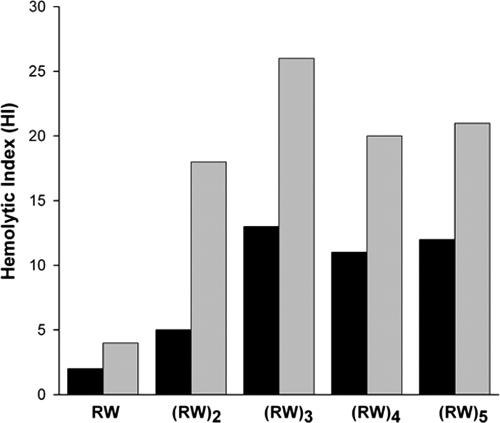
The calculated molecular weights and experimental determinations of the masses of the synthetic peptides indicate that the products correspond to the designed sequences (Table 1). All five peptides show antimicrobial activities (Table 2). Data for the shortest peptides are in agreement with data from Svendsen's group (14, 47). The chain length of the (RW)n peptides strongly correlates with antibacterial activity assayed by growth inhibition and colony formation: peptides with longer chains are much more effective in killing bacteria but increasingly stimulate hemolytic activity. The (RW)3, (RW)4, and (RW)5 peptides are potent antimicrobial agents, with all IC50s roughly in the μM range. Selectivity is measured by hemolytic index, defined as the ratio of HD50 to IC50. Figure 1 shows the hemolytic index as a function of chain length. While the longer chains are almost equally selective, (RW)3 offers an optimal choice in terms of efficiency of synthesis for both E. coli and S. aureus (Fig. 1).
TABLE 2.
Antimicrobial and hemolytic activities of (RW)n-NH2 peptides
| Peptide | IC50 (μM)a
|
MBC50 (μM)a
|
HD50 (μM)ab
|
||
|---|---|---|---|---|---|
| E. coli | S. aureus | E. coli | S. aureus | Red blood cells | |
| RW-NH2 | 4.3 × 103 | 2.1 × 103 | 4.7 × 103 | 2.3 × 103 | 8.9 × 103 |
| (RW)2-NH2 | 7.3 × 102 | 2.1 × 102 | 9.9 × 102 | 6.2 × 102 | 3.7 × 103 |
| (RW)3-NH2 | 16 | 8.0 | 29 | 15 | 2.1 × 102 |
| (RW)4-NH2 | 9.6 | 5.1 | 14 | 12 | 1.0 × 102 |
| (RW)5-NH2 | 6.2 | 3.6 | 7.2 | 6.9 | 76 |
Results are the means of three independent experiments performed in parallel.
HD50 values determined from dose-response curves are peptide concentrations corresponding to 50% hemolysis.
FIG. 1.
HI values. The HI, defined as the ratio of HD50 to IC50, indicates the selectivity of (RW)n-NH2 peptides (E. coli, black bars; S. aureus, gray bars).
Secondary structures of the peptides studied by CD.
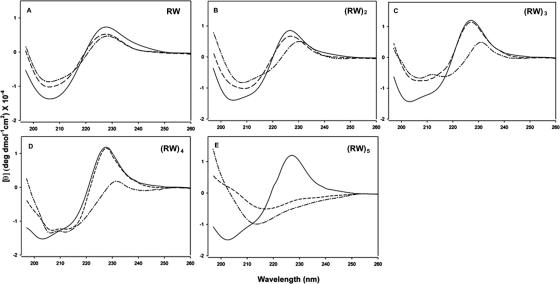
The CD spectra of the W-containing linear peptides (RW)n dissolved in buffer show negative bands in the region between 200 and 210 nm and a positive band between the 225- and 230-nm region (Fig. 2). The negative band around 200 nm is characteristic of small unfolded peptides, while the band at 225 nm is due to the indole side chain of W (21, 57), as seen in the CD spectrum of a peptide such as GGWGG containing a single chiral W residue (44). Each of the five peptides exhibits distinctive CD spectra in the presence of POPG and POPC. On binding a peptide to lipids, the decreased intensity at a low wavelength implies a loss of extended or unfolded conformations. The apparent conformational changes are a function of chain length and lipid composition. For example, while RW and (RW)2 have similar spectra and may be unfolded in the presence of water or lipids, the CD of (RW)3 and that of (RW)4 show differences in the presence of POPG but not in the presence of POPC (Fig. 2). (RW)5, interestingly, reveals large differences between water and both lipids. The presence of additional tryptophan side chains in the same molecule may affect the CD signal nonlinearly, and so these data cannot be interpreted more quantitatively. The trends with peptide length are hard to reconcile with the biological data in any case.
