Abstract
We mapped the coding single nucleotide polymorphisms in four toxin genes—exoS, exoT, exoU, and exoY—of the Pseudomonas aeruginosa type III secretion system among several clinical isolates. We then used this information to design a multiplex PCR assay based on the simultaneous amplification of fragments of these genes. Eight strains of known genotype were used to test our multiplex PCR method, which showed 100% sensitivity and specificity in this small sample size. This assay appears to be promising for the rapid and accurate genotyping of the presence of these genes in clinical strains of P. aeruginosa.
Pseudomonas aeruginosa is an opportunistic gram-negative bacillus that causes a variety of clinically important infections in compromised hosts and in the critically ill, commonly affecting patients with cystic fibrosis, severe burns, neutropenia, and the mechanically ventilated (3). It has emerged as one of the most problematic gram-negative bacteria in hospital settings, causing 15 to 20% of cases of hospital-acquired pneumonia (2). Infection is associated with crude mortality rates as high as 70% overall (32) and 90% in mechanically ventilated patients (7). Attributable mortality risks are about 40% (8).
P. aeruginosa utilizes a large number of secreted and cell-associated virulence factors that have been implicated in the pathogenesis of infection. These include exotoxin A, phospholipase, alkaline protease, elastase, pyocin, pili, flagella, and lipopolysaccharide (34). One important determinant of virulence is the type III secretion system (TTSS), which is present in several gram-negative bacilli, including Salmonella, Shigella, and Yersinia spp. (16). P. aeruginosa is able to produce and secrete virulence factors directly into the cytoplasm of host cells by the cell contact-mediated TTSS. The system consists of three separate protein complexes: the secretion apparatus itself, the translocation or targeting apparatus, and the secreted toxins (effector proteins) and their cognate chaperones (25). The effector proteins currently described include two ADP-ribosylating enzymes (ExoS and ExoT) (13, 14, 39), an acute cytolytic factor (ExoU) (11, 22), and an adenylate cyclase (ExoY) (40).
The genes that encode these proteins are characterized by variable traits, i.e., they are present in some isolates but not in others and they are scattered throughout the 6.3-Mb genome of P. aeruginosa (35). Several studies have shown that the TTSS is present in nearly all clinical and environmental isolates but that individual isolates and populations of isolates from distinct disease sites differ in their effector genotypes (9). Although exoY and exoT are present in nearly all clinical isolates, a significant number lack either exoS or exoU (9, 11, 22). Various authors have classified P. aeruginosa strains based on the genotypic expression of these toxins. For example, Fleiszig et al. (12) have classified isolates based on their ability to invade host cells. Others have shown that invasive strains tend to express ExoS, whereas noninvasive or cytotoxic strains express ExoU (11, 22). Clinical isolates that secrete type III secretion (TTS) toxins are associated with worse clinical outcomes in patients with ventilator-associated pneumonia (11, 12, 33) and the presence of ExoS, ExoT, or ExoU secretion correlates with a sixfold greater relative risk of mortality (33). ExoU, in particular, correlates with acute cytotoxicity and lung damage (11). Introduction of the exoU gene confers a cytotoxic phenotype on some otherwise-noncytotoxic P. aeruginosa strains and, for recombinant strains that could express ExoU, there was markedly increased virulence in a mouse model of acute pneumonia and systemic spread (1). Isogenic mutants that do not produce or secrete ExoU resulted in a loss of cytotoxicity and reduced virulence in a mouse model of acute lung infection (11), a finding also reported by Hauser et al. (22). Clinically, isolates that secrete ExoU in vitro were more likely to have caused severe pneumonia in mechanically ventilated patients (21).
Given the evidence that the toxins of the TTSS are important for the determination of strain virulence and prediction of more severe clinical outcomes, it would be advantageous to have a simple, rapid, and accurate system for the detection of these genes. We describe here the construction of single-nucleotide-polymorphism (SNP) maps for the four known effector proteins of the P. aeruginosa TTSS. Thus, we were able to determine the genetic heterogeneity among clinical strains and to identify conserved regions within each gene. We used this information to develop a reliable multiplex PCR assay for the simultaneous detection of these genes.
