Abstract
Virulence-associated genes and neutral DNA markers of Helicobacter pylori strains from the Santhal and Oroan ethnic minorities of West Bengal, India, were studied. These people have traditionally been quite separate from other Indians and differ culturally, genetically, and linguistically from mainstream Bengalis, whose H. pylori strains have been characterized previously. H. pylori was found in each of 49 study participants, although none had peptic ulcer disease, and was cultured from 31 of them. All strains carried the cag pathogenicity island and potentially toxigenic s1 alleles of vacuolating cytotoxin gene (vacA) and were resistant to at least 8 μg of metronidazole per ml. DNA sequence motifs in vacA mid-region m1 alleles, cagA, and an informative insertion or deletion motif next to cagA from these strains were similar to those of strains from ethnic Bengalis. Three mobile elements, IS605, IS607, and ISHp608, were present in 29, 19, and 10%, respectively, of Santhal and Oroan strains, which is similar to their prevalence in Bengali H. pylori. Thus, there is no evidence that the gene pools of H. pylori of these ethnic minorities differ from those of Bengalis from the same region. This relatedness of strains from persons of different ethnicities bears on our understanding of H. pylori transmission between communities and genome evolution.
Helicobacter pylori has been the focus of intense research efforts for some 20 years, motivated by recognition of its role as a major cause of peptic ulcer disease (46) and an early risk factor for gastric cancer (32), even though most infections are asymptomatic. It colonizes the gastric mucosa of more than half of all people worldwide (31). Infection tends to start in early childhood and to persist for years or decades once established (47). Much of the pathology associated with infection probably results from the host response rather than bacterial toxins or other virulence factors per se. The host response may be affected by bacterial genotype as well as by human host genotype and physiology and environmental conditions (6).
One of the most intriguing aspects of H. pylori is its great genetic diversity (1, 2, 13), which is at a level as yet unseen in other bacterial pathogens. Independent isolates generally differ from one another by some 3 to 5% base substitutions even in essential housekeeping genes, most of which, however, involve synonymous changes that do not affect amino acid sequence (20). Virulence-associated vacuolating cytotoxin gene (vacA) and cagA sequences are much more diverse, and much of this diversity reflects nonsynonymous base substitutions (1, 3, 12, 30). This pattern suggests selection for proteins with different structures, functions, or antigenic properties during different infections. The genetic diversity of H. pylori is postulated to reflect numerous factors: mutation (44), recombination between divergent lineages (42), selection based on differences among human hosts in traits that may be important to individual strains (14), and finally its non-epidemic (preferentially intrafamilial) mode of transmission, which lowers the chance of selection for any one or a few possibly most fit genotypes (13, 29). Clinical isolates from adults are also likely to have diverged genetically from strains that may have infected them years before (in infancy), the result of years of selection for derivatives that are better adapted to the particular person, coupled with changes in gastric physiology over time. There are also differences in H. pylori genotypes that predominate in different geographic regions or human populations (16, 23, 28). This may reflect a combination of selection for different motifs (as in vacA and cagA above) plus genetic drift resulting simply from the highly localized mode of H. pylori transmission and isolation by distance of H. pylori strains in different human populations. Studies of H. pylori genotypes from different distinct human populations may contribute to our understanding of bacterium-host interactions and the origin and evolution of this bacterium in humans.
H. pylori infection is very common in India (28; H. H. Gill, P. Majumdar, K. Shankaran, R. H. Kalro, and H. G. Desai, Abstr. Annu. Conf. Ind. Soc. Gastroenterol., Ind. J. Gastroenterol. 9[Suppl. 1]:A17, 1990), and duodenal ulcer which is H. pylori-associated is of particular importance here and is far more common than in most other geographic regions (27). We had characterized the genotypes of H. pylori strains from adult ethnic Bengalis in Calcutta, in the state of West Bengal (8, 28). However, the Indian population is diverse, with close-knit communities, each consisting of peoples of different ethnicities, linguistic groups, and/or social status often living quite close to one another. Despite possibilities for transmission of H. pylori between unrelated people (e.g., through unclean water, food, or utensils), it was not clear a priori that the characteristics of mainstream Bengali H. pylori strains will fully represent the range of genotypes of H. pylori in other Indian communities, especially given ideas of how H. pylori tracks with human migration (11, 16, 23).
