Abstract
The prevalence of new species of Pseudallescheria and Scedosporium in a collection of 46 clinical isolates was analyzed. Strain identification was done by morphological and molecular methods. Four Scedosporium aurantiacum isolates were detected among the panel of clinical strains. The susceptibility profile of S. aurantiacum was similar to that of Scedosporium apiospermum.
Pseudallescheria boydii is a pathogen able to cause asymptomatic colonization and localized and disseminated infections (11).
Recently, it has been demonstrated that high genetic variation exists in the P. boydii species complex (2, 4, 8). Two new species, Pseudallescheria minutispora and Scedosporium aurantiacum, are phylogenetically and morphologically separated from P. boydii. In addition, P. angusta, P. ellipsoidea, and P. fusoidea seem to present genetic differences and could be proposed as new species of Pseudallescheria (4).
We have analyzed the prevalence of these new species and their antifungal susceptibility profiles in a collection of clinical isolates of P. boydii.
Strains.
A total of 46 clinical isolates of P. boydii and S. apiospermum were included in this study. Twenty-four strains were isolated from respiratory sites, five from biopsies, six from ear samples, five from skin, four from ocular samples, one from blood culture, and one from an abscess.
Table 1 displays the identification of the 42 sequences obtained from the GenBank database that were used as controls.
TABLE 1.
Scedosporium reference strains used for comparison of ITS sequences and their GenBank accession numbers
| Species | Strain identification | GenBank accession no. |
|---|---|---|
| Pseudallescheria angusta | AJ888414 | |
| Pseudallescheria angusta | AJ888413 | |
| Pseudallescheria ellipsoidea | CBS 418,73 | AJ888426 |
| Pseudallescheria ellipsoidea | AJ888427 | |
| Pseudallescheria fusoidea | CBS 106,53 | AJ888428 |
| Pseudallescheria fusoidea | AJ888429 | |
| Pseudallescheria minutispora | AJ888424 | |
| Pseudallescheria minutispora | AY228119 | |
| Pseudallescheria minutispora | AJ888384 | |
| Scedosporium apiospermum | AJ888443 | |
| Scedosporium apiospermum | AJ888392 | |
| Scedosporium apiospermum | AF181558 | |
| Scedosporium apiospermum | AJ888385 | |
| Scedosporium apiospermum | AJ888438 | |
| Scedosporium apiospermum | AJ888391 | |
| Scedosporium apiospermum | AY213683 | |
| Scedosporium apiospermum | CBS 10854 | AY228112 |
| Scedosporium apiospermum | AJ888400 | |
| Scedosporium apiospermum | AY217658 | |
| Scedosporium apiospermum | AY228123 | |
| Scedosporium apiospermum | AY939802 | |
| Scedosporium apiospermum | AF455484 | |
| Scedosporium apiospermum | CBS 10122 | AY213680 |
| Scedosporium apiospermum | AY213681 | |
| Scedosporium apiospermum | CBS 59190 | AY228118 |
| Scedosporium apiospermum | AY228122 | |
| Scedosporium apiospermum | AY213682 | |
| Scedosporium apiospermum | AJ888398 | |
| Scedosporium apiospermum | AJ888436 | |
| Scedosporium apiospermum | AJ888386 | |
| Scedosporium apiospermum | AJ888387 | |
| Scedosporium apiospermum | AJ888394 | |
| Scedosporium apiospermum | AJ888397 | |
| Scedosporium apiospermum | AJ888405 | |
| Scedosporium apiospermum | AJ888410 | |
| Scedosporium apiospermum | AJ888411 | |
| Scedosporium apiospermum | CBS 101,22 | AJ888435 |
| Scedosporium apiospermum | AJ888437 | |
| Scedosporium aurantiacum | AJ888432 | |
| Scedosporium aurantiacum | AJ888439 | |
| Scedosporium aurantiacum | AJ888440 | |
| Scedosporium aurantiacum | AJ888441 |
Morphological identification.
All isolates were identified by conventional methods (3).
PCR and DNA sequencing of the internal transcribed spacer (ITS) region.
Molds were cultured in GYEP medium (0.3% yeast extract, 1% peptone [Difco, Madrid, Spain], 2% glucose [Sigma Aldrich Quimica, Madrid, Spain]) for 24 to 48 h at 30°C. Genomic DNA was isolated using a previously described extraction procedure (5).
DNA segments comprising the ITS1 and ITS2 regions were amplified with primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (1) in a GeneAmp 9700 PCR system (Applied Biosystems) (10). The reaction products were analyzed in a 0.8% agarose gel.
Sequencing reactions were done with 2 μl of reaction mixture from a sequencing kit (BigDye terminator cycle sequencing kit, Ready Reaction mixture; Applied Biosystems), 1 μM of the primers (ITS1 and ITS4), and 3 μl of the PCR product in a final volume of 10 μl.
Sequence analysis.