MATERIALS AND METHODS
Determination of gene heterogeneity.
In previous experiments (33), we collected 108 clinical isolates from blood and respiratory sources. We randomly chose 23 of these isolates for sequencing. Eight were cloned and sequenced for exoT, six were cloned and sequenced for exoU, and nine were cloned and sequenced for exoY (Table 1). Bacteria from frozen stock were grown overnight at 37°C in Luria-Bertani medium (Fisher Scientific, Pittsburgh, Pa.), and DNA was isolated by using a DNA purification kit according to the manufacturer's protocol (Clontech, Palo Alto, Calif.). Each gene was amplified in total by using a single set of PCR primers (Table 2), the proofreading DNA polymerase PFU (Stratagene, San Diego, Calif.), and an optimal protocol. The results were analyzed on a 0.9% agarose gel (Life Technologies, Carlsbad, Calif.) with ethidium bromide (0.5 mg/ml). A band of appropriate size was excised, and DNA was extracted by using a QIAquick gel extraction kit (Qiagen, Valencia, Calif.) according to the manufacturer's manual. Each gene was cloned into the plasmid vector pCR-Blunt II-TOPO (Invitrogen, Carlsbad, Calif.). Cloned DNA was sequenced by the University of California San Francisco Biomolecular Resource Center by using two to five primers. The entire gene sequence was reconstructed from these data by using DNA analysis software. The sequences of strains previously published in GenBank were used for the evaluation of exoS (10, 35). These included the following strains (accession number): 388 (L27629), PAK (AYO29248), CCU1 (AYO29240), CCU2 (AYO29241), CCU3 (AYO29252), CCU4 (AYO29243), CCU5 (AYO29246), CCU6 (AYO29250), CCU8 (AYO29249), CCU9 (AYO29239), DG1 (AYO29247), FRD1 (AYO29251), PA-U1 (AYO29245), PA-U3 (AYO29242), and ATCC 27853 (AYO29244).
TABLE 1.
P. aeruginosa isolates used in gene sequencing and multiplex PCR
| Isolate(s)a | Use | Reference |
|---|---|---|
| PAO1 | exoT and exoY sequencing, M-PCRb | 35 |
| PA103 | exoU sequencing, M-PCR | 11 |
| 2038, 2013, 2002, 2036, 1047, 1027, 2034, and 1061 | exoT sequencing | 33 |
| 1089, 2038, 2013, 2026, 2002, and 1105 | exoU sequencing | 33 |
| 2038, 2002, 1072, 2056, 1079, 1080, 1056, 1027, and 1105 | exoY sequencing | 33 |
| 6294, 6206, 6073, 6487, and PAK | M-PCR | 12 |
PAO1, PA103, 19960, and PAK are laboratory stock strains. Isolates 2002 and 1105 were obtained from blood culture; isolates 6294, 6206, 6073, and 6487 were obtained from ocular cultures. The remaining strains were obtained from respiratory culture.
M-PCR, multiplex PCR.
TABLE 2.
PCR primers used for gene amplification and multiplex assay
| PCR primer use and namea | Primer, DNA sequence |
|---|---|
| Gene amplification | |
| exoT (1,564-bp fragment) | |
| 5′-Primer (position −100) | ExoT5, 5′-CGA CGG CCG CCA ACA GTA AAA-3′ |
| 3′-Primer (position 1464) | ExoT3, 5′-AGG TAT CCT TGC CGC CCA TTG-3′ |
| exoU (2,103-bp fragment) | |
| 5′-Primer (position −38) | ExoU5, 5′-ATA TTT GCT CCG AAC CCT CG-3′ |
| 3′-Primer (position 2065) | ExoU3, 5′-CTG TGC CAG CCA TGT ATC AA-3′ |
| exoY (1,178-bp fragment) | |
| 5′-Primer (position −15) | ExoT5, 5′-GCG GGA AAA CGA ACC ATG-3′ |
| 3′-Primer (position 1163) | ExoT3, 5′-CGG GCT TTG CCA ACG ACC-3′ |
| Gene detection in multiplex assay | |
| exoS (118-bp fragment) | |
| 5′-Primer (position 686) | ExoS-MP5, 5′-GCG AGG TCA GCA GAG TAT CG-3′ |
| 3′-Primer (position 804) | ExoS-MP3, 5′-TTC GGC GTC ACT GTG GAT GC-3′ |
| exoT (152-bp fragment) | |
| 5′-Primer (position 624) | ExoT-MP5, 5′-AAT CGC CGT CCA ACT GCA TGC G-3′ |
| 3′-Primer (position 776) | ExoT-MP5, 5′-TGT TCG CCG AGG TAC TGC TC-3′ |
| exoU (134-bp fragment) | |
| 5′-Primer (position 1265) | ExoU-MP5, 5′-CCG TTG TGG TGC CGT TGA AG-3′ |
| 3′-Primer (position 1399) | ExoU-MP3, 5′-CCA GAT GTT CAC CGA CTC GC-3′ |
| exoY (289-bp fragment) | |
| 5′-Primer (position 755) | ExoY-MP5, 5′-CGG ATT CTA TGG CAG GGA GG-3′ |
| 3′-Primer (position 1044) | ExoY-MP3, 5′-GCC CTT GAT GCA CTC GAC CA-3′ |
Position 1 is at the translational start site.
