Abstract
We have identified and sequenced the genes encoding the quinolone-resistance determining region (QRDR) of ParC and GyrA in fluoroquinolone-susceptible and -resistant Streptococcus suis clinical isolates. Resistance is the consequence of single point mutations in the QRDRs of ParC and GyrA and is not due to clonal spread of resistant strains or horizontal gene transfer with other bacteria.
Streptococcus suis is a gram-positive bacterium distributed worldwide that causes meningitis, endocarditis, septicemia, septic arthritis, pneumonia, and abortion in humans and pigs (32). In intensive swine industry, S. suis infections are one of the major causes of bacterial infections and economic loss. Sporadically, human cases are described, being considered by the World Health Organization an occupational disease due to infection through direct contact with pigs or pig products (34). In Southeastern Asia transmission to humans usually occurs in a constant rate, but sudden outbreaks have also recently been reported (31). During the summer of 2005, 215 people were infected by S. suis in the Sichuan province, China; 39 (18%) of these infections led to a fatal outcome (35). Although deaths were also caused by meningitis, they were mainly due to a novel form of invasive toxic shock syndrome (29). In all cases, S. suis from pigs was the origin of the outbreak.
Along with aminopenicillins, quinolones (fluoroquinolones), such as enrofloxacin in pigs or ciprofloxacin in humans, were the preferred treatment for S. suis infections (17). Most importantly, these antimicrobials are currently used against gram-negative pathogens in pigs that are frequently carriers of S. suis. In recent years, we have observed an emergence of fluoroquinolone-resistant strains among clinical swine S. suis isolates. The emergence of this resistance in a zoonotic pathogen such as S. suis has unpredictable consequences for pig production and public health (1, 33).
Resistance to fluoroquinolones in streptococci is mainly due to specific point mutations in the quinolone resistance-determining regions (QRDRs) of the GyrA subunit of the DNA gyrase and in the ParC subunit of the DNA Topoisomerase IV, enzymes that control DNA topology (6). Acquisition of mutations in the coding genes of these subunits, gyrA and parC, has been related to the appearance of single amino acid substitutions at positions S79 and D83 in ParC or S81 and E85 in GyrA. Furthermore, the genes or gene fragments containing these mutations may be transferred to other streptococci, rendering them resistant to fluoroquinolones by recombining their susceptible gene with the resistant homologue (3, 18).
(An initial report of this study has been presented at the 16th European Congress for Clinical Microbiology and Infectious Diseases [J. A. Escudero, et al., Abstr. p1251, 2006].).
The Veterinary Health Surveillance Group (VISAVET) at the Veterinary School in Madrid diagnoses bacterial diseases in Spanish pig farms. Since 2003, 992 samples have been identified through the commercial biochemical Rapid ID32 system as being S. suis. From these samples, ∼1.2% (12 isolates) were highly resistant to enrofloxacin, a fluoroquinolone currently used for the treatment of insidious infections in pig farms as an alternative to aminopenicillins. To characterize fluoroquinolone resistance in these strains, a more complete quinolone and fluoroquinolone resistance profile with MIC determination of all resistant bacteria was assessed (Table 1) . Microdilutions with incubations at 37°C for 24 h with antimicrobials supplied by Sigma Aldrich (Sigma Chemical Co., St. Louis, MO) were performed. The breakpoints used for enrofloxacin were those recommended by the Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) in 2002 (24) for veterinary gram-positive microorganisms. All isolates were highly resistant to the three quinolones tested (nalidixic acid, flumequine, and oxolinic acid), with an MIC of ≥64 μg/ml. Further, all isolates resistant to enrofloxacin were highly resistant to ciprofloxacin, levofloxacin, and norfloxacin, with MICs ranging from 16 to ≥64 μg/ml. Six fluoroquinolone-susceptible strains, including type strain ATCC 43765, a clinical isolate from South America, and four epidemiologically unrelated isolates from our collection, were also analyzed for their antimicrobial profiles. All susceptible strains had MICs to ciprofloxacin of 0.5 μg/ml and were highly resistant to quinolones such as flumequine, oxolinic acid (data not shown), and nalidixic acid (MIC ≥ 16 μg/ml).
TABLE 1.