Santhals and Oroans are two distinct ethnic groups that had settled in the Birbhum district of West Bengal centuries ago (37-39). They constitute less than 5% of the overall population of West Bengal (Census of India, 2001). Ethnically, Santhals are proto-Australoids and speak the dialect Santhali of the Austro-Asiatic language family (37, 39), while the Oroans are ethnically Dravidian and speak the Khurukh dialect of the Dravidian linguistic family (38). That is, the two lineages to which they belong have been distinct for millennia. In contrast, the ethnic Bengalis have an Indo-European ancestry, and their Bengali language is derived from Sanskrit. Some scholars trace the migration and settlement of the Santhal communities in Bengal through the protohistoric and historic eras and link up their traditional homeland to Central India (initially known as the Dandakaranya area; in present-day Chattishgarh State). Later they came to the eastern flank of Dandakaranya (now in Bihar and Orissa States) and further migrated eastward to West Bengal (37). The original home of Oroans was in the western part of India, whence they came to the Kaimur hills and the plateau of Rohtas in Shahabad. Driven from Rohtas by Muhammadans, they followed the course of the Ganges and settled in Rajmahal hills and Chotonagpur plateau of Bihar, and from there migrated into adjacent West Bengal (38). Traditionally both Santhals and Oroans have been hunter-gatherers, but most have now become settled agriculturists, though they nevertheless continue to hunt and fish to supplement food. However, many of these peoples have begun to take other employment, typically as laborers in factories, mines, and farms of others (38). Nevertheless, they remain culturally, and linguistically distinct from most of Indian society and rarely intermarry with people of other ethnicities in India. Their separation from mainstream Bengalis and other Indians during much of human history is reflected in genetic differences in autosomal and mitochondrial DNA markers (7, 39).
These considerations and our interest in the population genetics and evolution of H. pylori motivated the present study of the prevalence and genotypes of strains from these two ethnic minorities in Bengal.
MATERIALS AND METHODS
Patient-derived samples.
A total of 49 healthy adults of both sexes (age 19 to 40 years) of the Santhal [31] and Oraon [18] ethnic communities of West Bengal underwent nonsedated upper gastrointestinal endoscopies using a GIF XQ 30 endoscope (Olympus Optical Company, Tokyo, Japan) under topical lignocaine anesthesia at the Institute of Post Graduate Medical Education and Research, Calcutta. A detailed history was taken, and a physical examination of each subject was carried out prior to endoscopy. The objectives of the study were explained to all, and written informed consent was obtained from them under protocols approved by the ethical committee of the Institute of Post Graduate Medical Education and Research, Calcutta. The endoscope was carefully cleaned and disinfected between patients by first immersing the tube in detergent solution for 10 min and then rinsing it with sterile distilled water. During gastroscopy, three biopsy samples were taken from the antrum and body of the stomach for rapid urease test (RUT), histology, and culture. For RUT, one biopsy sample was inoculated into urea broth containing a pH-sensitive marker (phenol red), and any color change was noted within 10 to 30 min. A positive result was recorded if the color changed from yellow to pink within 30 min. Another biopsy sample was fixed in buffered 10% formalin for histopathological examinations, and the third was placed in 0.5 ml of brucella broth (Difco Laboratories, Detroit, Mich.) containing 15% glycerol and transported to the microbiology laboratory on ice, where it was inoculated immediately onto culture medium (below) or stored at −70°C.
Histopathological examination.
Formalin-treated tissue biopsies were processed for paraffin embedding, followed by cutting thin (4-μm) sections, which were stained with modified Giemsa stain. Slides were examined microscopically using ×40 magnification. If H. pylori was observed, the bacterial density was graded semiquantitatively on an ordinal scale (ranging from 0 to 3) based on the Sydney system (36) by a single pathologist. All examinations were performed under code. If the two biopsies from each site (antrum or corpus) showed a different grade, then the higher grade was recorded.
H. pylori culture.
Biopsy samples in transport medium were vortexed vigorously for 2 min, and 100 μl of the media was plated on brain heart infusion (BHI) agar (Difco) supplemented with 7% horse blood, 0.4% IsoVitaleX, and H. pylori dent supplement (Oxoid, Basingstoke, Hampshire, England) (8). Plates were incubated at 37°C in an atmosphere of 5% O2 to 10% CO2 to 85% N2 for 3 to 6 days. H. pylori bacteria were identified based on characteristic colony morphology and appearance on Gram staining, positive urease, and gene-specific PCR tests. H. pylori cells that grew out of one biopsy on the primary culture plate were collected as a pooled population and preserved in sterile BHI broth with 15% glycerol at −70°C. In general only one such culture was analyzed per individual.
Determination of antimicrobial susceptibility and resistance.
Resistance or susceptibility to metronidazole (MTZ) (Sigma Chemical Co., St. Louis, Mo.) and clarithromycin (Sigma) were determined in this study. H. pylori strains growing exponentially on drug-free BHI agar were suspended in phosphate-buffered saline buffer, a series of 10-fold dilutions of these cell suspensions were prepared, and 10 μl of each dilution was spotted on freshly prepared BHI agar containing various concentrations of MTZ (0, 0.2, 0.5, 1.5, 3, 8, 16, 32, 64, and 128 μg/ml) (essentially as described in reference 22). The susceptibilities of strains to MTZ are described here in terms of MIC, defined operationally as the lowest of the MTZ concentrations listed above that reduces the efficiency of colony formation by at least 10-fold. We use this culture dilution protocol because it is more sensitive and reliable than traditional standard agar dilution or E-test methods for studying susceptibility to MTZ (MIC) with H. pylori, as discussed previously (23). For clarithromycin, the concentrations used ranged from 0.125 to 32 μg/ml. The plates were incubated for 3 to 4 days at 37°C under microaerophilic conditions. Mtzs and Clas transformant derivatives of H. pylori reference strain 26695, clarithromycin-resistant strains 5883 and 5898 (45), and three Mtzr Bengali H. pylori isolates (I-18, I-67, and I-86) (28) from our collection were included in each test as controls. For MTZ, resistance was defined as a MIC of >8 μg/ml, while for clarithromycin, resistance was defined as a MIC of >2 μg/ml (45).