Multiplex PCR assay.
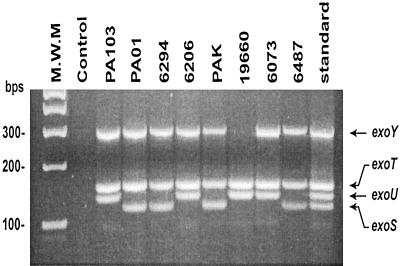
Multiplex PCR was performed on eight previously isolated strains of P. aeruginosa (Table 1). Mapping of the gene sequence variability was used to design PCR primers that were placed in conserved regions of each gene. They amplified a 118-bp fragment of exoS, a 134-bp fragment of exoU, a 153-bp fragment of exoT, and a 289-bp fragment of exoY. Bacteria were grown overnight at 37°C in Luria-Bertani medium (Fisher Scientific), and DNA was isolated by using a DNA purification kit according to the published protocol (Clontech). The PCR was set up as follows: 1 μl of DNA template (100 to 200 ng), 4 μl of total PCR primers (Operon Technologies, Alameda, Calif.), (a final 200 mM concentration of each primer), 12.5 μl of AccuPrime SuperMix II (Invitrogen), and 7.5 μl of sterile water. The negative control contained AccuPrime SuperMix II, no DNA, and 8.5 μl of sterile water. The standard reaction included 1 μl each of PAO1 and PA103 DNA, AccuPrime SuperMix II, and 6.5 μl of sterile water. The PCR was as carried out follows: initial denaturation at 94°C for 2 min; 36 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min; and a final extension step at 68°C for 7 min. The reaction was run in a 2% Metaphor agarose gel (FMC BioProducts, Rockland, Maine) with 0.5 mg of ethidium bromide/ml (Fig. 1).
FIG. 1.
Genotyping of exoS, exoT, exoU, and exoY in clinical and laboratory P. aeruginosa isolates by multiplex PCR. Agarose gel electrophoresis shows bands representing amplified DNA fragments of each gene. All isolates were genotype positive for exoT, but only strain 19660 was genotype negative for exoY. Strains positive for exoU were genotype negative for exoS. M.W.M., molecular weight marker; standard, standard DNA fragments representing each gene; bps, base pairs.