Susceptibility to selected quinolones and fluoroquinolones of S. suis strains
| Strain | MIC (μg/ml)a
|
Country or region | Source or reference | |||||
|---|---|---|---|---|---|---|---|---|
| CIP | CIP+R | ENR | NOR | LVX | NAL | |||
| ATCC 43765 | 0.5 | 0.5 | 0.5 | 4 | 1 | >64 | England | Collection |
| BB1001 | 0.5 | 0.5 | 0.5 | 2 | 1 | >64 | Spain | This study |
| BB1002 | 0.5 | 0.5 | 0.5 | 2 | 1 | >64 | South America | This study |
| BB1003 | 0.5 | 0.5 | 0.5 | 4 | 0.5 | >64 | Spain | This study |
| BB1004 | 0.5 | 0.5 | 0.5 | 2 | 1 | >64 | Spain | This study |
| BB1005 | 0.5 | 0.5 | 0.25 | 4 | 0.5 | >64 | Spain | This study |
| BB1006 | 2 | 1 | 1 | 32 | 1 | >64 | Spain | This study |
| BB1007 | 8 | 4 | 4 | 16 | 4/2 | >64 | Spain | This study |
| BB1008 | 64 | 32 | 32 | >64 | 16 | >64 | Spain | This study |
| BB1009 | 32 | 16 | 16 | 32 | 16 | >64 | Spain | This study |
| BB1010 | 32 | 16 | 32 | >64 | 32 | >64 | Spain | This study |
| BB1011 | 16 | 16 | 16 | 32 | 16 | >64 | Spain | This study |
| BB1012 | 32 | 16 | 8 | >64 | 16 | >64 | Spain | This study |
| BB1013 | 64 | 16 | 8 | >64 | 16 | >64 | Spain | This study |
| BB1014 | 64 | 16 | 16 | >64 | 32 | >64 | Spain | This study |
| BB1015 | 32 | 16 | 16 | >64 | 16 | >64 | Spain | This study |
| BB1016 | 32 | 16 | 8 | >64 | 32 | >64 | South America | This study |
| BB1017 | 64 | 16 | 16 | >64 | 64 | >64 | Spain | This study |
CIP, ciprofloxacin; ENR, enrofloxacin; LVX, levofloxacin; NAL, nalidixic acid; NOR, norfloxacin; R, reserpine (10 μg/ml).
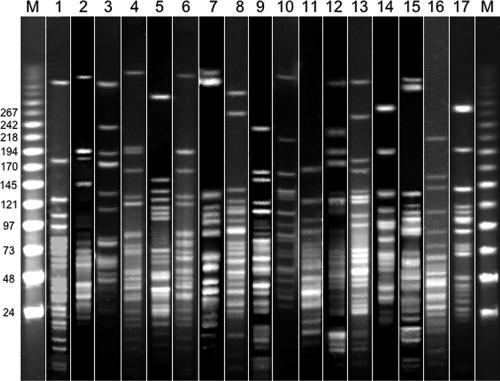
Before continuing characterization of these S. suis isolates, species identification was confirmed. For this purpose, the recently described PCR technique based on amplification of a 688-bp fragment from the glutamate dehydrogenase gene (gdh) from S. suis was used (25) (Table 2) . All isolates biochemically identified in our laboratory as S. suis gave a positive PCR signal (data not shown), showing that biochemical identification together with experienced laboratory personnel is a reliable method for the identification of S. suis (17). To assess the genetic variability of these strains, the 12 fluoroquinolone-resistant and the 6 fluoroquinolone-susceptible isolates were subjected to pulsed-field gel electrophoresis (PFGE) essentially as described previously (30). All isolates showed different PFGE patterns, confirming the high genetic diversity of S. suis shown in previous studies (5, 30) and implying that fluoroquinolone resistance in S. suis is not due to the clonal spread of a resistant isolate but rather to a characteristic independently acquired by each isolate (Fig. 1).
TABLE 2.
Primer sets used to amplify the gdh, gyrA, and parC genes of S. suis
| Gene | Primer | Sequence (5′-3′) | Position | GenBank accession no. |
|---|---|---|---|---|
| gdh | JP4 | GCAGCGTATTCTGTCAAACG | 1177-1196 | AF229683 |
| JP5 | CCATGGACAGATAAAGATGG | 508-527 | ||
| gyrA | gyrA-F | CGCCGTATTTTGTATGGGATG | 130-150 | DQ832724 |
| gyrA-R | GTTCCGTTAACCAGAAGGTT | 487-507 | ||
| parC | parC-F | AAGGACGGCAACACTTTTGAC | 151-171 | DQ832742 |
| parC-R | AGTGGGTTCTTTTTCCGTATC | 442-462 |
FIG. 1.