DNA methods.
Chromosomal DNA was prepared by the hexadecyltrimethyl ammonium bromide extraction method from confluent BHI agar plate cultures. Specific PCR was carried out in 20-μl reaction volumes using 10 pmol of each primer per reaction, 0.25 mM (each) deoxynucleoside triphosphates, 1 U of Taq polymerase (Takara, Shuzo, Tokyo, Japan), and 10 ng of DNA (from both the pool of colonies and single-colony isolates) in a standard PCR buffer (Takara). PCR was carried out for 30 cycles of the following: (i) denaturation at 94°C for 1 min, (ii) primer-template DNA annealing at 55°C for 1 min, and (iii) DNA synthesis at 72°C for a time based on expected fragment size (1 min/kb). The primers used are listed in Table 1. PCR products were purified with the QIA quick gel extraction kit (Qiagen Corporation, Chatsworth, Calif.) according to the manufacturer's instruction and were directly sequenced using the BigDye terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.) on an automated DNA sequencer (ABI Prism 310). DNA sequence editing and analysis were performed with programs in the GCG package (Genetics Computer Group, Madison, Wis.), programs and data in The Institute for Genomic Research H. pylori genome database (http://www.tigr.org/tdb/mdb/hpdb.html), and the Phylip program of J. Felsenstein (http://www.evolution.genetics.washington.edu/phylip.html). Phylogenetic trees were visualized using Treeview (version 1.61 [http://www.taxonomy.zoology.gla.ac.uk/rod/treeview.html]).
TABLE 1.
Primers used in this study
| Region | Primer | Nucleotide sequence | Reference |
|---|---|---|---|
| vacA s1 or vacA s2 | VA1-F | 5′-ATG GAAATACAACAAACACAC | 28 |
| VA1-R | 5′-CTGCTTGAATGCGCCAAAC | ||
| vacA m1b | Vam-F3 | 5′-GGCCCCAATGCAGTCATGGAT | 28 |
| Vam-R3 | 5′-GCTGTTAGTGCCTAAAGAAGCAT | ||
| vacA m2 | VA4-F | 5′-GGAGCCCCAGGAAACATTG | 28 |
| VA4-R | 5′-CATAACTAGCGCCTTGCAC | ||
| vacA 0.7-kb middle | VAm-F | 5′-GCTCATTACGGCTTCCACTAATGT | 28 |
| VAm-R | 5′-GCGGTTATTGTTGTTATAAAGGGCTA | ||
| cagA (5′ end) | cagA5 | 5′-GGCAATGGTGGTCCTGGAGCTAGGC | 28 |
| cag A2 | 5′-GGAAATCTTTAATCTCAGTTCGG | ||
| cag-PAI empty site | Luni 1 | 5′-ACATTTTGGCTAAATAAACGCTG | 28 |
| R5280 | 5′-GGTTGCACGCATTTTCCCTTAATC | ||
| iceA1 | iceA1F | 5′-TATTTCTGGAAC TTGCGCAACCTGAT | 28 |
| M.Hpy1R | 5′-GGCCTACAACCGCATGGATAT | ||
| iceA2 | cycSF | 5′-CGGCTGTAGGCACTAAAGCTA | 28 |
| IceA2R | 5′-TCAATCCTATGTGAAACAATGATCGTT | ||
| iceA1 Δ94 bp | A1F673 | 5′-GGTGAGTCGTTGGGTAAGCGTTACAGAATT | 28 |
| A1R1174 | 5′-CACAACCATCATATTCAGCCTCCCCCTCATA | ||
| IS605 orfA | ORF18F | 5′-CGCCTTGATCGTTTCAGGATTAGC | 28 |
| ORF18R | 5′-CAACCAACCGAAGCAAGCATAATC | ||
| IS605 orfB | ORF19F | 5′-GGCTGTTCTAGGGTCGTGTATAAC | 28 |
| ORF19R | 5′-CAAGCTAGATGCAATCTAGCTACC | ||
| IS607 | IS607 Flk.F | 5′-GGCTACAAACAGAAACTAAAAT | 25 |
| IS607 F2 | 5′-AGATTACCGAATTTATAGATACG | ||
| IS608 | 608-F1 | 5′-CCATAACGCCTTAATAGTGTGTC | 26 |
| 608-R | 5′-CAAGCTTTGGAGTGATGAAGTTC | ||
| Type III | fcn unk | 5′-TGGATTAAATCTTAATGAATTATCG | 23 |
| cagF4856 | 5′-GCGATGAGAAGAATATCTTTAGCG |
Statistical analysis of data was performed by using the chi-square test and Fisher's exact test with significance set at a P value of <0.05.
