Abstract
The aminoglycoside antibiotic hygromycin B was examined in Escherichia coli cells for inhibitory effects on translation and ribosomal-subunit formation. Pulse-chase labeling experiments were performed, which verified lower rates of ribosomal-subunit synthesis in drug-treated cells. Hygromycin B exhibited a concentration-dependent inhibitory effect on viable-cell numbers, growth rate, protein synthesis, and 30S and 50S subunit formation. Unlike other aminoglycosides, hygromycin B was a more effective inhibitor of translation than of ribosomal-subunit formation in E. coli. Examination of total RNA from treated cells showed an increase in RNA corresponding to a precursor to the 16S rRNA, while mature 16S rRNA decreased. Northern hybridization to rRNA in cells treated with hygromycin B showed that RNase II- and RNase III-deficient strains of E. coli accumulated 16S rRNA fragments upon treatment with the drug. The results indicate that hygromycin B targets protein synthesis and 30S ribosomal-subunit assembly.
Currently, only a limited number of antibiotics are available to treat multidrug-resistant strains of infectious bacteria, and resistance to even the newest antimicrobial agents is appearing (13). Increasing antibiotic resistance in microbial populations has necessitated the search for alternate cellular targets for new and existing antimicrobial agents. There are many antibiotics available that inhibit translation in bacterial cells by binding to the 30S or 50S ribosomal subunit (3). It has been observed that a number of ribosomal antibiotics possess a second inhibitory activity that prevents ribosomal-subunit assembly. The formation of a functional ribosome is a vital cellular process and is therefore also an important cellular target (4).
In order to broaden our understanding of subunit assembly inhibitors, it is important to examine new or poorly studied antibiotics for their possible effects on inhibition of subunit formation. Identifying and comparing these antimicrobial agents will aid in the search for more suitable targets for future antibiotics. Crystallographic data have been obtained for several antibiotics from the aminoglycoside class (2, 25). These studies have shown that binding occurs through unique base interactions in the decoding center of the 30S ribosome, which effectively halts translation by preventing movements necessary for protein synthesis to occur (8). Recent work with the aminoglycoside compounds neomycin and paromomycin has shown that these antimicrobial agents have a second inhibitory target, which is inhibition of 30S ribosomal-subunit formation (18, 19). These drugs are capable of inhibiting 30S ribosomal-subunit formation by stalling assembly at an intermediate phase of 30S biosynthesis.
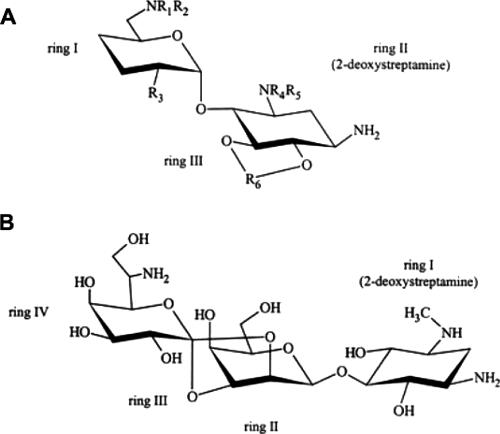
There are many antibiotics that target translation by binding to ribosomal subunits that have not been examined for possible inhibitory effects on ribosomal-subunit assembly. One of those antimicrobial agents is the aminoglycoside hygromycin B, a 30S subunit translational inhibitor. Most aminoglycosides have two variable amino sugar rings that are connected to a 2-deoxystreptamine ring (Fig. 1A). The position at which the two variable rings are attached determines the class of aminoglycoside to which the compound belongs. Hygromycin B is unusual in that its structure has two ether linkages connecting two of its three sugar residues, resulting in a fourth, five-membered ring (Fig. 1B). This extended structure has been shown to bind to the 30S ribosomal subunit near the A, P, and E sites and spans a 13-Å distance on the small subunit (2, 20).
FIG. 1.
Chemical structures of aminoglycoside antibiotics. (A) A generic 2-deoxystreptamine aminoglycoside. (B) Hygromycin B.