PFGE fingerprint patterns of S. suis isolates used in the present study using the ApaI endonuclease. Lanes 1 to 6, fluoroquinolone-susceptible strains; lanes 7 to 17, fluoroquinolone-resistant isolates. The first and last lanes are bacteriophage lambda ladder PFGE markers (Boehringer Mannheim, Germany); lane 1, S. suis ATCC 43765; lane 2, BB1001; lane 3, BB1002; lane 4, BB1003; lane 5, BB1004; lane 6, BB1005; lane 7, BB1006; lane 8, BB1007; lane 9, BB1008; lane 10, BB1009; lane 11, BB1011; lane 12, BB1012; lane 13, BB1013; lane 14, BB1014; lane 15, BB1015; lane 16, BB1016; lane 17, BB1017. Analysis was performed with QuantityOne software (Bio-Rad, Richmond, CA).
As stated above, the products of the parC and the gyrA genes are the primary and secondary targets of ciprofloxacin in other streptococci (2, 23). However, these genes have not been identified in S. suis. To determine the QRDRs of parC and gyrA in this species, the genes in the six unrelated fluoroquinolone-susceptible S. suis isolates were amplified and sequenced (Table 2). The predicted amino acid sequence of the parC and gyrA genes revealed, in all six fluoroquinolone-susceptible isolates, no single amino acid difference in the QRDRs, showing the high degree of conservation of this regions, even in geographically and epidemiologically unrelated strains (Fig. 2 and 3). The QRDR of ParC in S. suis is homologous to the ParC sequences of other streptococci, with identities ranging from 97% with the QRDRs from S. oralis and S. mitis to 88% with the QRDRs from S. agalactiae and S. mutans (Fig. 2). Analogously, the QRDRs of GyrA presented an identity from 95% with S. pyogenes to 92% with S. equi subsp. equi and S. equi subsp. equisimilis.
FIG. 2.
Amino acid sequence alignment of the QRDRs of GyrA and ParC in streptococci. Amino acids critical for fluoroquinolone resistance are marked with an arrowhead and standard E. coli numbering. White shading denotes different amino acids in relation to the S. suis sequence.
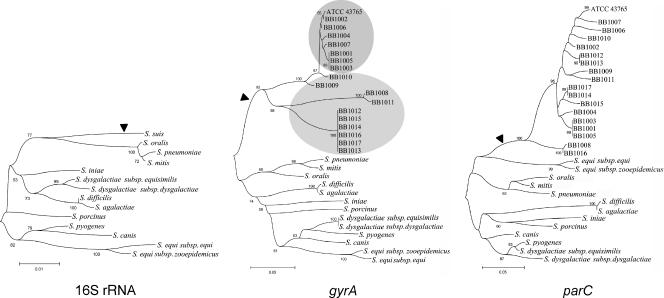
FIG. 3.
Dendrograms from the nucleotide sequences of 16S rRNA gene and the QRDRs of gyrA and parC of streptococci. Note that in the 16S rRNA gene tree the branch of S. suis (arrowhead) contains in the corresponding gyrA and parC trees all of the S. suis strains analyzed in the present study (arrowheads), implying that these genes from S. suis were not acquired from other streptococci by horizontal gene transfer. Further, the gyrA dendrogram clusters fluoroquinolone-susceptible and low-level resistant strains (upper oval) and high-level resistant strains (lower oval), whereas in the parC tree the strains are dispersed, indicating that parC is more prone to accumulating mutations than gyrA. In line with this, the 16S rRNA gene dendrogram has no significant differences with the gyrA tree, whereas streptococci are clustered differently in the dendrogram constructed with the parC sequences. The dendrograms were constructed with MEGA software version 3.1 (21) by using the neighbor-joining grouping procedure with the Kimura two-parameter distance measure (19). The bar denotes genetic distance. Bootstrap values are the result of 1,000 iterations.