Nucleotide sequence accession numbers.
The sequences obtained here were deposited in GenBank under the following accession numbers: AY162446 to AY162453 for 219 bp of cagA sequence and AY167579 to AY167584, AY168866 to AY168872, and AY169676 for 648 bp of vacA m1 alleles from Santhal and Oroan strains.
RESULTS
Incidence of H. pylori.
All 49 persons of the two ethnic communities (31 Santhal, 18 Oraon) who participated in this study were asymptomatic and appeared to have normal stomach and duodenal lining during visual examination during endoscopy. H. pylori was detected by RUT and histology in biopsies from every participant. Giemsa-stained tissue sections were carefully examined, and H. pylori colonization densities on gastric epithelia were graded on a four-point scale of none (grade 0), mild (grade 1), moderate (grade 2), or severe (grade 3), according to Sydney system guidelines (36). Of 49 samples, none belonged to grade 0, 13 (27%) were grade 1, 20 (41%) were grade 2, and 16 (33%) were grade 3. H. pylori was cultured from 20 of 31 biopsies from Santhals (64.5%) and 11 of 18 biopsies from Oroans (61%).
Genotypes of H. pylori strains. (i) cag PAI.
The presence or absence of the cag pathogenicity island (PAI) was scored by PCR with specific primers using DNA extracted from cultured strains. A 324-bp product indicative of the cag PAI was obtained with primers specific for the cagA gene from each of the 31 strains. None yielded a 550-bp product expected of a cag empty site, which would indicate complete absence of the cag PAI (Table 2).
TABLE 2.
Genotypic characteristics of H. pylori from Santhal and Oroan communities
| Trait or markera | No. of strains with trait or marker/no. tested
|
|
|---|---|---|
| Santhals (n = 20) | Oroans (n = 11) | |
| cag PAI+ onlyb | 20/20 | 11/11 |
| cag PAI− onlyb | 0/20 | 0/11 |
| cag Type III | 13/20 | 11/11 |
| vacAs1 only | 20/20 | 11/11 |
| vacAs2 | 0/20 | 0/11 |
| vacAm1c | 12/20 | 10/11 |
| vacAm2 only | 5/20 | 1/11 |
| vacAm1c, -m2 mixed | 3/20 | 0/11 |
| iceA1 | 10/20 | 3/11 |
| iceA2 | 9/20 | 4/11 |
| iceA1, iceA2 mixed | 1/20 | 4/11 |
| IS605 | 5/20 | 4/11 |
| IS606 | 1/20 | 0/11 |
| IS607 | 2/20 | 4/11 |
| ISHp608 | 2/20 | 0/11 |
Distribution of DNA markers was determined by PCR.
cag PAI+ only, infection with strains carrying cag PAI only; cag PAI− only, infection with strains lacking the cag PAI only.
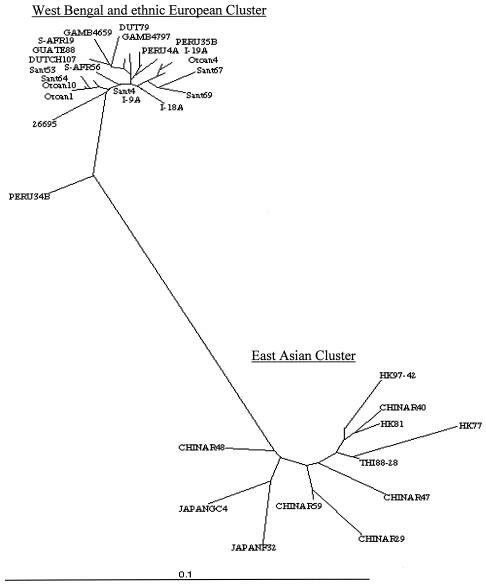
The 324-bp cagA PCR fragment is useful for assessing phylogenetic relationships (43) and was sequenced from eight representative strains (five Santhal, three Oroan). The eight sequences were closely related to one another and to corresponding cagA sequences of mainstream Bengali strains as well as those of ethnic European strains (28) and were distinct from those of East Asia (Fig. 1).
FIG. 1.
Phylogenetic tree based on an informative 220-bp segment at the 5′ end of cagA of H. pylori strains. The tree was generated using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein. The strains used are as follows (GenBank accession numbers are in parentheses): 26695 (AE000569); I-18 (Calcutta, India) (AF202224); I-9 (Calcutta, India) (AF202221); I-19 (Calcutta, India) (AF202225), Peru4A (AF198477), Peru34B (AF198475), Peru35B (AF198476), DUT79 (Dutch) (AJ252970), GAMB4659 (Gambia) (AF198468), GAMB4797 (Gambia) (AF198469), GUATE88 (Guatemala) (AF198472), S-AFR19 (South Africa) (AF198470), DUTCH107 (AJ252963), HK77 (Hong Kong) (AF198485), THI88 to 28 (Thailand) (AJ239722), HK97 to 42 (Hong Kong) (AJ239733), HK81 (Hong Kong) (AF198486), CHINAR47 (AJ252985), CHINAR40 (AJ252982), CHINAR59 (AJ252986), CHINAR29 (AJ252980), JAPANF32 (AJ239726), JAPANGC4 (AF198484), CHINAR48 (AJ252983), Sant 4 (Santhal) (AY162446), Sant 53 (Santhal) (AY162447), Sant 64 (Santhal) (AY162448), Sant 67 (Santhal) (AY162449), Sant 69 (Santhal) (AY162450), Oroan 1 (AY162451), Oroan 4 (AY162453), Oroan 10 (AY162452), and S-AFR56 (South Africa) (AF198471).