FIG. 2.
CD spectra of (RW)n-NH2 peptides in an aqueous solution (solid line) and in the presence of model lipid systems POPC (dashed line) and POPG (dashed-dotted line) in 20 mM phosphate buffer (pH 7.4; 100 mM NaCl). Data are expressed as mean residue ellipticities.
Peptide binding to lipids measured by tryptophan fluorescence spectroscopy.
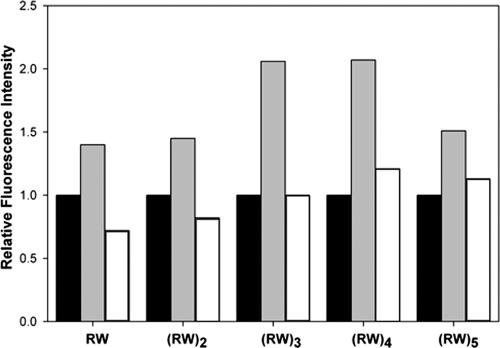
In order to monitor the extent to which the Trp side chains interact with lipids, we used fluorescence spectroscopy to probe the environment of Trp in the presence of lipids (20). Interactions between the Trp in our model peptides and the vesicle lipids result in a blue-shifted emission spectrum following excitation at 295 nm. The shift to shorter wavelengths was observed to varying degrees for the fluorescence emission spectra of all (RW)n peptides in the presence of phospholipids vesicles (Table 3). An increase in emission intensity is observed for the fluorescence emission when longer (RW)n-NH2 peptides (n, ≥3) bind to model lipids (Fig. 3). The observations of larger blue shifts and emission intensities for peptides binding to POPG suggest that the Trp side chain partitions preferentially into a more rigid, hydrophobic environment in POPG lipid bilayers than those of POPC (17). However, we do not detect any trends that correlate with the biological activity of the peptides: all the peptides show comparable blue shifts in the presence of POPG, consistent with a membrane interaction.
TABLE 3.
Fluorescence spectroscopy parameters measured for (RW)n-NH2 peptides in the presence and absence of POPG and POPC vesicles
| Peptides | λmax buffer (nm) | Blue shift (nm)
|
KSV (M−1)a
|
|||
|---|---|---|---|---|---|---|
| POPG | POPC | Buffer | POPG | POPC | ||
| RW-NH2 | 350 | 10 | 6.5 | 15 | 2.5 | 2.3 |
| (RW)2-NH2 | 350 | 11 | 6.0 | 14 | 2.7 | 2.5 |
| (RW)3-NH2 | 349 | 11 | 6.0 | 18 | 1.8 | 2.8 |
| (RW)4-NH2 | 350.5 | 10 | 6.0 | 17 | 2.3 | 5.8 |
| (RW)5-NH2 | 348 | 11 | 8.5 | 25 | 2.7 | 5.3 |
Stern-Vollmer constants, KSV (M−1) were determined from the Stern-Vollmer equation F0/F = 1 + KSV(Q), where Q is the concentration of quencher (acrylamide). Concentrations of the quencher varied from 0.01 to 0.40 M.
FIG. 3.
