Abstract
Twelve clonally related and multidrug-resistant Acinetobacter baumannii isolates were recovered during a 4-month period from 12 patients hospitalized at the Valenciennes Hospital in France. Antibiograms determined by the double-disk diffusion technique on cloxacillin-containing plates detected a clavulanic acid-inhibited extended-spectrum β-lactamase (ESBL). PCR and sequencing identified the gene encoding the Ambler class A ESBL VEB-1. This gene was located on the chromosome and was part of a class 1 integron identical to that previously identified in Pseudomonas aeruginosa isolates from Thailand. Additionally, seven clonally related blaVEB-1-positive A. baumannii strains were identified in the immediate environment of the hospitalized patients. This is the first report of the ESBL VEB-1 in Acinetobacter spp. and the first description of VEB-1-producing strains as a source of an outbreak occurring outside Southeast Asia. This report underlines the difficulty of the identification of ESBLs in A. baumannii.
Acinetobacter baumannii is an opportunistic pathogen involved in outbreaks occurring in intensive care units (ICUs) (2, 14). It is an important source of nosocomial septicemia, pneumonia, and urinary tract infections (4). Reports of multidrug-resistant isolates have increased during the last decade, probably as a result of the extensive use of broad-spectrum antibiotics (1). In many cases, these multidrug-resistant isolates are resistant to expanded-spectrum cephalosporins and carbapenems (1, 2, 28). Recent studies report that carbapenem-hydrolyzing β-lactamases of Ambler class B (metalloenzymes) and Ambler class D (oxacillinases) are sources of multidrug resistance in A. baumannii (3, 5, 24, 27, 30). However, most of the expanded-spectrum β-lactamases of gram-negative organisms are the clavulanic acid-inhibited extended-spectrum β-lactamases (ESBLs) of Ambler class A that have been reported extensively in members of the family Enterobacteriaceae (19). They usually confer resistance to cefotaxime, ceftriaxone, ceftazidime, and the monobactam aztreonam but do not confer resistance to imipenem (19).
Most of these ESBLs that are disseminated worldwide are structurally related to the narrow-spectrum TEM- and SHV-type β-lactamases, but none of them has been detected so far in A. baumannii (19).The non-TEM, non-SHV derivative ESBL PER-1 is the only known ESBL in A. baumannii and has been detected in Turkish and French isolates (21, 31, 32). An epidemiological survey performed in Turkey in 1996 identified the spread of PER-1-positive A. baumannii isolates (32), and infections with these PER-1-positive isolates have been associated with a higher risk of mortality (31).
Another non-TEM, non-SHV ESBL, VEB-1, has been detected in Enterobacteriaceae and Pseudomonas aeruginosa strains from Southeast Asia (7, 8, 15-17, 23). Unlike most of the ESBL genes, blaVEB-1 is part of a gene cassette and is located in class 1 integrons of various structures (7, 8, 15-17, 23). Integrons are genetic structures responsible for the expression of cassette-associated and mobile resistance genes (25).
The aim of the present study was to analyze the molecular mechanisms involved in the β-lactam resistance of multidrug-resistant and nosocomial A. baumannii isolates. This study identified for the first time the ESBL VEB-1 in A. baumannii. It shows also that (i) ESBL-positive A. baumannii isolates may be responsible for nosocomial outbreaks, (ii) ESBLs may contribute to the multidrug resistance of A. baumannii isolates, and (iii) that ESBL production may remain undetected in this species.
(This work was previously presented in part at the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, 2002 [O. Menuteau, L. Poirel, N. Agoli, C. Cattoen, and P. Nordmann, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1876, 2002].)
MATERIALS AND METHODS
Bacterial isolates.
A. baumannii clinical and environmental isolates (strains 1 to 20) were identified by using the API 32GN system (bioMérieux SA, Marcy l'Etoile, France). Electrocompetent Escherichia coli DH10B (GIBCO BRL, Life Technologies, Cergy Pontoise, France) was used as the recipient strain in transformation experiments. Rifampin-resistant E. coli HB101 was used as the host in conjugation experiments (23). E. coli MG-1, which carries the blaVEB-1 gene, was used as a VEB-1-producing reference strain (23). E. coli K-12 DNA, which generates seven I-CeuI restricted fragments of 2,460, 700, 670, 530, 130, 92, and 44 kb, was used as a size marker in pulsed-field gel electrophoresis (PFGE) experiments. Reference strain A. baumannii CIP7034T (Institut Pasteur Strain Collection, Paris, France) was used as a nonclonally related strain in the PFGE experiments, and an in vitro-obtained rifampin-resistant derivative was used in conjugation experiments.