(ii) vacA alleles.
The presence of potentially toxigenic vacAs1 versus nontoxigenic vacAs2 alleles at the 5′ end of vacA was determined based on sizes of PCR products (259 versus 286 bp, respectively) generated with vacAs region-specific primers. All 31 Santhal and Oroan strains yielded a 259-bp fragment, indicating that they carried s1 alleles; no s2 alleles were found (Table 2). The frequency of vacA s1 alleles observed here (100%) is higher (P < 0.02) than that seen for strains from mainstream Bengalis (8, 28).
The alleles of the vacA middle (m) region, which determines the cell type specificity of vacuolating cytotoxin action, were also studied by PCR. Products were obtained only with vacA m1b/c primers with 12 of 20 Santhal strains and 10 of 11 Oraon strains; only with vacA m2 primers with five Santhal strains and one Oraon strain; and with both m1b/c and m2 primers with three Santhal strains (indicating mixed infection; these cultures, although free of non-H. pylori bacteria, had not been purified by single-colony isolation before PCR analysis).
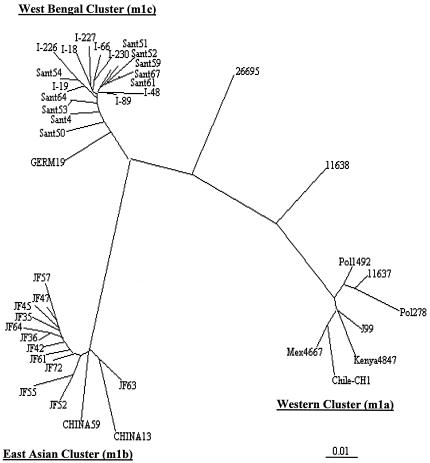
A 0.7-kb PCR fragment containing vacA mid-region alleles was sequenced from single-colony isolates of 10 Santhal strains and four Oroan strains that had yielded products with vacA m1b/c-specific primers. Analyses of sequences revealed that all were closely related to one another and to the vacA m1c alleles that are characteristic of mainstream Bengali H. pylori strains (28) and that each was distinct from European m1a alleles and East Asian m1b alleles (Fig. 2).
FIG. 2.
Phylogenetic tree based on an informative 650-bp segment of vacA gene containing vacAm1 alleles of H. pylori strains. The tree was generated using PHYLIP (Phylogeny Inference Package), version 3.5c, of J. Felsenstein. The strains used are as follows (GenBank accession numbers are in parentheses): I-19 (Calcutta, India) (AF220111), I-226 (Calcutta, India) (AF220115), I-89 (Calcutta, India) (AF220114), I-48 (Calcutta, India) (AF220112), I-18 (Calcutta, India) (AF220110), I-230 (Calcutta, India) (AF220117), GERM19 (Germany) (AJ006967), I-66 (Calcutta, India) (AF220113), I-227 (Calcutta, India) (AF220116), JF52 (Japan) (AF049631), JF55 (Japan) (AF049632), CHINAR59 (AF035611), JF63 (Japan) (AF049635), CHINAR13 (AF035610), JF42 (Japan) (AF049626), JF72 (Japan) (AF049651), JF45 (Japan) (AF049628), JF47 (Japan) (AF049629), JF57 (Japan) (AF049634), JF35 (Japan) (AF049625), JF61 (Japan) (AF049645), JF36 (Japan) (AF049462), JF64 (Japan) (AF049647), JF45 (Japan) (AF0496628), Pol1492 (Poland) (AF097570), Pol278 (Poland) (AF097571), NCTC11637 (AF049653), J99 (AE001511), NCTC11638 (U07145), 26695 (AE000598), Kenya AFN4847 (AF191644), Chile-CH1 (AF479031),and Mex 4467 9 (Mexico) (AF159855).
(iii) iceA alleles.
PCR was used to test for iceA1, which is virulence associated in some populations, and the completely unrelated iceA2 gene, which occupies the same chromosomal locus in strains lacking iceA1 (34). The iceA1 gene was found alone in 10 of 20 Santhal cultures and in 3 of 11 Oroan cultures, iceA2 was found alone in nine Santhal cultures and four Oroan cultures, and a mixture of iceA1 and iceA2 alleles (again, indicating mixed infection) was found in one Santhal culture and four Oroan cultures (Table 2). None of the 18 iceA1 alleles found here contained the 94-bp deletion near the 3′ end of iceA1, which was found in about one-fifth of mainstream Bengali strains (28).