Relative fluorescence intensity of (RW)n-NH2 peptides in aqueous solution and the presence of SUVs (buffer, black bars; POPG, gray bars; POPC, white bars). Fluorescence intensity in aqueous solution was used as the reference. The peptide and lipid concentrations were 10 μM and 500 μM, respectively, for a lipid-to-peptide ratio of 50.
The relative accessibility of Trp residues to solvent can be compared using Stern-Vollmer plots of the decrease in fluorescence as a function of an added soluble quencher. A decrease in quenching (smaller KSV values) reflects a more protected Trp residue. However, it should be noted that KSV values for longer peptides do not resolve individual Trp residues (37). Experimental results show that POPG and POPC offer the same protection to both RW and (RW)2 and therefore a similar extent of Trp side-chain burial. However, increased protection in the presence of POPG vesicles is observed for longer (RW)n (n, ≥3) peptides, while the Trp residues of these peptides appear to be less accessible in POPG than in POPC (Table 3). For example, the KSV for (RW)3 in an aqueous solution is 18 M−1 compared to 1.8 and 2.8 M−1 in POPG and POPC vesicles, respectively. These values indicate that the Trp side chains in free (RW)3 are substantially more accessible to quencher than in the presence of lipids and that (RW)3 partitions more effectively into POPG vesicles than into POPC vesicles. While this trend is consistent with the biological data, the presence of multiple Trp side chains in longer peptides precludes a strict quantitative interpretation of these data. In fact, all peptides are more protected from the quencher in the presence of POPG than they are in the presence of POPC.
DISCUSSION
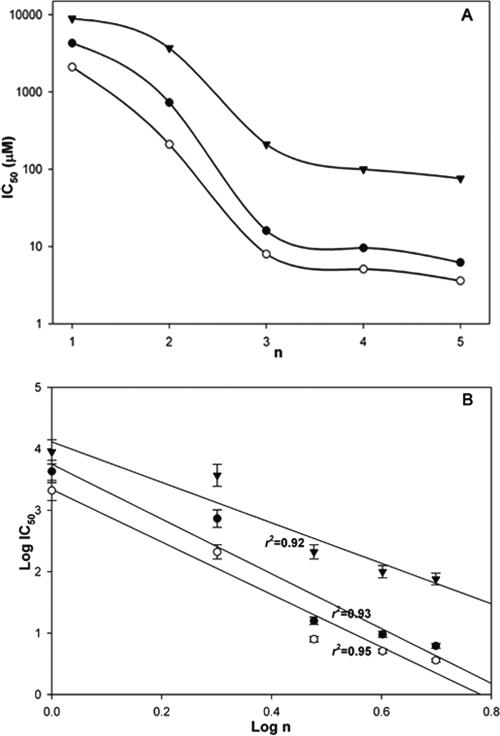
It is not unexpected that the antibacterial activities of peptides vary with their chain lengths (9, 10, 11, 31, 51). One previous study has reported that antibacterial activity decreases with an increase in the chain length of the peptides, while the reverse situation was found for hemolytic activity (31). Here we find that longer-chain linear peptides (RW)n-NH2 are more effective in killing both gram-negative and gram-positive bacteria. The dependence of activity data on chain length suggests an almost biphasic behavior (Fig. 4A): the two shortest chains are relatively inactive, while the three longer chains approximate a similar activity level. These data suggest the possibility of a threshold in biological response, attained at (RW)3. It is worth noting that this nonlinear biological response still obeys a power law with respect to the chain length [log (IC50) ∞ log(n)], with a slope of −4.5 (r2 = 0.93) and −4.3 (r2 = 0.95) for E. coli and S. aureus, respectively (Fig. 4B). The longer chains also show increasing hemolytic activity on red blood cells with a slope of −3.3 (r2 = 0.92). Power law behavior underlies a wide range of complex system responses, indicative of stronger effects arising from the longer chains. Regardless of how we interpret the detailed length dependence, (RW)3-NH2 is found to have the optimal selectivity for bacteria over blood cells relative to that of synthetic cost.