Susceptibility testing and screening for ESBL-producing strains.
The antibiotic susceptibilities of the A. baumannii isolates were first determined by the disk diffusion method on Mueller-Hinton (MH) agar plates with antibiotic-containing disks (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France), and the results were interpreted according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (www.sfm.fr). The double-disk synergy test was performed with cefepime, ceftazidime, and ticarcillin-clavulanic acid disks on MH agar plates; and the results were interpreted as described previously (10). To counteract the effect of high-level expression of the naturally produced AmpC-type β-lactamase of A. baumannii, double-disk synergy tests were also performed on cloxacillin (200 μg/ml)-containing plates (6).
The MICs of the β-lactams were determined by an agar dilution technique on MH agar plates with or without cloxacillin (200 μg/ml) with an inoculum of 104 CFU per spot, as described previously (20). MIC results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (18).
IEF analysis.
Cultures of A. baumannii isolates were grown overnight at 37°C in 10 ml of Trypticase soy (TS) broth. Analytical isoelectric focusing (IEF) was performed with an ampholine polyacrylamide gel, as described previously (20).
PCR-based amplification of β-lactamase genes and class 1 integrons and sequencing.
Under standard PCR conditions (26), a series of primers was used for detection of Ambler class A β-lactamase genes. Detection of genes coding for β-lactamases TEM, SHV, PER-1 or PER-2, VEB-1, and GES-1 was performed as described previously (7, 8). Detection of an ampC-type gene of A. baumannii was performed with primers preAB-1 (5′-ACAGAGGAGCTAATCATGCG-3′) and preAB-2 (5′-GTTCTTTTAAACCATATACC-3′), which hybridized to the internal part of this β-lactamase gene. For each reaction, 0.5 μg of whole-cell DNA of the A. baumannii isolates was used. The primers used for detection of class 1 integrons (primers 5′-CS and 3′-CS [12]) hybridized to sequences located in the 5′ and 3′ conserved segments. Combinations of primer 5′-CS or 3′-CS and blaVEB-1-specific primers (primers 5′-CS and VEB-B or primers 3′-CS and VEB-F [15, 17]) were also used for determination of the genetic contents of the class 1 integrons. Additionally, since the oxa-10 and arr-2 genes had been associated with the blaVEB-1 gene in E. coli MG-1 and P. aeruginosa isolates (7, 16), their positions relative to that of the blaVEB-1 gene were determined by PCR with primers specific for blaVEB-1 (primers VEB-1A and VEB-1B and primers VEB-INV4F and VEB-INFV3B), for blaOXA-10 (primers OXA-10promB, OPR-1, and OPR-2), and for arr-2 (primers ARR-2-F and ARR-2-B) (7, 8). Finally, since a blaVEB-1-containing integron had been reported to be bracketed by two IS26 insertion sequences in E. coli MG-1 (15), PCRs with blaVEB-1-specific primers and IS26-specific primers were also performed (16). Sequencing reactions were performed with the same blaVEB-1-specific primers and an automated sequencer (ABI 377; Applied Biosystems, Foster City, Calif.). The nucleotide and deduced amino acid sequences were analyzed with software available over the Internet (22, 23).
PFGE.
PFGE analysis was done according to the instructions of the manufacturer (Bio-Rad). In brief, whole-cell DNA of the A. baumannii isolates was digested with the ApaI restriction enzyme overnight at 25°C (9). Electrophoresis was performed with a CHEF DRII apparatus (Bio-Rad) through a 1% agarose gel in 0.5× Tris-borate-EDTA buffer. Migration conditions were as follows: temperature, 14°C; voltage, 5 V/cm; and switch angle, 120°, with one linear switch ramp of 5 to 20 s for 24 h. The ethidium bromide-stained gel was photographed under UV illumination. A bacteriophage λ DNA ladder (Bio-Rad) was used as a DNA molecular weight marker. The chromosomal fingerprints were compared by eye and assigned to PFGE types and subtypes (29).
Hybridizations.