(iv) cag Right junction motifs.
The type III motif at the extreme right end of cag PAI, which predominates in H. pylori strains from mainstream Bengali strains (8, 23, 28), was found by PCR with diagnostic primers in 13 of 20 Santhal strains and in all 11 Oroan strains.
(v) Neutral markers.
PCR tests for insertion sequences, which may be considered neutral markers, identified IS605 in 5 of 20 Santhal strains and 4 of 11 Oroan strains, IS606 in just one Santhal strain and no Oroan strains, IS607 in two Santhal strains and four Oraon strains, and ISHp608 in two Santhal strains and no Oroan strains. Only 1 of the 31 cultures contained two IS elements (IS605 and IS607), and 14 lacked each of these elements (Table 2). Thus, each element may be carried independently of the others, and the prevalence is similar to that for Bengali H. pylori (25, 26, 28).
Antimicrobial susceptibility.
All Santhal and Oroan H. pylori strains were sensitive to even 0.125 μg of clarithromycin/ml, as is typical of mainstream Bengali H. pylori strains (28), whereas known resistant control strains 5883 and 5898 (45), tested in parallel, were scored as resistant to 8 μg of clarithromycin per ml, as expected. In contrast, all strains were resistant to MTZ, with MICs of 16 μg/ml (18 strains), 32 μg/ml (10 strains), or 64 μg/ml (3 strains). Control strain 26695 tested in parallel was scored as sensitive to MTZ (MIC of 4 μg/ml). An extremely high prevalence of MTZ resistance (>90% of strains) is also typical of mainstream Bengali strains (9, 28).
DISCUSSION
Here we report that H. pylori infection is common in the Santhal and Oroan ethnic minorities of West Bengal, if anything even more common than in mainstream Bengali society, whereas overt H. pylori-associated disease is rare in these people, even though the genotypes of the strains they carry are similar to those for mainstream Bengalis. The near-universality of H. pylori infection can be ascribed to relatively low levels of sanitation, hygiene, and education (37), conditions that contribute to a high risk of infection and superinfection, even in adulthood (21, 41). A continuing risk of infection throughout life may explain occasional findings of mixed infection even in analyses of strains from single biopsies. Such results probably underestimate true frequencies of mixed infection for the following reasons: (i) H. pylori infections are often patchy (4), such that any single biopsy may often contain only a subset of strains that may be carried by a mixedly infected person, and (ii) mixed infections were detected here by PCR for conserved markers and insertion/deletion motifs, not point mutation differences, which are the most common polymorphisms in H. pylori populations.
The indication that Santhal and Oraon strains are closely matched to those of mainstream Bengalis is based most critically on the intermingling of phylogenetic trees of sequences from cagA and vacA genes from the strains obtained from these three communities and PCR data indicating similar frequencies of various markers, in particular, the four IS elements, iceA1 versus iceA2 genes, and polymorphic sequences downstream of cagA (8, 23, 28). Since these ethnic minorities and other Bengalis are of very different ancestries, the observed strain similarities suggest transmission among the populations, presumably from the far more numerous mainstream Bengali population to these ethnic minorities. Transmission between communities might have (i) involved displacement of a preexisting distinct strain population (assuming that H. pylori had been endemic in these peoples before extensive contact with Bengalis, as some would predict (5, 11, 16), due simply to the far great abundance of Bengalis and thus of their strains in water supplies and other possible environmental sources or due to a possibly greater vigor of Bengali strains; or (ii) resulted from easy transmission to and epidemic spread within a highly susceptible population if the hunter-gatherer ancestors of these ethnic minorities were H. pylori free, as predicted by a model in which H. pylori infection became widespread in humans only after the start of agriculture (23). It is striking that each of the 31 strains tested was MTZ resistant, since MTZ is used far less in Santhal and Oraon communities than in mainstream Bengali society (where some 90% of strains are also MTZ resistant) (9). Since MTZ only became widely available in India in the 1970s (9), this suggests that much of the postulated transmission from Bengalis to ethnic minorities may have been quite recent.
The carriage of the cag PAI and of putatively toxigenic s1 alleles of vacA, which contribute to virulence when active, is in accord with the high abundance (about 90%) of this strain type in mainstream Indian populations (8, 28), the high overall risk of H. pylori infection in these populations, and the idea that frequent transmission tends to select for more virulent strains of a pathogen (15). This abundance is also remarkable, in light of the apparent absence of H. pylori-associated gastrointestinal disease in these populations. This suggests that on average, H. pylori infections are less virulent in these ethnic minorities than in mainstream Indians. Such avirulence might be due to subtle features of bacterial strains or aspects of the human host environment. For example, lesser virulence of the H. pylori strains themselves might be ascribed to subtle (e.g., point) mutations that affect levels of expression of virulence genes or the potencies of their products, which do not, however, affect outcomes of diagnostic PCR tests used here. At least two reports of decreased virulence apparently being selected in vivo have appeared: one during human infection (24) and another during adaptation of human strains to mice (35). Avirulence might also reflect features of the host. One possibility entails concurrent infection with particular parasites that may downregulate inflammatory responses to infection, as has been documented in a mouse infection model (19). Resistance to pathogenic effects of putatively virulent H. pylori strains might also be determined by features of human host genotype. For example, studies using different inbred mouse lines have shown that the severity of inflammation, and thus potentially the risk of overt disease, that a given H. pylori strain can elicit is affected by the host genotype (17, 18). Indeed, the historic importance of cholera and other diarrheal diseases as a major cause of death in the Indian subcontinent and the importance of H. pylori infection in increasing susceptibility to enteric pathogens (10, 33, 40) might have helped select for human genotypes that result in decreased virulence during chronic infection.