FIG. 4.
Correlation between antimicrobial activity and chain length of (RW)n-NH2 peptides. (A) Semilog plot of the data suggests a threshold effect in which short chains are inactive while chains longer than (RW)3 have roughly similar activities. (B) The data can be fit to a power law as well; y = cxa (where a and c are constants). Here we have log (IC50) = log (c) + a log(n) (n = 1, 2, 3, 4, or 5; IC50 is replaced by HD50 in the correlation for red blood cells). The slopes are −4.5 (r2 = 0.93), −4.3 (r2 = 0.95), and −3.3 (r2 = 0.92), for E. coli (solid circles), S. aureus (open circles), and red blood cells (triangles), respectively.
CD spectroscopy is widely used to analyze the secondary structure of proteins and peptides because it is extremely sensitive to conformational changes. (RW)n in buffer appears to be unfolded due to the negative band around 200 nm that is characteristic of unfolded model peptides (44); the positive band around 225 nm due to the indole side chain is Trp specific (21, 57). Peptide interactions with liposomes are accompanied by distinct CD spectral changes. Amphipathic AMPs such as indolicidin are thought to undergo an ordering transition on interaction with membranes (22). However, as we have noted, it is hard to interpret the CD spectra (or fluorescence data) simply in terms of secondary structure because of their multiple Trp contents. Changes in spectra are detected at shorter chain lengths in the presence of negatively charged POPG than in neutral POPC (Fig. 2). For example, the CD spectrum of (RW)3 indicates the presence of a structure that is ordered in POPG but that remains disordered in POPC. This interpretation is supported by our nuclear magnetic resonance studies of (RW)3 peptide conformation in negatively charged sodium dodecyl sulfate (SDS) and neutral dodecylphosphocholine (DPC) micelles. The structural ensemble of (RW)3 peptides appears to be more ordered in SDS than in DPC (data not shown).
Trp-rich AMPs preferentially bind to model membranes containing a negatively charged headgroup relative to those containing a zwitterionic headgroup (8, 17, 37). We recognize that the presence of multiple Trp residues does not allow for a simple interpretation of some of the spectroscopic data reported here (37). Our results are in accordance with previous studies, in which blue shifts and KSV values (Table 3) demonstrate that the Trp residues in all five peptides partition to a greater extent in SUVs that contain negatively charged phosphatidylglycerol (POPG) (a major component of bacterial membranes) (17). However, there is no obvious trend in the blue shifts with respect to chain length, suggesting that the interactions with synthetic vesicles are likely to model those with intact bacterial membranes only imperfectly, given the complexity of the biological system. Similarly, we find that even the shortest peptide is fully inaccessible to the acrylamide quencher in POPG. The picture from these experiments is that all the peptides of the series interact effectively with negatively charged POPG vesicles but that the spectroscopic measurements do not correlate with the biological results in any simple way.
Membrane perturbation by cationic antimicrobial peptides has been proposed to proceed via membrane destabilization and/or transmembrane pore formation (42). Because of their short lengths, it seems reasonable that (RW)n peptides attack bacteria via membrane disruption rather than by end-to-end pore assembly as in the case of gramicidins, although this is admittedly speculative. Arg residues provide electrostatic interactions that attract cationic peptides to negatively charged membrane surfaces (19, 43). The indole ring of tryptophan in other RW-rich AMPs (50) appears to partition into the interfacial region of membranes flanking the hydrophobic core (16, 36, 56). Linear RW chains might then interact preferentially with the polar headgroups, such that the peptides intercalate between the core leaflet of the bilayer and the polar headgroups. Peptides have been found to decrease the temperature of the main phase transition (17) consistent with destabilization of the bilayer. Quenching studies with internal lipid spin labels report that related RW peptides associate with the hydrocarbon core of neutral bilayers and locate near the polar headgroups in negatively charged model membranes. Their precise positions appear to be independent of peptide structure (37). The spectroscopic behavior of the (RW)n peptides seems consistent with an intercalation model (1, 18). Intercalation of peptides into a membrane could indeed produce an increase in the lateral pressure near the interface, resulting in local disruption in the packing of lipid chains. The differences observed in the activities of (RW)n could be associated with different spatial arrangements of the charged residues in the lipid head group region with increasing local density of RW motifs, resulting in different peptide surface areas and differently organized peptide-lipid clusters.