Hygromycin B has been in use since 1957 and was the first additive in livestock feed to be approved by the Food and Drug Administration. It has been used extensively since then as a growth aid and parasite control agent in the production of swine and poultry. It removes parasites by disrupting the egg-laying process and then killing the adults (1, 17). Hygromycin B has also been used in the laboratory to select for recombinant cells containing a hygromycin B resistance marker (12).
The structure and mechanism of binding of hygromycin B sets the compound apart from other aminoglycosides, and therefore it is of interest to examine the effects these differences have on the inhibition of subunit assembly and its effects on turnover of 16S rRNA in hygromycin B-treated cells. Previous work has shown that RNases II and III and polynucleotide phosphorylase (PNPase) are involved in the turnover of 23S rRNA in the precursor that forms in the presence of the 50S inhibitor azithromycin (23). The accumulation of 16S rRNA fragments during treatment with 30S ribosomal-subunit inhibitors has not been examined. One goal of this study was to determine if the 30S ribosomal-subunit inhibitor hygromycin B had any effect on the accumulation of 16S rRNA fragments in E. coli cells with RNase mutations. The results indicate that hygromycin B also has two inhibitory targets, preventing protein synthesis and 30S ribosomal-subunit formation with a stimulation of 16S rRNA turnover.
MATERIALS AND METHODS
Studies were conducted with Escherichia coli strain D10-1 (11). The characteristics of the RNase mutant strains have been described recently (23). Hygromycin B was purchased from Sigma. [3H]uridine (45 mCi/mmol) and [35S]methionine (Trans 35S-Label; 1.175 Ci/mmol) were purchased from Perkin-Elmer and MP Biomedical, respectively.
Measurements of cell growth, viability, subunit assembly, and translation rates.
The MIC for hygromycin B was determined by a dilution method as described previously (6). Cells were grown at 37°C in tryptic soy broth (TSB) in the presence and absence of the antibiotic. Growth rates were measured by following the increase in cell density using a Klett-Summerson colorimeter. Cell viability was measured by colony counting after serial dilution. RNA was labeled by treating the cells with 1 μCi/ml [3H]uridine (1 μg/ml) and allowing the cultures to grow for two doublings in the presence or absence of hygromycin B. Isotope incorporation was halted by adding uridine to a final concentration of 50 μg/ml, followed by a 15-min chase period. Cells were collected by centrifugation at 6,500 rpm for 10 min and held at −70°C until lysis. The cells were lysed with a lysozyme freeze-thaw method as described previously (7). The lysates were spun on 5 to 20% sucrose gradients in S buffer (10 mM Tris-HCl, pH 8.0, 0.5 mM Mg acetate, 50 mM NH4Cl) to separate ribosome particles. Fractions were collected from the gradients and mixed with 3 ml of Scintisafe Gel (Fisher), and the incorporation of [3H]uridine was measured by liquid scintillation counting.
Protein synthesis rates were determined by adding [35S]methionine (1 μCi/ml) to the cell culture before the uridine chase period. Samples of 0.4 ml were collected at 5-min intervals for 15 min. The collected samples were precipitated with 20% trichloroacetic acid (TCA). The TCA-precipitated material was collected on Whatman GF/A glass fiber filters, washed with 10% TCA, and measured by liquid scintillation counting.
Uridine pulse-chase labeling.
Cells were grown in 25 ml of TSB at 27°C in the presence or absence of the drug. At a Klett reading of 10, hygromycin B (50 μg/ml) was added to the culture. The control sample was grown without antibiotic. After growth had continued for one cell doubling, the RNA was pulse-labeled with [3H]uridine (1 μCi/ml) for 3 min and then chased with uridine at 50 μg/ml. Samples of 4 ml were taken at six time intervals. Cells were collected by centrifugation, washed in S buffer, and stored at −70°C before lysis and sucrose gradient centrifugation.
Analysis of total RNA via the Aligent Bioanalyzer.
E. coli cells were grown in TSB with various concentrations of hygromycin B. RNA was extracted from 1 ml of cells using the AquaPure RNA isolation kit (Bio-Rad). Total RNA was examined using an Aligent Bioanalyzer 2100 and the RNA 6000 “lab on a chip”: 5 μl (200 ng/μl) of sample from the total RNA was loaded into each well of the RNA 6000 chip. Sample preparation and the loading procedure and run were carried out according to the manufacturer's recommendations for total-RNA analysis.