Sequences were assembled and edited using the SeqMan II and EditSeq software packages (Lasergene; DNAstar, Inc., Madison, WI). Sequence analysis was performed by comparison of the DNA sequences with 42 ITS sequences of Scedosporium and Pseudallescheria strains obtained from the GenBank database (http://www.ncbi.nih.gov/GenBank/). Full information about these strains is displayed in Table 1.
Phylogenetic analysis.
All phylogenetic analyses were conducted with Fingerprinting II Informatix software, version 3.0 (Bio-Rad Laboratories, Madrid, Spain). The methodology used was maximum parsimony clustering. Phylogram stability was assessed by parsimony bootstrapping with 2,000 simulations. The ITS sequence of Scedosporium prolificans CNM-CM-3571 (Mold Collection of the Spanish National Center for Microbiology) was used as the outgroup.
Antifungal susceptibility testing.
Microdilution testing was performed following the CLSI (formerly NCCLS) reference method (6) with the following minor modifications. (i) RPMI 1640 was supplemented with glucose to reach a 2% concentration. (ii) Inoculum size was between 1 × 105 to 5 × 105 CFU/ml. Inoculum preparations were performed by means of counting spores in a hematocytometer (1, 7, 9). Aspergillus fumigatus ATCC 2004305 and Aspergillus flavus ATCC 2004304 were used as quality control strains (6).
The antifungal agents used in the study were amphotericin B (range, 16 to 0.03 mg/liter) (Sigma Aldrich Química), itraconazole (range, 8 to 0.015 mg/liter) (Janssen S.A., Madrid, Spain), and voriconazole (range, 8 to 0.015 mg/liter) (Pfizer S.A.). The endpoint was the antifungal concentration that produced a complete inhibition of visual growth at 48 h.
Morphological identification.
All strains were identified as Scedosporium apiospermum by conventional methods (3). We were not able to identify by morphological characteristics the new proposed species (4, 8).
Molecular identification.
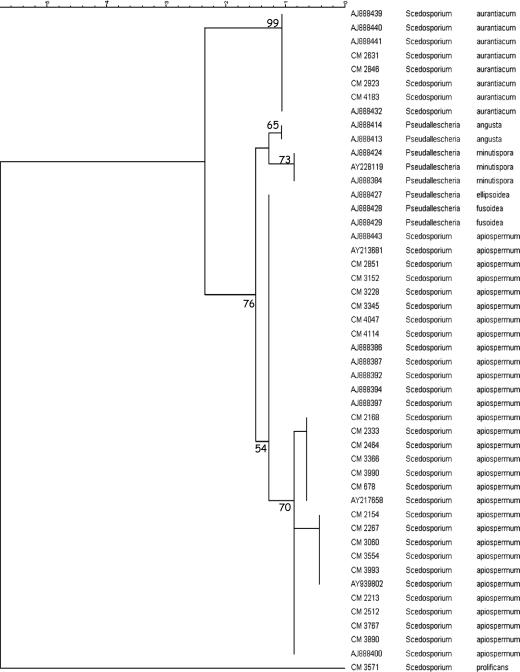
Figure 1 shows the phylogenetic analysis by means of maximum parsimony of a subset of the strains included in the study. Among 46 clinical strains analyzed, four S. aurantiacum strains were identified with a bootstrap value of ≥99. They were isolated from sputum, bronchial aspirate, a corneal sample, and an ear swab. In this collection of clinical isolates, we did not found any strain of P. minutispora or P. angusta, but the sequences obtained from GenBank were supported by bootstrap values of 73 and 65, respectively. On the other hand, the analyses of ITS sequences indicate that Pseudallescheria ellipsoidea and P. fusoidea are indistinguishable from S. apiospermum isolates (Fig. 1).
FIG. 1.
Phylogenetic tree of the subset of isolates included in the study obtained by using maximum parsimony phylogenetic analyses and 2,000 bootstrap simulations based on ITS sequences. Scedosporium prolificans CNM-CM-3751 was used as the outgroup to root the tree.
Antifungal susceptibility testing.
The MICs of antifungal agents for the collection of clinical isolates are shown in Table 2. They are sorted by species identification by means of ITS sequencing.
TABLE 2.
Antifungal susceptibility results for clinical isolates of Scedosporium aurantiacum and geometric mean and range of the 42 Scedosporium apiospermum isolates
| Isolate | Strain no. | Sample | MIC (mg/liter)
|
||
|---|---|---|---|---|---|
| Amphotericin B | Itraconazole | Voriconazole | |||
| S. aurantiacum | CNM-CM-2923 | Corneal | 8 | 16 | 16 |
| CNM-CM-2631 | Sputum | 4 | 2 | 0.5 | |
| CNM-CM-2846 | Ear swab | 16 | 8 | 1 | |
| CNM-CM-4183 | Bronchial aspirate | 8 | 16 | 0.5 | |
| Geometric mean | 6.96 | 6.06 | 1.15 | ||
| S. apiospermum | |||||
| Geometric mean | 6.21 | 2.62 | 0.73 | ||
| Range | 0.25-32 | 0.25-16 | 0.125-16 | ||
It has always been suspected that Pseudallescheria boydii was a complex of species. Hitherto, morphological analyses of Pseudallescheria boydii isolates did not allow for a clear separation among different species. However, the analysis of DNA sequences has permitted the proposal of new species from Pseudallescheria, such as S. aurantiacum, P. minutispora, P. angusta, P. ellipsoidea, and P. fusoidea (4, 8).