A second point to consider is the extent to which bound peptides associate once they lie at the surface of the nonpolar core. Studies by Chen et al. have shown clear evidence for concentration dependence in the interaction of peptides such as alamethicin with vesicles (6). A concentration-dependent process in which effectively neutralized peptides associate more strongly with neighboring peptides as the chain length increases seems plausible, since the indole rings apparently do not bury themselves in the core but tend to orient along the interface (55, 56).
The CD spectra that reveal structural changes in peptides upon interaction with negatively charged bilayers can be interpreted as evidence for intermolecular as well as intramolecular ordering. CD spectral changes are frequently observed when disordered peptides in solution partition into lipid bilayers (54). The high cost of partitioning peptide bonds into the membrane interface is a major driving force for secondary structure formation in membrane environments (23, 55, 56). However, for a peptide as short as RW, formation of intermolecular structure seems more favorable than intramolecular structure. Thus, we argue that the length effect reflects greater facility with interpeptide interactions rather than the exclusive formation of internal structure. How assembled peptides act to disrupt membranes remains unclear. In this work, we have identified (RW)3 as a sequence with potential to serve as a relatively nontoxic antimicrobial that can be produced economically on a large scale.
In summary, we have analyzed the relationship between the antibacterial activity of peptides in the series (RW)n-NH2 and their spectral properties in two model membrane systems. The results confirm that the RW combination is an active element for antimicrobial activity: longer chains are more effective in killing both gram-negative and gram-positive bacteria, although at the same time hemolytic activity increases. The (RW)n peptides show distinctive CD and fluorescent spectral changes upon interacting with model membranes. They preferentially interact with a phosphoglycerol headgroup than those with phosphocholine.
Acknowledgments
This work was supported by a grant from ONR (N00014-03-1-0129). We acknowledge the NCRR/NIH for a Research Facilities Improvement Grant (C06 RR-16572) at NYU.
We thank Steven Gu for helpful discussions.
Footnotes
Published ahead of print on 4 December 2006.
REFERENCES
- 1.Aliste, M. P., and D. P. Tieleman. 2005. Computer simulation of partitioning of ten pentapeptides Ace-WLXLL at the cyclohexane/water and phospholipid/water interfaces. BMC Biochem. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellamy, W., M. Takase, H. Wakabayashi, K. Kawase, and M. Tomita. 1992. Antibacterial spectrum of lactoferricin-B, a potent bactericidal peptide derived from the N-terminal region of bovine lactoferrin. J. Appl. Bacteriol. 73:472-479. [DOI] [PubMed] [Google Scholar]
- 3.Blondelle, S. E., E. Takahashi, K. T. Dinh, and R. A. Houghten. 1995. The antimicrobial activity of hexapeptides derived from synthetic combinatorial libraries. J. Appl. Bacteriol. 78:39-46. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197-215. [DOI] [PubMed] [Google Scholar]
- 5.Brown, K. L., and R. E. W. Hancock. 2006. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 18:24-30. [DOI] [PubMed] [Google Scholar]