Northern analysis of 16S rRNA.
RNA oligonucleotides were isolated from the top fractions of sucrose gradients and were denatured and separated on 1.5% gels as described previously (23). RNA from the gels was blotted onto Nytran nylon membranes using a Turbo blot apparatus (S&S). Construction of biotinylated 16S and 23S rRNA-specific DNA probes and the hybridization of membranes with biotin-labeled probe were performed as described previously (23). The probe was detected on the membranes using the North2South chemiluminescent hybridization kit (Pierce Chemical Co.) according to the manufacturer's directions. A low-range, ready-to-use RNA ladder (Fermentas) was biotin labeled using the Label-It kit (Mirus) according to the manufacturer's directions and used to estimate the size of the RNA.
RESULTS
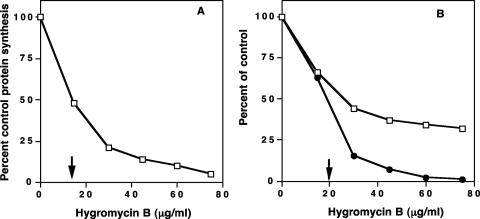
Aminoglycoside antibiotics are well-known inhibitors of translation in bacterial cells (3). The MIC for hygromycin B in E. coli cells growing in TSB was found to be 150 μg/ml. Cells were grown at a subinhibitory concentration to examine the effects of the antibiotic on cellular activities. The rate of protein synthesis in growing E. coli cells was determined by measuring the incorporation of [35S]methionine into cellular proteins. Figure 2A shows the relative rate of incorporation of [35S]methionine with increasing concentrations of hygromycin B as a percentage of control [35S]methionine incorporation. The 50% inhibitory concentration (IC50) for inhibition of translation was 16 μg/ml.
FIG. 2.
Inhibition of protein synthesis, growth rate, and cell viability in E. coli cells with increasing concentrations of hygromycin B. (A) Inhibition of protein synthesis by hygromycin B. The arrow indicates the IC50. (B) Percent decrease in growth rate (□) and viable-cell numbers (•) with increasing concentrations of hygromycin B. The arrow indicates the IC50 for the cell numbers. The results are the means of two determinations.
The inhibition of cell viability and of the cellular growth rate was consistent with the demonstrated inhibitory effects of hygromycin B on translation. As shown in Fig. 2B, hygromycin B diminished the number of viable cells and increased the doubling time in a concentration-dependent fashion. Hygromycin B inhibited viable-cell numbers by 50% at a concentration of 20 μg/ml, and the growth rate was reduced by half at 25 μg/ml.
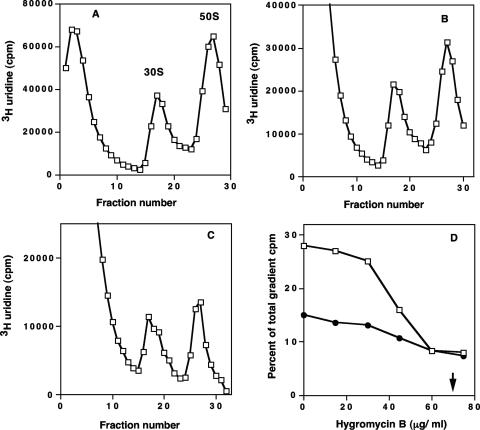
The effect of hygromycin B on ribosomal-subunit formation in growing E. coli cells was also determined. The concentration dependence of ribosomal-subunit assembly was measured by examining sucrose gradient profiles of ribosomal subunits labeled with [3H]uridine during growth in the presence and absence of hygromycin B. Figure 3A to C shows sucrose gradient profiles of lysates from cells grown with and without the antibiotic. The 30S ribosomal-subunit amounts were reduced in proportion to the drug concentration (Fig. 3D). An IC50 of 65 μg/ml for inhibition of ribosomal-subunit formation was seen. There was also increased accumulation of fragmented RNA in the top fractions of gradients from cells treated with hygromycin B. The amounts of 50S ribosomal subunits were also reduced at higher concentrations of the antibiotic.
FIG. 3.