RESULTS
For each gene considered, the sequences of several isolates were aligned and analyzed for deviation from the sequence of the reference strain (Table 3). Each gene ranged from about 1.1 to 2.0 kb. Although almost all strains of P. aeruginosa possess a set of genes for the TTSS itself (25), not all strains carry genes for all of the TTS toxins. For instance, PAO1, the strain sequenced by the Pseudomonas Genome Project (35), has a negative genotype for exoU, and strain PA103 has a negative genotype for exoS. Therefore, PAO1 was the reference strain for exoS, exoT, and exoY, and PA103 was the reference strain for exoU. The total number of SNPs was ascertained and then further divided into those which were nonsynonymous (i.e., resulted in a change in the amino acid sequence) and those that were synonymous (i.e., resulted in the same amino acid). The number of base pairs per nonsynonymous SNP (NSS) was calculated to represent the frequency of sequence variability. The largest gene, exoU, had the smallest number of SNPs (14 total), while the other genes had similar total SNPs (27 to 34 each). When only NSSs were considered, exoU remained the most conserved (5 versus 9 to 18 variations). Despite being the smallest gene, exoY had the highest total number of SNPs (34 SNPs) and almost two to three times as many amino acid-altering SNPs as the other genes (18 versus 5 to 10). The number of base pairs per NSS was lowest in exoY and highest in exoU (63 versus 413). The genes exoS and exoT had similar values for total SNPs, NSS, and the number of base pairs per NSS. The position and frequency of the SNPs and amino acid substitutions resulting from the NSSs in ExoS, ExoT, and ExoU is shown in Fig. 2, 3, and 4, respectively.
TABLE 3.
Summary of the gene sequence heterogeneity of exoS, exoT, exoU, and exoY
| Gene | n | Size (bp) | Reference sequence | Total no. of SNPs | Size (bp)/SNP | No. of NSSs | Size (bp)/NSS | Deletion(s) |
|---|---|---|---|---|---|---|---|---|
| exoS | 13 | 1,361 | PAO1 | 30 | 45 | 9 | 151 | None |
| exoT | 8 | 1,374 | PAO1 | 27 | 50 | 10 | 137 | None |
| exoU | 6 | 2,064 | PA103 | 14 | 147 | 5 | 413 | None |
| exoY | 9 | 1,137 | PAO1 | 34 | 33 | 18 | 63 | Three isolates |
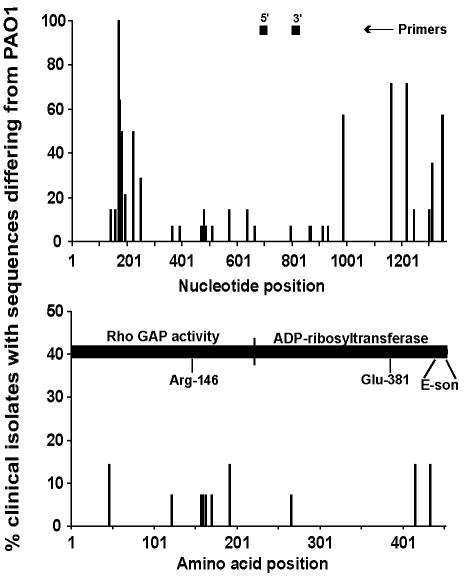
FIG. 2.
Position and frequency of exoS DNA and amino acid sequence variations (n = 13) and position of PCR primers, GAP, and ADP-ribosyltransferase functional domains. The x axis represents the nucleotide or amino acid position; the y axis represents the percentage of P. aeruginosa isolates that differed in sequence from PAO1 at each position. Arg-146 is essential for Rho GAP activity, whereas Glu-381 is essential for ADP-ribosyltransferase activity. E-son in the ADP-ribosyltransferase domain is the binding site of a 14-3-3 protein (factor for activating ExoS [FAS]). The exoS sequence of various isolates reported by Ferguson et al. (10) was used in this analysis.
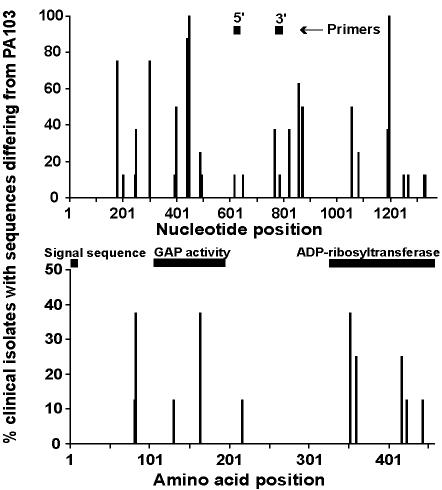
FIG. 3.
Position and frequency of exoT DNA and amino acid sequence variations (n = 8) and the position of PCR primers and signal sequence, GAP, and ADP-ribosyltransferase domains. The x axis represents the nucleotide or amino acid position; the y axis represents the percentage of P. aeruginosa isolates that differed in sequence from PAO1 at each position.