- 6.Chen, F. Y., M. T. Lee, and H. W. Huang. 2002. Sigmoidal concentration dependence of antimicrobial peptide activities: a case study on alamethicin. Biophys. J. 82:908-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P. W., C. L. Shyu, and F. C. Mao. 2003. Antibacterial activity of short hydrophobic and basic-rich peptides. Am. J. Vet. Res. 64:1088-1092. [DOI] [PubMed] [Google Scholar]
- 8.Dathe, M., H. Nikolenko, J. Klose, and M. Bienert. 2004. Cyclization increases the antimicrobial activity and selectivity of arginine- and tryptophan-containing hexapeptides. Biochemistry 43:9140-9150. [DOI] [PubMed] [Google Scholar]
- 9.Dathe, M., M. Schumann, T. Wieprecht, A. Winkler, M. Beyermann, E. Krause, K. Matsuzaki, O. Murase, and M. Bienert. 1996. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 35:12612-12622. [DOI] [PubMed] [Google Scholar]
- 10.Deslouches, B., S. M. Phadke, V. Lazarevic, M. Cascio, K. Islam, R. C. Montelaro, and T. A. Mietzner. 2005. De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob. Agents Chemother. 49:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epand, R., R. I. Lehrer, A. Waring, W. Wang, R. Maget-Dana, D. Lelievre, and R. M. Epand. 2003. Direct comparison of membrane interactions of model peptides composed of only Leu and Lys residues. Biopolymers 71:2-16. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E. 1999. Host defence (cationic) peptides: what is their future clinical potential? Drugs 57:469-473. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haug, B. E., W. Stensen, T. Stiberg, and J. S. Svendsen. 2004. Bulky nonproteinogenic amino acids permit the design of very small and effective cationic antibacterial peptides. J. Med. Chem. 47:4159-4162. [DOI] [PubMed] [Google Scholar]
- 15.Houston, M. E., L. H. Kondejewski, Jr., D. N. Karunaratne, M. Gough, S. Fidai, R. S. Hodges, and R. E. Hancock. 1998. Influence of preformed alpha-helix and alpha-helix induction on the activity of cationic antimicrobial peptides. J. Pept. Res. 52:81-88. [DOI] [PubMed] [Google Scholar]
- 16.Hu, W., K. C. Lee, and T. A. Cross. 1993. Tryptophans in membrane proteins: indole ring orientations and functional implications in the gramicidin channel. Biochemistry 32:7035-7047. [DOI] [PubMed] [Google Scholar]
- 17.Jing, W., H. N. Hunter, J. Hagel, and H. J. Vogel. 2003. The structure of the antimicrobial peptide Ac-RRWWRF-NH2 bound to micelles and its interactions with phospholipid bilayers. J. Pept. Res. 61:219-229. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. E., N. M. Rao, S. W. Hui, and R. B. Cornell. 1998. Conformation and lipid binding properties of four peptides derived from the membrane-binding domain of CTP: phosphocholine cytidylyltransferase. Biochemistry 37:9509-9519. [DOI] [PubMed] [Google Scholar]
- 19.Juffer, A. H., C. M. Shepherd, and H. J. Vogel. 2001. Protein-membrane electrostatic interactions: application of the Lekner summation technique. J. Chem. Phys. 114:1892-1905. [Google Scholar]
- 20.Ladokhin, A. S. 1999. Evaluation of lipid exposure of tryptophan residues in membrane peptides and proteins. Anal. Biochem. 276:65-71. [DOI] [PubMed] [Google Scholar]
- 21.Ladokhin, A. S., M. E. Selsted, and S. H. White. 1997. Bilayer interactions of indolicidin, a small antimicrobial peptide rich in tryptophan, proline, and basic amino acids. Biophys. J. 72:794-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladokhin, A. S., M. E. Selsted, and S. H. White. 1999. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry 38:12313-12319. [DOI] [PubMed] [Google Scholar]
- 23.Ladokhin, A. S., and S. H. White. 1999. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J. Mol. Biol. 285:1363-1369. [DOI] [PubMed] [Google Scholar]