Sucrose gradient profiles of E. coli grown in the presence and absence of hygromycin B. (A) Gradient profile of control cell lysate. (B) Lysate from cells grown in 30 μg/ml hygromycin B. (C) Lysate from cells grown in 60 μg/ml hygromycin B. (D) Concentration-dependent inhibition of ribosomal-subunit assembly in E. coli cells. The percentages of the total gradient cpm were calculated for the 30S and 50S regions for the control and antibiotic-treated gradient samples. Inhibition of 30S assembly (•) and inhibition of 50S assembly (□) are shown. The arrow indicates the IC50 for 30S ribosomal-subunit formation. The results are the means of two determinations.
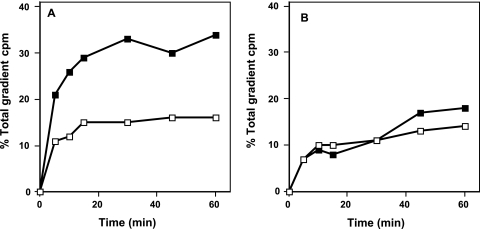
Pulse-chase labeling kinetic analysis was also used to measure the rates of ribosomal-subunit synthesis in growing cells. Figure 4A and B shows the pulse-chase kinetic analysis of ribosomal-subunit formation in control and hygromycin B-treated cells. In the absence of the antibiotic, 30S ribosomal-subunit formation was complete in 15 min and 50S subunit formation reached a plateau in 30 min. When cells were treated with hygromycin B, 30S subunit formation did not reach control levels until 60 min and 50S ribosomal subunit amounts were estimated to reach control levels in 120 min. 30S and 50S ribosomal-subunit formation rates were inhibited equally relative to the control rates. These results are comparable to those observed in previous assembly studies with the aminoglycosides neomycin and paromomycin (18).
FIG. 4.
[3H]uridine pulse-chase labeling kinetic analysis of ribosomal-subunit formation. (A) Assembly of 30S subunits (□) and 50S subunits (▪) without hygromycin B. (B) Assembly of 30S subunits (□) and 50S subunits (▪) with hygromycin B. The results are the means of two determinations.
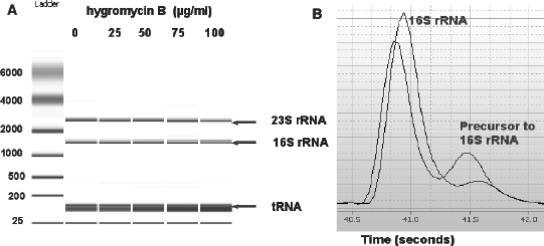
Total RNA was examined in E. coli cells grown with hygromycin B at various concentrations of antibiotic and in control cells using the RNA 6000 lab on a chip kit and Bioanalyzer 2100 (Agilent). Figure 5A shows the virtual gel from the Agilent Bioanalyzer analysis. With increasing concentrations of hygromycin B, there was an increase in the amount of RNA accumulation corresponding to a 16S rRNA precursor (Fig. 5B). There was also a concentration-dependent decrease in the amount of 16S rRNA present in the antibiotic-treated cells, along with an increase in the number of small fragments of RNA. A small decrease in 23S rRNA was also seen at the highest concentration of hygromycin B used. Table 1 lists the percentage of the total area for each peak, corresponding to small RNA, 16S rRNA, its precursor, and 23S rRNA.
FIG. 5.
Agilent bioanalyzer analysis of total RNA from E. coli cells treated with hygromycin B. (A) Virtual gel produced by the Agilent software based on the results from a fluorescent chromatograph. 23S and 16S rRNAs and tRNA are indicated. (B) Tracing of lane 4 (75 μg/ml) showing a 16S rRNA precursor peak.
TABLE 1.
Relative amounts of RNA from Agilent RNA analysis
| Amt of hygromycin B (μg/ml) | % of total areaa
|
|||
|---|---|---|---|---|
| Small RNA | 16S RNA | 16S precursor | 23S RNA | |
| 0 | 60.9 | 9.6 | 0.3 | 13.0 |
| 25 | 59.0 | 10.4 | 0.5 | 13.5 |
| 50 | 62.8 | 10.4 | 0.75 | 12.5 |
| 75 | 64.5 | 7.3 | 1.1 | 12.7 |
| 100 | 68.8 | 4.8 | 1.2 | 10.0 |
The values were calculated from duplicate RNA analyses like that shown in Fig. 5.