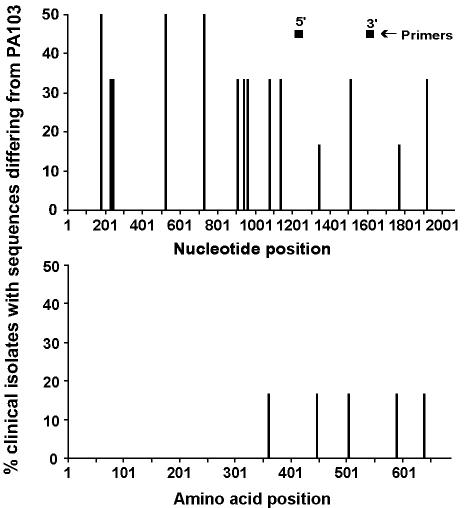
FIG. 4.
Position and frequency of exoU DNA and amino acid sequence variations (n = 6) and position of PCR primers. The x axis represents the nucleotide or amino acid position; the y axis represents the percentage of P. aeruginosa isolates that differed in sequence from PA103 at each position.
ExoS contains a C-terminal ADP-ribosyltransferase (amino acids 232 to 453), which modifies and blocks activation of RAS in vivo (17, 23, 28, 31, 37) and Rho GAP (GTPase-activating protein) activity (amino acids 1 to 234) in vitro (20). Although the SNPs were scattered throughout the gene, the NSSs were concentrated in the amino terminus of ExoS in the strains investigated (Fig. 2). The GAP domain contains a catalytic arginine (Arg-146) in the arginine finger domain, which is necessary for the stimulation of GAP by Rho (41). In addition to this amino acid, several other residues are conserved between the bacterial GAP of Yersinia (YopE) and Salmonella (SptP) spp.: Ala-139, Gly-141, Gly-143, Leu-145, and Thr-150 (38). These residues were conserved in the clinical isolates sequenced. The consensus leucine between eukaryotic and bacterial GAP (Leu-171) and the catalytic glutamate of the ADP-ribosyltransferase domain (Glu-381) were also conserved.
The last 27 amino acids of ExoS form the binding site for the eukaryotic protein FAS (factor for activating ExoS), a member of the 14-3-3 family, which is necessary for the ADP-ribosylating activity of both ExoT (39) and ExoS (4, 15). Deletion of this binding site results in a protein that is unable to efficiently inactivate Ras and displays reduced lethal activity (24). There was a single amino acid substitution in this region in the ExoS sequence of one (10%) of the clinical isolates sequenced (Fig. 2), but this single point mutation (residue 443) was not sufficient to prevent infection by this strain.
The N terminus of ExoT contains an ADP-ribosyltransferase that has only 0.2 to 1% of the catalytic activity of ExoS in vitro (29, 39). It also has GAP activity for Rho GTPases at its C terminus and inhibits bacterial internalization by eukaryotic cells (5, 18, 26). Although the 9-amino-acid signal sequence (amino acids 1 to 9) was conserved, both the region of GAP activity (amino acids 78 to 237) (26) and ADP-ribosyltransferase activity (amino acids 235 to 457) (29) contained protein sequence-altering SNPs (Fig. 3).
ExoU, the largest of the toxin genes, was the most highly conserved on both the nucleic acid and amino acid levels (Table 4). When compared to the reference strain PA103, the SNPs were scattered throughout the ExoU gene, but there were no changes in the 359-amino-acid N-terminal sequence of the protein (Fig. 4). Furthermore, the five amino acid sequence alterations found in the C-terminal of the protein occurred in only 16% of the strains sequenced.
TABLE 4.