The I-CeuI restriction enzyme (Ozyme; New England Biolabs), which digests a 26-bp sequence in the rrn genes for the 23S large-subunit rRNA, was used to search the chromosome to determine whether the β-lactamase gene has a chromosomal location (13); and the fragments were separated by PFGE. The sizes of the I-CeuI-generated fragments of A. baumannii clinical isolates AYE and 13 and of reference strain CIP7034T were determined by comparison with those of E. coli K-12. After Southern transfer onto a nylon membrane (Hybon N+; Amersham Pharmacia Biotech, Orsay, France) (26), the DNAs were UV cross-linked (Stratalinker; Stratagene) and hybridized successively with two probes: a 1,504-bp PCR-generated probe specific for the 16S and 23S rRNA genes (8) and a 650-bp probe specific for blaVEB-1 (23). Labeling and signal detection were carried out according to the instructions of the manufacturer (Amersham Pharmacia Biotech).
Conjugation, electroporation, and plasmid DNA analysis.
Conjugation experiments were performed between A. baumannii clinical isolate AYE and E. coli HB101, which were resistant to rifampin, and in vitro-obtained rifampin-resistant A. baumannii CIP7034T in solid and liquid media at 37°C, as reported previously (20). Transconjugants were selected on TS agar plates containing 300 μg of rifampin per ml and 150 μg of ticarcillin per ml. The plasmid DNA of the A. baumannii isolates was extracted as described previously (7, 22). Plasmid extracts were electroporated into E. coli DH10B, and recombinant strains were selected on ceftazidime (2 μg/ml)-containing TS agar plates.
RESULTS
Preliminary PCR detection of β-lactamase gene.
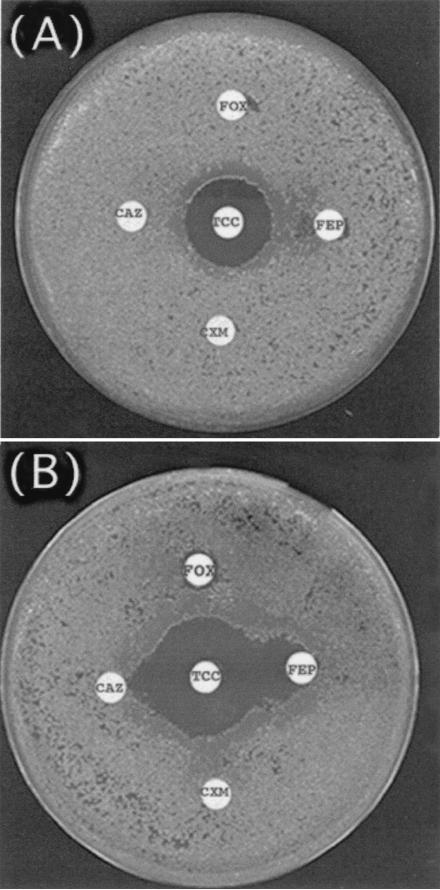
A multidrug-resistant A. baumannii isolate, isolate AYE, was recovered from a patient with a urinary tract infection. The patient was a 61-year-old man who was hospitalized at the Bicêtre Hospital (Le Kremlin-Bicêtre, France) in October 2001. He had been transferred directly from the ICU of the Valenciennes Hospital (a 1,000-bed facility in the north part of France), where he had been hospitalized for pneumonia and where an antibiotic regimen containing imipenem and gentamicin had been given. Preliminary antibiotic susceptibility testing by disk diffusion showed that A. baumannii isolate AYE was resistant to all β-lactams except imipenem, piperacillin-tazobactam, and ticarcillin-clavulanate. It was resistant to other antibiotics such as fluoroquinolones, aminoglycosides, chloramphenicol, and tetracycline and was of intermediate susceptibility to rifampin (data not shown). A synergy between ceftazidime- or cefepime- and clavulanate-containing disks was evidenced only when cloxacillin-containing agar plates were used (Fig. 1). PCR experiments with primers specific for blaTEM, blaSHV, blaVEB-1, blaGES-1, blaPER-1, and blaCTX-M-1 were performed and gave a positive result for a blaVEB-1-like gene. Sequence analysis of this blaVEB-1-like gene revealed a 100% identity of its DNA with that of blaVEB-1. In addition and as expected, PCR with primers located at the ends of the ampC-type β-lactamase gene of A. baumannii gave a 1,242-bp DNA fragment, indicating the presence of this naturally occurring β-lactamase gene in A. baumannii AYE.