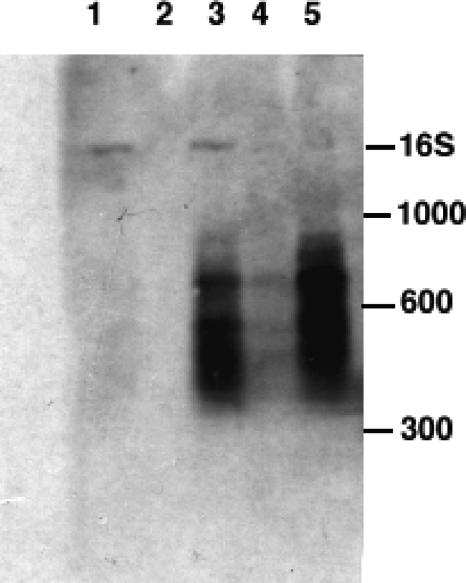
Previous work with the 50S ribosomal-subunit inhibitor azithromycin showed that 23S rRNA fragments accumulate upon drug treatment (23). They were present in greater numbers in E. coli strains containing one or more mutations in specific RNase genes. RNases II and III and PNPase were shown to be involved in the turnover of 23S rRNA in 50S subunit precursors. Analysis by Northern hybridization with 16S and 23S rRNA-specific probes was performed on RNAs isolated from the top fractions of sucrose gradients of lysates from wild-type and RNase mutant strains of E. coli grown with hygromycin B. The RNase mutant strains examined in this study included strains defective in RNase I, RNase II, RNase III, and PNPase. Figure 6 shows a Northern blot hybridized with a 16S rRNA-specific probe. RNase II and III mutant strains contained substantial numbers of 16S fragments in comparison with the control and an RNase I mutant. The sizes of RNA fragments were estimated to be between 200 and 900 bases. No accumulation of 23S RNA fragments was visualized in the top gradient fractions examined with a 23S rRNA-specific probe (data not shown).
FIG. 6.
Northern hybridization analysis of rRNA from control and RNase mutant strains. RNA was isolated from the top regions of sucrose gradients from cells grown with and without hygromycin B. Lane 1, control RNA from an RNase I strain; lane 2, RNA from an RNase I mutant grown with hygromycin B; lane 3, RNA from an RNase II mutant grown with hygromycin B; lane 4, RNA from a PNPase mutant grown with hygromycin B; lane 5, RNA from an RNase III mutant grown with hygromycin B. The positions of RNA size standards are indicated, along with 16S rRNA added as a marker in lanes 1 and 3.
DISCUSSION
It is known that aminoglycoside antibiotics inhibit translation by binding to the 30S ribosomal subunit. Recent studies have shown that the aminoglycoside antibiotics neomycin and paromomycin possess a secondary inhibitory target, preventing the formation of functional 30S ribosomal subunits (18, 19). These previous studies showed that the antibiotics prevent translation and ribosomal-subunit formation with equal effectiveness in growing E. coli cells. A difference was seen between the effects of hygromycin B on protein synthesis and ribosomal-subunit assembly. Hygromycin B was a better inhibitor of translation than of 30S ribosomal-subunit formation, with IC50s of 16 μg/ml and 65 μg/ml, respectively. Inhibition of the growth rate and total viable-cell numbers was in accordance with the reduction of protein synthesis rates.
Hygromycin B has been shown to affect a ribosomal ATPase (RbbA) that is required for protein synthesis (10). RbbA is a protein that binds to the 30S subunit near the E site and functions to aid ejection of tRNA from the subunit (26). Hygromycin B binds near the binding site for RbbA, disrupting the binding of the ATPase and inhibiting 70 to 80% of its activity. Other aminoglycoside antibiotics did not have this strong inhibitory effect. The effects of hygromycin B on this enzyme may contribute to the marked difference seen in its inhibitory effect on translation compared with the effect on subunit formation.