To assess the involvement of GyrA and ParC of S. suis in fluoroquinolone resistance, the QRDRs of the gyrA and parC genes were determined in 12 fluoroquinolone-resistant clinical isolates (Table 3). In all cases, an amino acid change in a position known to be related to fluoroquinolone resistance in other streptococci (S79 and D83 in ParC or S81 and E85 in GyrA) was detected. Most isolates had one or two modifications in both GyrA and ParC, indicating that, as in many other species, both proteins are involved in fluoroquinolone resistance in S. suis. One isolate, BB1006, had a single nucleotide substitution in the parC gene (TCC→TTC), giving rise to an S79F replacement, implying that one mutation in parC is sufficient to confer resistance to these antimicrobials. The finding suggests that topoisomerase IV is a primary target of fluoroquinolones in S. suis, as in other gram-positive bacteria such as S. aureus (9) and S. pneumoniae (23). However, it has been shown that mutations in either gyrase or topoisomerase IV can occur depending on the structure of the fluoroquinolone (26). The isolates analyzed in the present study were probably selected through treatment with enrofloxacin. Studies from our laboratory with other fluoroquinolones show that single mutations in GyrA, but not in ParC, may be selected in vitro, indicating that the primary target of fluoroquinolones in S. suis may also depend on the type of molecule selecting resistance (data not shown). Ten other isolates carried identical amino acid substitution at the same codon of ParC, S79Y, albeit combined with either a single (S81Y, S81K, S81I, or S81F) or double (S81K E85D) amino acid change in GyrA. One strain, BB1007, presented a unique combination, with substitutions D83H in ParC and E85K in GyrA (Table 3). Nonetheless, it is worth mentioning that these data do not fully characterize fluoroquinolone resistance in S. suis. The reduction of the MIC down to 16 μg/ml in three resistant isolates when the pump inhibitor reserpine was added to ciprofloxacin shows that active efflux pumps, such as PmrA in S. pneumoniae (11) or Lde in Listeria monocytogenes (12), may play a role in resistance in S. suis. Further, mutations in GyrB or ParE may also contribute to resistance to fluoroquinolones in S. suis, as is the case in viridans group streptococci (13) and pneumococci (15).
TABLE 3.
Amino acid and codon changes in critical positions of the gyrA and parC QRDRs in S. suisa
| Strain | CIP MIC (μg/ml) |
gyrA
|
parC
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Position 81
|
Position 85
|
Position 79
|
Position 83
|
||||||
| Codon | Aa | Codon | Aa | Codon | Aa | Codon | Aa | ||
| ATCC | 0.5 | AGT | Ser | GAA | Glu | TCC | Ser | GAT | Asp |
| BB1001 | 0.5 | - | - | - | - | - | - | - | - |
| BB1002 | 0.5 | - | - | - | - | - | - | - | - |
| BB1003 | 0.5 | - | - | - | - | - | - | - | - |
| BB1004 | 0.5 | - | - | - | - | - | - | - | - |
| BB1005 | 0.5 | - | - | - | - | - | - | - | - |
| BB1006 | 2 | - | - | - | - | TTC | Phe | - | - |
| BB1007 | 8 | - | - | AAA | Lys | - | - | CAT | His |
| BB1008 | 64 | TAT | Tyr | - | - | TAC | Tyr | - | - |
| BB1009 | 32 | AAG | Lys | - | - | TAC | Tyr | - | - |
| BB1010 | 32 | ATT | Ile | - | - | TAC | Tyr | - | - |
| BB1011 | 16 | TTT | Phe | - | - | TAC | Tyr | - | - |
| BB1012 | 32 | AAG | Lys | GAC | Asp | TAC | Tyr | - | - |
| BB1013 | 64 | AAG | Lys | GAC | Asp | TAC | Tyr | - | - |
| BB1014 | 64 | AAG | Lys | GAC | Asp | TAT | Tyr | - | - |
| BB1015 | 32 | AAG | Lys | GAC | Asp | TAC | Tyr | - | - |
| BB1016 | 32 | AAG | Lys | GAC | Asp | TAC | Tyr | - | - |
| BB1017 | 64 | AAG | Lys | GAC | Asp | TAT | Tyr | - | - |
CIP, ciprofloxacin. -, identical to the ATCC strain. Sequence differences are indicated in boldface. Aa, amino acid.