Deletion mutations found in exoY of P. aeruginosa isolates
| Isolate | Nucleotide position(s) | Deletion length (bp) | Translational result |
|---|---|---|---|
| PA103 | 140-150 | 11 | The protein was truncated at amino acid 63 |
| 2038 | 727 | 1 | The protein was truncated at amino acid 249 |
| 2002 | 1121 | 1 | The last five amino acids were altered |
The only gene with a deletion in the clinical strains studied was exoY (Table 4). It is an adenylate cyclase that elevates the intracellular cyclic AMP levels in eukaryotic cells and causes rounding of certain cell types (36, 40). Although PA103 contains this gene, it does not secrete the protein; thus, this strain is not included in the analysis of nucleotide or amino acid sequence variability. Figure 5 shows the position of the SNPs and deletions in exoY. There was an 11-bp deletion in PA103 at positions 140 to 150. There was also a single-base-pair deletion in strain 2038 at position 727 and in strain 2002 at position 1121. These deletions resulted in a frameshift mutation and a predicted protein product truncated at amino acid positions 63 and 249 for strains PA103 and 2038, respectively. The deletion in strain 2002 resulted in an alteration in the last five amino acids. This alteration was not sufficient to prevent infection by this strain.
FIG. 5.
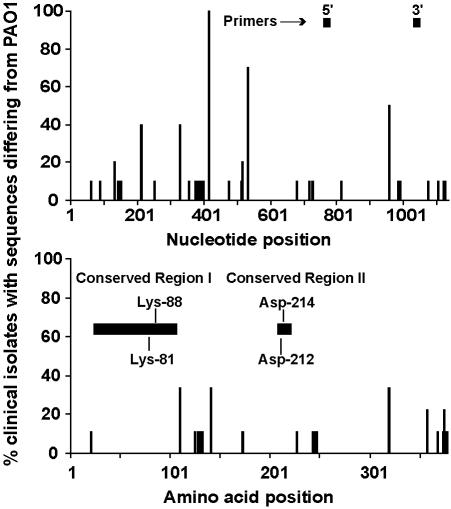
Position and frequency of exoY DNA and amino acid sequence variations (n = 9) and position of PCR primers and conserved regions between P. aeruginosa, B. pertussis, and B. anthracis adenylate cyclases. The x axis represents the nucleotide or amino acid position; the y axis represents the percentage of P. aeruginosa isolates that differed in sequence from PAO1 at each position. Two lysine residues (Lys-81 and Lys-88) in conserved region I and two arginine residues (Arg-212 and Arg-214) in conserved region II are essential for the adenylate cyclase activity of ExoY.
Two areas of homology exist between ExoY and the calmodulin-activated adenylate cyclases of Bordetella pertussis (CyaA) and Bacillus anthracis (edema factor) (6, 30, 40). In ExoY, conserved region I (amino acids 41 to 107) contains an ATP/GTP-binding site A motif, which is thought to play a role in contacting the α-phosphate of bound nucleotide (19). Conserved region II (amino acids 209 to 221) is proposed to participate in contacting the β- and γ-phosphates of bound nucleotide (19). Four residues are essential for the adenylate cyclase activity of ExoY. Two in conserved region I (Lys-81 and Lys-88) are predicted to be directly involved in ATP binding. Two in conserved region II (Asp-212 and Asp-214) are thought to play a role in contacting bound nucleotide (19). These residues were conserved in the clinical isolates sequenced (Fig. 5).
Purified DNA from eight strains of P. aeruginosa previously classified as either cytotoxic (PA103, 6206, 6073, and 19660) or noncytotoxic and invasive (PAO1, PAK, 6294, and 6487) (11, 12) were used in the multiplex PCR assay (Fig. 1). All of the strains tested have exoT. Also, all of the strains except strain 19660 have exoY. Four of the strains (PA103, 6206, 19960, and 6073) have exoU but not exoS, and the remainder had exoS but not exoU (PAO1, 6294, PAK, and 6487). The accuracy of the amplification of each gene fragment was checked against the results of Southern blot analysis of exoS, exoT, and exoU previously published, in collaboration, by members of our lab (33). The results of the multiplex system completely matched the results obtained by Southern blot. Furthermore, when the multiplex PCR was repeated four times with the same samples, the patterns remained the same. These results indicate that this multiplex system is both accurate and reproducible.