Inhibition of 50S ribosomal-subunit assembly by hygromycin B was also observed. Most 50S protein biosynthesis inhibitors do not affect 30S subunit synthesis (4). However, 50S ribosomal-subunit formation is affected by inhibitors of 30S ribosomal-subunit assembly. The aminoglycosides neomycin and paromomycin have been shown to cause a reduction in 50S ribosomal-subunit formation in E. coli at higher concentrations of these drugs (18). Hygromycin B demonstrated a similar effect on the formation of the 50S ribosomal subunit. The indirect action of these antibiotics on the 50S subunit is likely due to a downstream polar effect on 23S rRNA processing. 30S particle biosynthesis precedes 50S ribosomal-subunit synthesis and is dependent on RNase III processing from a common precursor RNA (24). Therefore, an inhibitory effect on 30S formation could have a coupled downstream action of slowing or halting 50S biosynthesis. A generalized inhibitory effect on ribosomal-protein synthesis is a less likely explanation, because with inhibitors like the streptogramin A compounds, a parallel decline in the formation of both subunits is observed (5).
Pulse-chase kinetic analysis of ribosomal-subunit formation in cells treated with hygromycin B supports this linkage effect on 50S particle synthesis. In control cells, the time required to synthesize a 50S particle was twice as long as 30S synthesis. In drug-treated cells, the rates of synthesis of the 30S and 50S subunits were decreased fourfold compared with control rates. The results for the pulse-chase kinetic analysis suggested equal inhibition of the rates of formation of 30S and 50S ribosomal subunits, consistent with a linkage of 30S particle formation with 50S synthesis.
An examination of total RNA from hygromycin B-treated E. coli cells also indicated a specific inhibition of 16S rRNA processing. Total-RNA analysis showed an increase in small fragments of RNA and a decrease in the amount of RNA corresponding to 16S rRNA. In addition, the amount of RNA corresponding to the 17S rRNA precursor increased (Table 1 and Fig. 5). The accumulation of the 17S precursor has been observed in ribosomal-subunit chaperone mutants (14-16, 22). 23S rRNA amounts were reduced only at the highest antibiotic concentration used. This is consistent with an indirect effect of the stalling of 30S subunit formation on 50S particle synthesis.
Northern blot analysis with a 16S rRNA-specific probe showed increased accumulation of fragmented 16S rRNA in several RNase mutants (Fig. 6). RNA from RNase II and RNase III mutant strains treated with drug contained larger amounts of 16S fragments than an RNase I mutant. Previous work with the 50S assembly inhibitors has also shown these enzymes to be important in the turnover of 23S rRNA in a stalled intermediate (23). This accumulation of rRNA fragments is the result of antibiotic binding to an intermediate particle in ribosomal-subunit assembly. These particles are degraded by RNases in wild-type cells. In drug-treated RNase-defective strains, this process is retarded, and a buildup of rRNA fragments occurs.
The accumulation of 16S rRNA fragments in the RNase II and RNase III mutants is an indication of the importance of these enzymes in the breakdown of the ribosomal-subunit precursor. RNase III is an important endoribonuclease that is essential for the separation of 16S and 23S rRNAs from a common precursor (24). Hybridization with a 23S-specific probe showed no accumulation of 23S fragments in any of the drug-treated samples, indicating that hygromycin B specifically targets only the 30S ribosome in assembly and translation.
Hygromycin B binds with specificity only to the 30S ribosomal subunit and inhibits translation by interfering with the A, P, and E sites only on this subunit (2, 20). Hygromycin B has been shown to bind to a part of 16S rRNA helix 44 that changes position during translocation, and it is believed that hygromycin B binding to helix 44 restricts the movement of the helix during protein synthesis (9). It is possible that hygromycin B binding to the helix could also restrict movements necessary to form functional 30S subunits and in so doing prevent conformational rearrangement required for processing of the 16S rRNA transcript and subsequent 30S ribosomal-subunit maturation.
This work is a continuation of studies examining possible ribosomal-subunit assembly inhibitors. The formation of novel hybrid aminoglycoside compounds between neomycin and hygromycin is an indication of the importance of these drugs in controlling infections (21, 27). More studies such as this one are needed to elucidate the potential of ribosomal-subunit assembly as an antibiotic target and to increase our understanding of the mechanism behind specific inhibition of this vital cellular function.
Footnotes
Published ahead of print on 16 October 2006.