- 24.Lejon, T., J. S. Svendsen, and B. E. Haug. 2002. Simple parameterization of non-proteinogenic amino acids for QSAR of antibacterial peptides. J. Pept. Sci. 8:302-306. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Z., H. Deshazer, J. R. Amanda, K. Chen, C. Zhou, and N. R. Kallenbach. 2006. Multivalent antimicrobial peptides from a reactive polymer scaffold. J. Med. Chem. 49:3436-3439. [DOI] [PubMed] [Google Scholar]
- 26.Maeda, M., R. A. Melnyk, A. W. Partridge, L. P. Liu, and C. M. Deber. 2003. Transmembrane segment peptides with double d-amino acid replacements: helicity, hydrophobicity, and antimicrobial activity. Biopolymers 71:77-84. [DOI] [PubMed] [Google Scholar]
- 27.Mavri, J., and H. J. Vogel. 1996. Ion pair formation of phosphorylated amino acids and lysine and arginine side chains: a theoretical study. Proteins 24:495-501. [DOI] [PubMed] [Google Scholar]
- 28.Metlitskaia, L., J. E. Cabralda, D. Suleman, C. Kerry, J. Brinkman, D. Bartfeld, and M. M. Guarna. 2004. Recombinant antimicrobial peptides efficiently produced using novel cloning and purification processes. Biotechnol. Appl. Biochem. 39:339-345. [DOI] [PubMed] [Google Scholar]
- 29.Morrissey, J. H. 2001. Morrissey laboratory protocol for preparing phospholipid vesicles (SUV) by sonication. http://www.avantilipids.com/pdf/MorrisseyLabProtocolForPrepSuvBySonication.pdf.
- 30.National Committee for Clinical Laboratory Standards. 2004. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M100-S14. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 31.Niidome, T., N. Matsuyama, M. Kunihara, T. Hatakeyama, and H. Aoyagi. 2005. Effect of chain length of cationic model peptides on antibacterial activity. Bull. Chem. Soc. Jpn. 78:473-476. [Google Scholar]
- 32.Oren, Z., and Y. Shai. 1997. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: structure-function study. Biochemistry 36:1826-1835. [DOI] [PubMed] [Google Scholar]
- 33.Patch, J. A., and A. E. Barron. 2003. Helical peptoid mimics of magainin-2 amide. J. Am. Chem. Soc. 125:12092-12093. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini, A., and R. von Fellenberg. 1999. Design of synthetic bactericidal peptides derived from the bactericidal domain P(18-39) of aprotinin. Biochim. Biophys. Acta 1433:122-131. [DOI] [PubMed] [Google Scholar]
- 35.Porter, E. A., X. Wang, H. S. Lee, B. Weisblum, and S. H. Gellman. 2000. Non-haemolytic beta-amino-acid oligomers. Nature 404:565. [DOI] [PubMed] [Google Scholar]
- 36.Reithmeier, R. A. F. 1995. Characterization and modeling of membrane-proteins using sequence-analysis. Curr. Opin. Struct. Biol. 5:491-500. [DOI] [PubMed] [Google Scholar]
- 37.Schibli, D. J., R. F. Epand, H. J. Vogel, and R. M. Epand. 2002. Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem. Cell Biol. 80:667-677. [DOI] [PubMed] [Google Scholar]
- 38.Schibli, D. J., P. M. Hwang, and H. J. Vogel. 1999. Structure of the antimicrobial peptide tritrpticin bound to micelles: a distinct membrane-bound peptide fold. Biochemistry 38:16749-16755. [DOI] [PubMed] [Google Scholar]
- 39.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 40.Selsted, M. E., M. J. Novotny, W. L. Morris, Y. Q. Tang, W. Smith, and J. S. Cullor. 1992. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 267:4292-4295. [PubMed] [Google Scholar]