DISCUSSION
The aim of the present study was to develop a simple, rapid, and reliable technique for the detection of the genes of four effector proteins of the P. aeruginosa TTSS. As the evidence for the importance of these toxins accumulates, the use of a less cumbersome alternative to Southern blot for the detection of these genes will become increasingly desirable. Rapid identification of the bacterial genotype would greatly aid clinical studies correlating genes and gene expression with clinical outcome. The first step was to sequence the genes of a reasonable but arbitrary number of clinical strains to determine the sequence variability of each gene. This step was necessary in order to design primers in conserved regions of the gene. This placement greatly increases the likelihood that the primers will successfully amplify the sequence of any isolate containing that gene. The data presented here indicate that this multiplex PCR technique is very sensitive and specific, providing results within 3 h. Strategies to improve the sensitivity of this assay include sequencing more clinical strains to possibly capture more genetic variations. This would increase our confidence that the primers designed are in conserved regions. Also, we could use multiple sets of primers for each toxin gene. With this strategy, we would be more likely to detect the gene even if one set of primers were inadvertently in a polymorphic region.
The relationship between the genotype and phenotype differs for each toxin gene. One study looking at P. aeruginosa isolated from the lower respiratory tracts of patients with ventilator-associated pneumonia found that, although every isolate examined harbored TTS genes, only 77% were capable of secreting detectable amounts of TTS proteins in vitro (21). The genotype and phenotype matched in 94% of the isolates for exoU but in only 80% of the isolates for exoS. ExoU, in particular, has been associated with cell death in tissue culture systems and more severe disease in animal models of acute pneumonia (11, 22, 27). In the clinical setting, ExoU-secreting isolates more frequently caused severe disease than did isolates not secreting TTS toxins. Others have reported that the vast majority of P. aeruginosa isolates contain either exoS or exoU but not both (9, 11, 12). This was also seen in our small sample, and the multiplex PCR was able to accurately distinguish the genotypes. Also, despite having 75% amino acid sequence homology, we were able to consistently differentiate exoS from exoT by this PCR method (Fig. 1). This is especially important since exoT is almost ubiquitous in the P. aeruginosa genome (9). PA103 is phenotype negative for exoY (40). We found a deletion mutation in this isolate, as well as two others, which produced an early stop codon and, therefore, a truncated protein product. This accounts for the discrepancy between the exoY genotype and phenotype seen in PA103.
Given the success of this multiplex system, we would be able to use the same strategy to develop a genotyping system for other virulence factors of the TTSS. Because an association between the expression of TTS proteins and morbidity and mortality in patients infected with P. aeruginosa has been described, the identification of the TTSS phenotype in clinical isolates may be useful in the determination of a patient's prognosis or help distinguish colonization from infection. In the future, we may be able to combine this simple genotyping system with an equally elegant phenotyping system, such as real-time PCR or microarray, to develop a system to fully characterize P. aeruginosa strains.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants HL59239 and AI44101 to J.P.W.-K., NIH grant HL07185 to T.A., and NIH grant HL67600 and American Lung Association RG004N grant to T.S.
REFERENCES
- 1.Allewelt, M., F. T. Coleman, M. Grout, G. P. Priebe, and G. B. Pier. 2000. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1996. National Nosocomial Infections Surveillance System surveillance report: data summary from October 1986 to April 1996. Am. J. Infect. Control 24:380-388. [PubMed] [Google Scholar]
- 3.Bodey, G., V. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 4.Coburn, J., A. V. Kane, L. Feig, and D. M. Gill. 1991. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J. Biol. Chem. 266:6438-6446. [PubMed] [Google Scholar]
- 5.Cowell, B. A., D. Y. Chen, D. W. Frank, A. J. Vallis, and S. M. J. Fleiszig. 2000. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect. Immun. 68:403-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escuyer, V., E. Duflot, O. Sezer, A. Danchin, and M. Mock. 1988. Structural homology between virulence-associated bacterial adenylate cyclases. Gene 71:293-298. [DOI] [PubMed] [Google Scholar]