Horizontal gene transfer has been shown to be responsible for the development of resistance to fluoroquinolones in S. pneumoniae, which acquires gyrA or parC with resistance-conferring mutations from viridans streptococci (3, 8, 27), and in S. pyogenes, which can acquire resistance from a mutated parC gene from S. dysgalactiae (28). This would be especially problematic in the case of S. suis, which could serve as donor of fluoroquinolone resistance for other pathogenic bacteria more widely distributed in humans than S. suis. In our strains, gyrA and parC showed an identity of ≥96% and no trace of non-S. suis DNA in either gene, as revealed by the dendrograms (Fig. 3). The GenBank database was screened for the presence of DNA sequences of gyrA and parC from S. suis in other bacterial species. This was performed by launching sequential 10-bp-overlapping 30-bp fragments of the parC or gyrA genes encoding fluoroquinolone resistance against the GenBank database, estimating 100% identity as horizontal gene transfer. Neither acquisition nor donation of gyrA or parC fragments was observed, and resistance is due to the emergence of mutations in each isolate, as previously shown in enterococci (7, 16) and staphylococci (10, 22). Thus, these genes from S. suis do not represent an antimicrobial resistance reservoir for other animal or human pathogens, at least in our strains. This analysis also revealed that evolution of gyrA in S. suis elicits a clear clustering of bacteria highly resistant on one branch and susceptible or low-level resistant to fluoroquinones in another branch (Fig. 3). This indicates that gyrases are genetically more stable than topoisomerases IV and reflect species microevolution (14), as shown by the recent use of these genes in multilocus sequence typing protocols in Yersinia and Acinetobacter spp. (4, 20). The analysis of gyrA and parC in more S. suis isolates would be a significant step forward to further understand the mechanisms and spread of fluoroquinolone resistance in this emerging zoonotic pathogen.
Nucleotide sequence accession numbers.
The nucleotide sequences for the gyrA and parC QRDRs have been deposited in GenBank under the following respective accession numbers: ATCC 43765, DQ832724 and DQ832742; BB1001, DQ832725 and DQ832743; BB1002, DQ832726 and DQ832744; BB1003, DQ832727 and DQ832745; BB1004, DQ832728 and DQ832746; BB1005, DQ832729 and DQ832747; BB1006, DQ832730 and DQ832748; BB1007, DQ832731 and DQ832749; BB1008; DQ832732 and DQ832750; BB1009, DQ832733 and DQ832751; BB1010, DQ832734 and DQ832752; BB1011, DQ832735 and DQ832753; BB1012, DQ832736 and DQ832754; BB1013, DQ832737 and DQ832755; BB1014, DQ832738 and DQ832756; BB1015, DQ832739 and DQ832757; BB1016, DQ832740 and DQ832758; and BB1017, DQ832741 and DQ832759.
Acknowledgments
We thank the National Ramon y Cajal Program from the Spanish Ministry of Education and Science for support of B.G.-Z., the Universidad Complutense de Madrid for the Ph.D. scholarship of J.A.E., and the Spanish Ministry of Education and Science for supporting the Ph.D. scholarships of A.S.M. and A.C. This study was partially financed by project PR1/06-14475-B from the Universidad Complutense de Madrid, S-0505/AGR/000265 (Vigilancia Sanitaria Program) from the Consejeria de Educacion, Comunidad de Madrid, and BIO2005-02189 from the Direccion General de Investigación, Ministerio de Educacion y Ciencia, Spain.
The VISAVET Group is acknowledged for the clinical isolates. We also thank A. Casamayor for excellent technical assistance with the PFGE.
Footnotes
Published ahead of print on 20 November 2006.