FIG. 1.
Double-disk synergy test with blaVEB-1-positive A. baumannii strain AYE. MH agar plates contained either no cloxacillin (A) or 200 μg of cloxacillin per ml (B). The disks tested contained ceftazidime (CAZ), cefuroxime (CXM), ticarcillin-clavulanic acid (TCC), cefoxitin (FOX), and cefepime (FEP).
MICs of β-lactams and IEF analysis.
The MICs of ceftazidime and cefotaxime for A. baumannii AYE were lowered when clavulanic acid was added, which is consistent with expression of the ESBL VEB-1 (Table 1). The contribution of VEB-1 to resistance to expanded-spectrum cephalosporins was demonstrated by using cloxacillin-containing MH agar plates (Table 1).
TABLE 1.
MICs of β-lactams for A. baumannii AYE determined on MH agar and cloxacillin-containing MH agar plates
| β-Lactam(s)a | MIC (μg/ml) on:
|
|
|---|---|---|
| MH agar plates | Cloxacillin-containing MH agar plates | |
| Amoxicillin | >512 | >512 |
| Amoxicillin + CLA | >512 | 8 |
| Ticarcillin | >512 | >512 |
| Ticarcillin + CLA | 32 | 16 |
| Piperacillin | 256 | 64 |
| Piperacillin + TZB | 8 | 0.06 |
| Cephalothin | >512 | >512 |
| Cefuroxime | >512 | 256 |
| Cefotaxime | 512 | 256 |
| Cefotaxime + CLA | 64 | 16 |
| Ceftazidime | >512 | >512 |
| Ceftazidime + CLA | 64 | 16 |
| Cefoxitin | >512 | 64 |
| Cefepime | 512 | 64 |
| Cefepime + CLA | 256 | 4 |
| Cefpirome | 512 | 256 |
| Cefpirome + CLA | 256 | 16 |
| Moxalactam | 64 | 64 |
| Aztreonam | >512 | >512 |
| Imipenem | 1 | 0.5 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
IEF analysis showed that A. baumannii AYE expressed several β-lactamases with pI values of 6.3, 7.4, and 8.5. The pI value of 6.3 likely corresponded to that of OXA-10 (7, 8), and the pI value of 7.4 corresponded to that of VEB-1, whereas the pI value of 8.5 corresponded to that of an AmpC-type cephalosporinase (3).
Identification of blaVEB-1-positive integron.
Fragments were obtained by PCR with primers 5′-CS and 3′-CS for detection of class 1 integrons and blaVEB-1-specific primers, indicating that blaVEB-1 is part of a class 1 integron. Taking into account the sizes of the amplified fragments, the structure of the integron of A. baumannii AYE was identical to that found in most of the P. aeruginosa isolates from Thailand, which also contained blaOXA-10 and rifampin resistance-encoding arr-2 genes (Fig. 2) (7). As in most of the Thai P. aeruginosa isolates (7), insertion sequence IS1999 was also located inside the class 1 integron structure, interrupting the integron recombination site attI. A PCR experiment also identified an IS26-like element upstream of the blaVEB-1-positive integron in A. baumannii AYE.
FIG. 2.
Schematic representation of the veb-1 gene cassette-containing integron found in A. baumannii AYE. Gene cassettes are shown as boxes, with arrows indicating the orientation of transcription and black circles indicating the 59-base element. The 5′ conserved segment contains the intI1 gene, which encodes integrase; and the 3′ conserved segment, downstream of the gene cassettes, includes the qacEΔ1 gene, an antiseptic resistance determinant. The IS1999 inverted repeats are indicated by filled and empty triangles.
Genetic location of blaVEB-1 gene.