- 7.Fagon, J. Y., J. Chastre, Y. Domart, J. L. Trouillet, J. Pierre, C. Carne, and C. Gilbert. 1989. Nosocomial pneumonia in patients receiving continuous mechanical ventilation. Am. Rev. Respir. Dis. 139:877-884. [DOI] [PubMed] [Google Scholar]
- 8.Fagon, J. Y., J. Chastre, A. Hance, P. Montravers, A. Novara, and C. Gibert. 1993. Nosocomial pneumonia in ventilated patients: a cohort study evaluating attributable mortality and hospital stay. Am. J. Med. 94:281-288. [DOI] [PubMed] [Google Scholar]
- 9.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson, M. W., J. A. Maxwell, T. S. Vincent, J. DaSilva, and J. C. Olson. 2001. Comparison of the exoS gene and protein expression in soil and clinical isolates of Pseudomonas aeruginosa. Infect. Immun. 69:2198-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. W. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 12.Fleiszig, S. M. J., J. P. Wiener-Kronish, H. Miyazaki, V. Vallas, K. E. Mostov, D. Kanada, T. Sawa, T. S. B. Yen, and D. W. Frank. 1997. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 65:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 14.Frithz-Lindsten, E., Y. Du, R. Rosqvist, and A. Forsberg. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aerugniosa via type III dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol. Microbiol. 25:1125-1139. [DOI] [PubMed] [Google Scholar]
- 15.Fu, H., J. Coburn, and R. J. Collier. 1993. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 90:2320-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 17.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 18.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. R. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser, P., H. Munier, A. M. Gilles, E. Krin, T. Porumb, O. Barzu, R. Sarfati, C. Pellecuer, and A. Danchin. 1991. Functional consequences of single amino acid substitutions in calmodulin-activated adenylate cyclase of Bordetella pertussis. EMBO J. 10:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goehring, U. M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 21.Hauser, A. R., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 22.Hauser, A. R., P. Kang, and J. N. Engel. 1998. PepA, a novel secreted protein of Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 23.Henriksson, M. L., R. Rosqvist, M. Telepnev, H. Wolf-Watz, and B. Hallberg. 2000. Ras effector pathway activation by epidermal growth factor is inhibited in vivo by exoenzyme S ADP-ribosylation of Ras. Biochem. J. 347:217-222. [PMC free article] [PubMed] [Google Scholar]
- 24.Henriksson, M. L., U. Troller, and B. Hallberg. 2000. 14-3-3 proteins are required for the inhibition of Ras by exoenzyme S. Biochem. J. 349:697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hueck, C. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a Rho GTP-ase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 104:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S., S. M. Kulich, and J. T. Barbieri. 1996. Identification of glutamic acid 381 as a candidate active site residue of Pseudomonas aeruginosa exoenzyme S. Biochemistry 35:2751-2758. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S., T. L. Yahr, D. W. Frank, and J. T. Barbieri. 1997. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J. Bacteriol. 179:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mock, M., E. Labruyere, P. Glaser, A. Danchin, and A. Ullmann. 1988. Cloning and expression of the calmodulin-sensitive Bacillus anthracis adenylate cyclase in Escherichia coli. Gene 64:277-284. [DOI] [PubMed] [Google Scholar]
- 31.Pederson, K. J., and J. T. Barbieri. 1998. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30:751-759. [DOI] [PubMed] [Google Scholar]
- 32.Pennington, J. E. 1995. Nosocomial respiratory infections, p. 2599-2607. In G. Mandell, J. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingtone, New York, N.Y.
- 33.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 34.Salyers, A. A., and D. D. Whitt. 2002. Bacterial pathogenesis: a molecular approach, p. 251-259. American Society for Microbiology Press, Washington, D.C.
- 35.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 36.Vallis, A. J., V. Fink-Barbancon, T. L. Yahr, and D. W. Frank. 1999. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect. Immun. 67:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. 1999. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol. 32:1054-1064. [DOI] [PubMed] [Google Scholar]
- 38.Wurtele, M., E. Wolf, K. J. Pederson, G. Buchwald, M. R. Ahmadian, J. T. Barbieri, and A. Wittinghofer. 2001. How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat. Struct. Biol. 8:23-26. [DOI] [PubMed] [Google Scholar]
- 39.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, B., Y. Zhang, C. C. Collins, D. I. Johnson, and Y. Zheng. 1999. A built-in arginine finger triggers the self-stimulatory GTPase-activating activity of Rho family GTPases. J. Biol. Chem. 274:2609-2612. [DOI] [PubMed] [Google Scholar]