Acknowledgments
We thank K. Rajendran for his help in statistical analysis.
This work was supported by grants from the Department of Biotechnology, Government of India (no. BT/MB/VAP/3/2/98), U.S. Public Health Service (AI 38166, AI 49161, DK 53727, and P30 DK 52574), and Indian Council of Medical Research (no. 5/8-1 (161)/H. pylori/2000/ECD-II).
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-54. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1986. Rapid identification of pyloric Campylobacter in Peruvians with gastritis. Dig. Dis. Sci. 31:1089-1094. [PubMed] [Google Scholar]
- 5.Blaser, M. J. 1998. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut 43:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1999. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J. Infect. Dis. 179:1523-1530. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli-Sforza, L. L., P. Menozzi, and A. Piazza. 1994. History and geography of human genes. Princeton University Press, Princeton, N.J.
- 8.Chattopadhyay, S., S. Datta, A. Chowdhury, S. Chowdhury, A. K. Mukhopadhyay, K. Rajendran, S. K. Bhattacharya, D. E. Berg, and G. B. Nair. 2002. Virulence genes in Helicobacter pylori strains from West Bengal residents with overt H. pylori-associated disease and healthy volunteers. J. Clin. Microbiol. 40:2622-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury, A., D. E. Berg, J. Y. Jeong, A. K. Mukhopadhyay, and G. B. Nair. 2002. Metronidazole resistance in Helicobacter pylori: magnitude, mechanism and implications for India. Ind. J. Gastroenterol. 21:23-28. [PubMed] [Google Scholar]
- 10.Clemens, J., M. J. Albert, M. Rao, F. Qadri, S. Huda, B. Kay, F. P. van Loon, D. Sack, B. A. Pradhan, and R. B. Sack. 1995. Impact of infection by Helicobacter pylori on the risk and severity of endemic cholera. J. Infect. Dis. 171:1653-1656. [DOI] [PubMed] [Google Scholar]
- 11.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 12.Cover, T. L., M. K. R. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566-10573. [PubMed] [Google Scholar]
- 13.Datta, S., A. Chowdhury, A. K. Mukhopadhyay, S. K. Bhattacharya, D. E. Berg, and G. B. Nair. 2002. Molecular and evolutionary genetics and drug resistance of the gastric pathogen, Helicobacter pylori. Ind. J. Med. Res. 115:71-99. [PubMed] [Google Scholar]
- 14.Dubois, A., D. E. Berg, E. T. Incecik, N. Fiala, L. M. Heman-Ackah, J. Del Valle, M. Yang, H. P. Wirth, G. I. Perez-Perez, and M. J. Blaser. 1999. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology 116:90-96. [DOI] [PubMed] [Google Scholar]
- 15.Ewald, P. W., J. B. Sussman, M. T. Distler, C. Libel, W. P. Chammas, V. J. Dirita, C. A. Salles, A. C. Vicente, I. Heitmann, and F. Cabello. 1998. Evolutionary control of infectious disease: prospects for vector-borne and water-borne pathogens. Mem. Inst. Oswaldo Cruz 93:567-576. [DOI] [PubMed] [Google Scholar]
- 16.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 17.Ferrero, R. L., and J. G. Fox. 2001. In vivo modeling of Helicobacter-associated gastrointestinal diseases, p. 565-582. In: H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. American Society for Microbiology, Washington, D.C.
- 18.Ferrero, R. L., and P. J. Jenks. 2001. In vivo adaptation to the host, p. 583-592. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. American Society for Microbiology, Washington, D.C.