REFERENCES
- 1.Biehl, L. 1986. Anthelmintics for swine. Vet. Clin. N. Am. Food Anim. Pract. 2:481-487. [DOI] [PubMed] [Google Scholar]
- 2.Brodersen, D., W. Clemons, A. Carter, R. Morgan-Warren, B. Wimberly, and V. Ramakrishanan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 3.Bryskier, A. 2005. Antimicrobial agents, antibacterials, and antifungals. ASM Press, Washington, DC.
- 4.Champney, W. 2003. Bacterial ribosomal subunit assembly is an antibiotic target. Curr. Top. Med. Chem. 3:929-947. [DOI] [PubMed] [Google Scholar]
- 5.Champney, W., and C. Tober. 2000. Specific inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide and streptogramin B antibiotics. Curr. Microbiol. 41:126-135. [DOI] [PubMed] [Google Scholar]
- 6.Champney, W., C. Tober, and R. Burdine. 1998. A comparison of the inhibition of translation and 50S subunit formation in Staphylococcus aureus cells by nine different macrolide antibiotics. Curr. Microbiol. 37:412-417. [DOI] [PubMed] [Google Scholar]
- 7.Champney, W. S., and R. Burdine. 1995. Macrolide antibiotics inhibit 50S ribosomal subunit assembly in Bacillus subtilis and Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourmy, D., M. Recht, S. Blanchard, and J. Puglisi. 1996. Structure of the A site of Escherichia coli 16S rRNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 9.Frank, J., and R. Agrawal. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318-322. [DOI] [PubMed] [Google Scholar]
- 10.Ganoza, M., and M. Kiel. 2001. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 45:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gesteland, R. 1966. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J. Mol. Biol. 16:67-84. [DOI] [PubMed] [Google Scholar]
- 12.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, R. 2005. Mechanisms of action of newer antibiotics for Gram-positive pathogens. Lancet Infect. Dis. 5:209-218. [DOI] [PubMed] [Google Scholar]
- 14.Himeno, H., K. Hanawa-Suetsugu, T. Kimura, K. Takagi, W. Sugiyama, S. Shirata, T. Mikami, F. Odagiri, Y. Osanai, D. Wantanabe, S. Goto, L. Kalachnyuk, C. Ushida, and A. Muto. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 32:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue, K., J. Alsina, J. Chen, and M. Inouye. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48:1005-1016. [DOI] [PubMed] [Google Scholar]
- 16.Kaczanowska, M., and M. Ryden-Aulin. 2005. The YrdC protein—a putative ribosome maturation factor. Biochim. Biophys. Acta 1727:87-96. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, G., and L. Olsen. 1960. Critical tests for hygromycin B as an ascaricide of swine. Cornell Vet. 50:60-65. [PubMed] [Google Scholar]
- 18.Mehta, R., and W. Champney. 2002. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob. Agents Chemother. 46:1546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta, R., and W. Champney. 2003. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 47:237-243. [DOI] [PubMed] [Google Scholar]
- 20.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 21.Murray, J., S. Meroueh, R. Russell, G. Lentzen, J. Haddad, and S. Mobashery. 2006. Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site. Chem. Biol. 13:129-138. [DOI] [PubMed] [Google Scholar]
- 22.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgF is involved in ribosome maturation. Genes Cells 10:393-408. [DOI] [PubMed] [Google Scholar]
- 23.Silvers, J., and W. Champney. 2005. Accumulation and turnover of 23S ribosomal RNA in azithromycin-inhibited ribonuclease mutant strains of Escherichia coli. Arch. Microbiol. 184:66-77. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava, A., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 25.Vicens, Q., and E. Westhof. 2003. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure 9:647-658. [DOI] [PubMed] [Google Scholar]
- 26.Xu, J., C. Kiel, A. Golshani, J. Chosay, H. Aoki, and M. Ganoza. 2006. Molecular localization of a ribosome-dependent ATPase on Escherichia coli ribosomes. Nucleic Acids Res. 34:1158-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, Y., E. Vlad, Z. Sun, B. Ayida, G. Winters, D. Murphy, K. Simonsen, D. Vourloumis, S. Fish, J. Froelich, D. Wall, and T. Hermann. 2005. Structure-guided discovery of novel aminoglycoside mimetics as antibacterial translation inhibitors. Antimicrob. Agents Chemother. 49:4942-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]