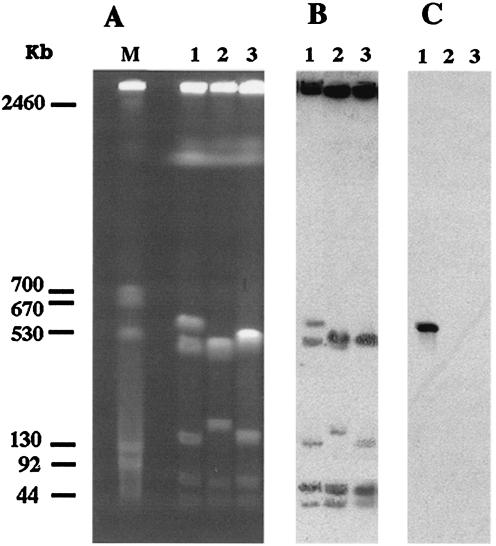
Transfer of the ceftazidime resistance marker by conjugation failed with the E. coli and A. baumannii recipient strains. Although no plasmid was detected in A. baumannii AYE, a suspension containing putative plasmid DNA of A. baumannii AYE was used to electroporate E. coli DH10B, but no ampicillin-resistant transformant was obtained. The location of blaVEB-1 was then determined by using the technique with the endonuclease I-CeuI. Five DNA fragments (2,200, 500, 150, 60, and 40 kb) were generated from the A. baumannii CIP7034T reference strain, whereas six fragments (the five fragments mentioned above plus a 600-kb fragment) were obtained from A. baumannii AYE (Fig. 3A). The probe for the rRNA genes hybridized to all DNA fragments except the 2,200-kb fragment for both A. baumannii strains (Fig. 3B). Hybridization of restricted DNA of A. baumannii AYE with the blaVEB-1-specific probe gave a single signal corresponding to the additional 600-kb fragment, indicating a chromosomal location of blaVEB-1 (Fig. 3C).
FIG. 3.
(A) PFGE profiles of I-CeuI-digested whole-cell DNA of three A. baumannii strains. Lanes 1, A. baumannii AYE (blaVEB-1 positive); lanes 2, A. baumannii isolate 13 (blaVEB-1 negative); lanes 3, A. baumannii reference strain CIP7034T (blaVEB-1 negative); lane M, bacteriophage lambda DNA ladder. Southern hybridization was done with a 16S-23S rRNA gene-specific probe (B) and a blaVEB-1-specific internal probe (C).
Retrospective epidemiological survey.
No other multidrug-resistant A. baumannii isolate was recovered in the ICU of Bicêtre Hospital when A. baumannii AYE was isolated. Thus, a retrospective survey was performed at the ICU of the hospital where the patient came from. Twelve additional multidrug-resistant A. baumannii isolates from 12 patients were recovered during a 4-month period that surrounded the date of isolation of A. baumannii AYE (isolates 1, 2, and 4 to 12 [Table 2] and isolate 13). Most of them suffered from pneumonia. In addition, systematic screening performed during this period gave seven additional multidrug-resistant A. baumannii isolates (isolates 14 to 20). These isolates were recovered from ventilators in several ICU rooms (data not shown). PCR with blaVEB-1-specific primers and sequencing of the PCR products revealed that all but one isolate (A. baumannii strain 13) possessed a blaVEB-1 gene (data not shown). A similar blaVEB-1-positive class 1 integron was identified by PCR analysis with whole-cell DNA of each blaVEB-1-positive strain as the template. The blaVEB-1-positive isolates had the same multiple antibiotic resistance patterns as strain AYE (data not shown).
TABLE 2.
Clinical features of the blaVEB-1-positive A. baumannii isolates
| Isolate | Date of isolation (mo-day-yr) | Dates of hospitalization (mo-day-yr) | Source | Underlying disease | Treatment |
|---|---|---|---|---|---|
| 1 | 07-01-01 | 06-08-01 to 07-04-01 | Blood | Sepsis | Cefotaxime, ciprofloxacin |
| 2 | 08-24-01 | 08-07-01 to 09-12-01 | Blood | Pneumonia | Ceftriaxone, ofloxacin |
| 3 (AYE) | 10-02-01 | 09-14-01 to 09-25-01 | Urine | Pneumonia | Imipenem, gentamicin |
| 4 | 10-10-01 | 09-14-01 to 10-14-01 | Blood | Intestinal obstruction | Amoxicillin-CLA,a ofloxacin |
| 5 | 10-10-01 | 09-28-01 to 10-15-01 | Tracheal aspirate | Pneumonia | Imipenem, teicoplanin |
| 6 | 10-11-01 | 10-01-01 to 10-18-01 | Tracheal aspiration | Sepsis | Piperacillin-tazobactam |
| 7 | 10-15-01 | 09-24-01 to 11-02-01 | Urine | Pneumonia | Amoxillin-CLA, ofloxacin |
| 8 | 10-15-01 | 10-06-01 to 10-15-01 | Catheter | Pneumonia | Amoxicillin-CLA, ofloxacin |
| 9 | 10-15-01 | 08-01-01 to 03-01-02 | Tracheal aspirate | Guillain-Barrè syndrome | Ceftazidime, amikacin |
| 10 | 10-15-01 | 10-02-01 to 10-22-01 | Urine | Pneumonia | Amoxicillin-CLA, ofloxacin |
| 11 | 10-22-01 | 09-16-01 to 10-28-01 | Peritonitis fluid | Pneumonia | Imipenem |
| 12 | 10-26-01 | 10-04-01 to 10-31-01 | Rectal swab | Pneumonia | Teicoplanin, amikacin |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml.