- 41.Shafer, W. M., F. Hubalek, M. Huang, and J. Pohl. 1996. Bactericidal activity of a synthetic peptide (CG 117-136) of human lysosomal cathepsin G is dependent on arginine content. Infect. Immun. 64:4842-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 43.Shepherd, C. M., K. A. Schaus, H. J. Vogel, and A. H. Juffer. 2001. Molecular dynamics study of peptide-bilayer adsorption. Biophys. J. 80:579-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi, Z. S., K. Chen, Z. Liu, A. Ng, W. C. Bracken, and N. R. Kallenbach. 2005. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proc. Natl. Acad. Sci. USA 102:17964-17968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staubitz, P., A. Peschel, W. F. Nieuwenhuizen, M. Otto, F. Gotz, G. Jung, and R. W. Jack. 2001. Structure-function relationships in the tryptophan-rich, antimicrobial peptide indolicidin. J. Pept. Sci. 7:552-564. [DOI] [PubMed] [Google Scholar]
- 46.Strom, M. B., B. E. Haug, O. Rekdal, M. L. Skar, W. Stensen, and J. S. Svendsen. 2002. Important structural features of 15-residue lactoferricin derivatives and methods for improvement of antimicrobial activity. Biochem. Cell Biol. 80:65-74. [DOI] [PubMed] [Google Scholar]
- 47.Strom, M. B., B. E. Haug, M. L. Skar, W. Stensen, T. Stiberg, and J. S. Svendsen. 2003. The pharmacophore of short cationic antibacterial peptides. J. Med. Chem. 46:1567-1570. [DOI] [PubMed] [Google Scholar]
- 48.Strom, M. B., O. Rekdal, and J. S. Svendsen. 2002. Antimicrobial activity of short arginine- and tryptophan-rich peptides. J. Pept. Sci. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 49.Subbalakshmi, C., E. Bikshapathy, N. Sitaram, and R. Nagaraj. 2000. Antibacterial and hemolytic activities of single tryptophan analogs of indolicidin. Biochem. Biophys. Res. Commun. 274:714-716. [DOI] [PubMed] [Google Scholar]
- 50.Subbalakshmi, C., V. Krishnakumari, N. Sitaram, and R. Nagaraj. 1998. Interaction of indolicidin, a 13-residue peptide rich in tryptophan and proline and its analogues with model membranes. J. Biosci. 23:9-13. [Google Scholar]
- 51.Tam, J. P., Y. A. Lu, and J. L. Yang. 2002. Antimicrobial dendrimeric peptides. Eur. J. Biochem. 269:923-932. [DOI] [PubMed] [Google Scholar]
- 52.Tinoco, L. W., A. da Silva, A. Leite, A. P. Valente, and F. C. L. Almeida. 2002. NMR structure of PW2 bound to SDS micelles. A tryptophan-rich anticoccidial peptide selected from phage display libraries. J. Biol. Chem. 277:36351-36356. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, H. J., D. J. Schibli, W. G. Jing, E. M. Lohmeier-Vogel, R. F. Epand, and R. M. Epand. 2002. Towards a structure-function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides. Biochem. Cell Biol. 80:49-63. [DOI] [PubMed] [Google Scholar]
- 54.White, S. H., W. C. Wimley, A. S. Ladokhin, and K. Hristova. 1998. Protein folding in membranes: determining energetics of peptide-bilayer interactions. Methods Enzymol. 295:62-87. [DOI] [PubMed] [Google Scholar]
- 55.Wimley, W. C., K. Hristova, A. S. Ladokhin, L. Silvestro, P. H. Axelsen, and S. H. White. 1998. Folding of beta-sheet membrane proteins: a hydrophobic hexapeptide model. J. Mol. Biol. 277:1091-1110. [DOI] [PubMed] [Google Scholar]
- 56.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842-848. [DOI] [PubMed] [Google Scholar]
- 57.Woody, R. W. 1994. Contributions of tryptophan side-chains to the far-ultraviolet circular-dichroism of proteins. Eur. Biophys. J. 23:253-262. [DOI] [PubMed] [Google Scholar]
- 58.Yau, W. M., W. C. Wimley, K. Gawrisch, and S. H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713-14718. [DOI] [PubMed] [Google Scholar]
- 59.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]