We examined the prevalence of these new species in a collection of clinical isolates. ITS sequence analyses revealed the presence of four S. aurantiacum strains, obtained from an ocular sample, an ear swab, bronchial aspirate, and sputum, and no strains of P. minutispora. Although Gilgado et al. (4) have defined morphological characteristics to identify S. aurantiacum and P. minutispora, great experience in classical taxonomy is required to perform this task. Therefore, it is not expected that many clinical microbiology laboratories are able to identify those new species unless they use molecular methodology. Regarding susceptibility to antifungal drugs, S. aurantiacum seems slightly more resistant than S. apiospermum to amphotericin B and itraconazole, although a higher number of isolates should be analyzed before any conclusion is drawn (Table 2).
Because little is know about the prevalence of these new species and therefore there are no studies regarding epidemiology, pattern of disease, risk factors, antifungal susceptibility testing, etc., we strongly recommend sending all strains of Scedosporium species involved in human infections to reference laboratories where those isolates can be properly identified to the species level and antifungal susceptibility testing can be performed. In this way, the importance of these new species can be ascertained. Meanwhile, and especially for clinical use, it would be better to maintain the use of the S. apiospermum name. From a practical point of view and for clinical microbiology laboratories, we suggest performing antifungal susceptibility testing rather than applying molecular methods for the identification of these species.
Acknowledgments
Ana Alastruey-Izquierdo has a predoctoral fellowship from Fondo de Investigaciones Sanitarias (grant FI05/00856). This work was supported in part by research project PY05/32 from the Instituto de Salud Carlos III.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Aberkane, A., M. Cuenca-Estrella, A. Gomez-Lopez, E. Petrikkou, E. Mellado, A. Monzon, and J. L. Rodriguez-Tudela. 2002. Comparative evaluation of two different methods of inoculum preparation for antifungal susceptibility testing of filamentous fungi. J. Antimicrob. Chemother. 50:719-722. [DOI] [PubMed] [Google Scholar]
- 2.Defontaine, A., R. Zouhair, B. Cimon, J. Carrere, E. Bailly, F. Symoens, M. Diouri, J. N. Hallet, and J. P. Bouchara. 2002. Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J. Clin. Microbiol. 40:2108-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Hoog, G. S., J. Guarro, J. Gené, and M. J. Figueras. 1995. Hyphomycetes, p. 301-302 and 899-901. In G. S. de Hoog and J. Guarro (ed.), Atlas of clinical fungi. Universitat Rovira i Virgili, Reus, Spain.
- 4.Gilgado, F., J. Cano, J. Gene, and J. Guarro. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J. Clin. Microbiol. 43:4930-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden, D. W. 1994. DNA mini prep method for Aspergillus fumigatus (and other filamentous fungi), p. 3-4. In B. Maresca and G. S. Kobayashi (ed.), Molecular biology of pathogenic fungi, a laboratory manual. Telos Press, New York, NY.
- 6.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility. Testing of filamentous fungi: approved standard document M38-A. Reference method for broth dilution antifungal susceptibility 22. NCCLS, Wayne, Pa.
- 7.Petrikkou, E., J. L. Rodriguez-Tudela, M. Cuenca-Estrella, A. Gomez, A. Molleja, and E. Mellado. 2001. Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. J. Clin. Microbiol. 39:1345-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rainer, J., G. S. de Hoog, M. Wedde, Y. Graser, and S. Gilges. 2000. Molecular variability of Pseudallescheria boydii, a neurotropic opportunist. J. Clin. Microbiol. 38:3267-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Tudela, J. L., E. Chryssanthou, E. Petrikkou, J. Mosquera, D. W. Denning, and M. Cuenca-Estrella. 2003. Interlaboratory evaluation of hematocytometer method of inoculum preparation for testing antifungal susceptibilities of filamentous fungi. J. Clin. Microbiol. 41:5236-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Tudela, J. L., T. M. Diaz-Guerra, E. Mellado, V. Cano, C. Tapia, A. Perkins, A. Gomez-Lopez, L. Rodero, and M. Cuenca-Estrella. 2005. Susceptibility patterns and molecular identification of Trichosporon species. Antimicrob. Agents Chemother. 49:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinbach, W. J., and J. R. Perfect. 2003. Scedosporium species infections and treatments. J. Chemother. 15(Suppl. 2):16-27. [DOI] [PubMed] [Google Scholar]