- 19.Fox, J. G., P. Beck, C. A. Dangler, M. T. Whary, T. C. Wang, H. N. Shi, and C. Nagler-Anderson. 2000. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nat. Med. 6:536-542. [DOI] [PubMed] [Google Scholar]
- 20.Ge, Z., and D. E. Taylor. 1999. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu. Rev. Microbiol. 53:353-387. [DOI] [PubMed] [Google Scholar]
- 21.Hildebrand, P., P. Bardhan, L. Rossi, S. Parvin, A. Rahman, M. S. Arefin, M. Hasan, M. M. Ahmad, K. Glatz-Krieger, L. Terracciano, P. Bauerfeind, C. Beglinger, N. Gyr, and A. K. Khan. 2001. Recrudescence and reinfection with Helicobacter pylori after eradication therapy in Bangladeshi adults. Gastroenterology 121:792-798. [DOI] [PubMed] [Google Scholar]
- 22.Jeong, J. Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Y. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H.-K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (hp0954) and frxA (hp0642) nitroreductase genes cause moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 25.Kersulyte, D., A. K. Mukhopadhyay, M. Shirai, T. Nakazawa, and D. E. Berg. 2000. Functional organization and insertion specificity of IS 607, a chimeric element of Helicobacter pylori. J. Bacteriol. 182:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersulyte, D., B. Velapatino, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: non-random geographic distribution, functional organization and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam, S. K. 2000. Differences in peptic ulcer between East and West. Baillieres Best Pract. Res. Clin. Gastroenterol. 14:41-52. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, A. K., J. Y. Jeong, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2003. The fdxA ferredoxin gene can down-regulate frxA nitroreductase gene expression and is essential in many strains of Helicobacter pylori. J. Bacteriol. 185:2927-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, Z. J., D. E. Berg, R. W. van der Hulst, W. W. Su, A. Raudonikiene, S. D. Xiao, J. Dankert, G. N. Tytgat, and A. van der Ende. 1998. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J. Infect. Dis. 178:220-226. [DOI] [PubMed] [Google Scholar]
- 31.Parsonnet, J. 1995. Incidence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 9:45-51. [PubMed] [Google Scholar]
- 32.Parsonnet, J. 1999. Helicobacter and gastric adenocarcinoma, p. 372-408. In J. Parsonnet (ed.), Microbes and malignancy: infection as a cause of human cancers. Oxford University Press, New York, N.Y.
- 33.Passaro, D. J., D. N. Taylor, R. Meza, L. Cabrera, R. H. Gilman, and J. Parsonnet. 2001. Acute Helicobacter pylori infection is followed by an increase in diarrheal disease among Peruvian children. Pediatrics 108:E87. [DOI] [PubMed] [Google Scholar]
- 34.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Phys. 110:531-544. [PubMed] [Google Scholar]
- 35.Philpott, D. J., D. Belaid, P. Troubadour, J. M. Thiberge, J. Tankovic, A. Labigne, and R. L. Ferrero. 2002. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell Microbiol. 4:285-296. [DOI] [PubMed] [Google Scholar]
- 36.Price, A. B. 1991. The Sydney system: histological division. J. Gastroenterol. Hepatol. 6:209-222. [DOI] [PubMed] [Google Scholar]
- 37.Ray, U. K., A. K. Das, and S. K. Basu. 1982. To be with Santhals. Cultural Research Institute, Scheduled Castes and Tribes Welfare Department, Government of West Bengal, Calcutta, India.
- 38.Risley, H. H. 1891. The tribes and castes of Bengal, vol. 2. Bengal Secretariat Press, Calcutta, India.
- 39.Roychoudhury, S., S. Roy, A. Basu, R. Banerjee, H. Vishwanathan, M. V. Usha Rani, S. K. Sil, M. Mitra, and P. P. Majumder. 2001. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum. Genet. 109:339-350. [DOI] [PubMed] [Google Scholar]
- 40.Shahinian, M. L., D. J. Passaro, D. L. Swerdlow, E. D. Mintz, M. Rodriguez, and J. Parsonnel. 2000. Helicobacter pylori and epidemic Vibrio cholerae O1 infection in Peru. Lancet 355:377-378. [DOI] [PubMed] [Google Scholar]
- 41.Soto, G., C. T. Bautista, R. H. Gilman, D. E. Roth, B. Velapatio, M. Ogura, G. Dailide, M. Razuri, R. Meza, U. Katz, T. P. Monath, D. E. Berg, D. N. Taylor, and the Gastrointestinal Physiology Working Group in Peru. Helicobacter pylori reinfection is common in Peruvian adults following antibiotic eradication therapy. J. Infect. Dis., in press. [DOI] [PubMed]
- 42.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Ende, A., Z. J. Pan, A. Bart, R. W. van der Hulst, M. Feller, S. D. Xiao, G. N. Tytgat, and J. Dankert. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, G., M. Z. Humayun, and D. E. Taylor. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7:488-493. [DOI] [PubMed] [Google Scholar]
- 45.Wang, W. H., B. C. Y. Wong, A. K. Mukhopadhyay, D. E. Berg, C. H. Cho, K. C. Lai, W. H. C. Hu, F. M. Y. Fung, W. M. Hui, and S. K. Lam. 2000. High prevalence of Helicobacter pylori infection with dual resistance to metronidazole and clarithromycin in Hong Kong. Aliment. Pharmacol. Ther. 14:901-910. [DOI] [PubMed] [Google Scholar]
- 46.Westblom, T. U., S. J. Czinn, and J. G. Nedrud. 1999. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Curr. Top. Microbiol. Immunol. 241:1-33.10087653 [Google Scholar]
- 47.Xia, H. H., and J. N. Talley. 1997. Natural acquisition and loss of Helicobacter pylori. Am. J. Gastroenterol. 10:1780-1787. [PubMed] [Google Scholar]