REFERENCES
- 1.Aarestrup, F. M., S. R. Rasmussen, K. Artursson, and N. E. Jensen. 1998. Trends in the resistance to antimicrobial agents of Streptococcus suis isolates from Denmark and Sweden. Vet. Microbiol. 63:71-80. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, R., M. Galimand, and P. Courvalin. 2002. parC mutation conferring ciprofloxacin resistance in Streptococcus pyogenes BM4513. Antimicrob. Agents Chemother. 46:3686-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, L., M. J. Ferrándiz, J. Liñares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartual, S. G., H. Seifert, C. Hippler, M. A. Luzon, H. Wisplinghoff, and F. Rodriguez-Valera. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelot-Herault, F., C. Marois, M. Gottschalk, and M. Kobisch. 2002. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el Amin, N. A., S. Jalal, and B. Wretlind. 1999. Alterations in GyrA and ParC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 43:947-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrándiz, M. J., A. Fenoll, J. Linares, and A. G. de la Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 10.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill, M. J., N. P. Brenwald, and R. Wise. 1999. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godreuil, S., M. Galimand, G. Gerbaud, C. Jacquet, and P. Courvalin. 2003. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob. Agents Chemother. 47:704-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González, I., M. Georgiou, F. Alcaide, D. Balas, J. Liñares, and A. G. de la Campa. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, W. M. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30:79-107. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen, J. H., L. M. Weigel, M. J. Ferraro, J. M. Swenson, and F. C. Tenover. 1999. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob. Agents Chemother. 43:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanematsu, E., T. Deguchi, M. Yasuda, T. Kawamura, Y. Nishino, and Y. Kawada. 1998. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 42:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka, Y., C. Sugimoto, M. Nakazawa, and M. Kashiwazaki. 1991. Detection of Streptococcus suis type 2 in tonsils of slaughtered pigs using improved selective and differential media. Vet. Microbiol. 28:335-342. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura, Y., H. Fujiwara, N. Mishima, Y. Tanaka, A. Tanimoto, S. Ikawa, Y. Itoh, and T. Ezaki. 2003. First Streptococcus agalactiae isolates highly resistant to quinolones, with point mutations in gyrA and parC. Antimicrob. Agents Chemother. 47:3605-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Kotetishvili, M., A. Kreger, G. Wauters, J. G. Morris, Jr., A. Sulakvelidze, and O. C. Stine. 2005. Multilocus sequence typing for studying genetic relationships among Yersinia species. J. Clin. Microbiol. 43:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:50-163. [DOI] [PubMed] [Google Scholar]
- 22.Li, Z., T. Deguchi, M. Yasuda, T. Kawamura, E. Kanematsu, Y. Nishino, S. Ishihara, and Y. Kawada. 1998. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV in quinolone-resistant clinical isolates of Staphylococcus epidermidis. Antimicrob. Agents Chemother. 42:3293-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz, R., and A. G. de la Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2004. Performance Standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals: informational supplement M31-S1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Okwumabua, O., M. O'Connor, and E. Shull. 2003. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 218:79-84. [DOI] [PubMed] [Google Scholar]
- 26.Pan, X. S. And L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pletz, M. W., L. McGee, B. Beall, C. G. Whitney, and K. P. Klugman. 2005. Interspecies recombination in type II topoisomerase genes is not a major cause of fluoroquinolone resistance in invasive Streptococcus pneumoniae isolates in the United States. Antimicrob. Agents Chemother. 49:779-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pletz, M. W., L. McGee, C. A. Van Beneden, S. Petit, M. Bardsley, M. Barlow, and K. P. Klugman. 2006. Fluoroquinolone resistance in invasive Streptococcus pyogenes isolates due to spontaneous mutation and horizontal gene transfer. Antimicrob. Agents Chemother. 50:943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, J., C. Wang, Y. Feng, W. Yang, H. Song, Z. Chen, H. Yu, X. Pan, X. Zhou, H. Wang, B. Wu, H. Wang, H. Zhao, Y. Lin, J. Yue, Z. Wu, X. He, F. Gao, A. H. Khan, J. Wang, G. P. Zhao, Y. Wang, X. Wang, Z. Chen, and G. F. Gao. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vela, A. I., J. Goyache, C. Tarradas, I. Luque, A. Mateos, M. A. Moreno, C. Borge, J. A. Perea, L. Domínguez, and J. F. Fernández-Garayzabal. 2003. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:2498-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wangkaew, S., R. Chaiwarith, P. Tharavichitkul, and K. Supparatpinyo. 2006. Streptococcus suis infection: a series of 41 cases from Chiang Mai University Hospital. J. Infect. 52:455-460. [DOI] [PubMed] [Google Scholar]
- 32.Willenburg, K. S., D. E. Sentochnik, and R. N. Zadoks. 2006. Human Streptococcus suis meningitis in the United States. N. Engl. J. Med. 354:1325. [DOI] [PubMed] [Google Scholar]
- 33.Wisselink, H. J., K. T. Veldman, C. Van den Eede, S. A. Salmon, and D. J. Mevius. 2006. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet. Microbiol. 113:73-82. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. August. 2005, posting date. Fact sheets: Streptococcus suis. World Health Organization, Geneva, Switzerland. [Online.] http://www.wpro.who.int/media_centre/fact_sheets/fs_20050802.htm.
- 35.Yu, H., H. Jing, Z. Chen, H. Zheng, X. Zhu, H. Wang, S. Wang, L. Liu, R. Zu, L. Luo, N. Xiang, H. Liu, X. Liu, Y. Shu, S. S. Lee, S. K. Chuang, Y. Wang, J. Xu, W. Yang, and Streptococcus suis study groups. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]