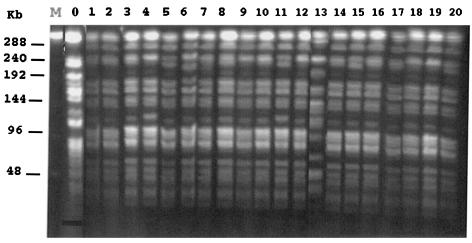
The PFGE profiles of ApaI-restricted DNA of all A. baumannii isolates except isolate 13 and the reference strain were identical (Fig. 4). An additional 220-kb fragment was identified in five isolates (strains 5, 6, 11, 15, and 20). Thus, all blaVEB-1-positive isolates seemed to be clonally related. They had the same antibiotic resistance pattern, according to disk diffusion susceptibility testing (data not shown).
FIG. 4.
PFGE profiles of ApaI-digested whole-cell DNA of 21 A. baumannii isolates. Lane 0, A. baumannii reference strain CIP7034T; lanes 1 to 20, A. baumannii isolates 1 to 20 (clinical isolates 1 to 13 and environmental isolates 14 to 20, respectively). Clinical isolate 3 is also referred as AYE throughout the text. Lane M, bacteriophage lambda DNA ladder.
DISCUSSION
This work describes multidrug-resistant A. baumannii isolates as the source of an outbreak in an ICU. Production of an ESBL was demonstrated, whereas dissemination of resistant isolates had already occurred. A 2-week closure of the ICU was the only way to control the outbreak, as demonstrated previously (11).
The clonally related A. baumannii isolates produced an unusual Ambler class A ESBL, VEB-1, whereas this enzyme has been identified extensively from members of the family Enterobacteriaceae and P. aeruginosa isolates in Southeast Asia (7, 8, 15-17, 23). Thus, this work identified this ESBL in A. baumannii isolates for the first time (and is the second ESBL found in A. baumannii) and represents the first report of an outbreak due to VEB-1-positive gram-negative organisms outside Southeast Asia.
Interestingly, this ESBL gene was identified in a class 1 integron of A. baumannii that was encoded by the chromosome, as reported for most of the blaVEB-1-positive P. aeruginosa isolates (7). The structure of this blaVEB-1-containing integron was identical to that found in most of the P. aeruginosa isolates in a study performed in Thailand (7). The locations of other antibiotic resistance genes in the same integron may contribute to the multidrug resistance of A. baumannii. Interestingly, this blaVEB-1-containing integron was bracketed, at least upstream, by insertion sequence IS26, as was found for the blaVEB-1-positive integron in E. coli MG-1 (16). Thus, the chromosomal location of blaVEB-1 in A. baumannii may result from a transposition event.
Analysis of this outbreak due to ESBL-producing A. baumannii strains raises the threat of endemicity of multidrug-resistant A. baumannii isolates, as in Turkey (32). Indeed, a nation-based study performed in that country found that 46% of A. baumannii strains produce another ESBL, PER-1 (32).
The spread of ESBL-producing A. baumannii isolates may be enhanced by underdetection and underreporting. We show here that the use of cloxacillin-containing plates that inhibit cephalosporinase activity may enhance the ability to detect these organisms in a routine laboratory. Those plates should be used in investigations of outbreaks due to multidrug-resistant A. baumannii isolates. Such investigations should be performed, since outbreaks due to A. baumannii are usually more difficult to control than those due to members of the family Enterobacteriaceae (2, 4). Indeed, the reservoirs of A. bauman-nii are difficult to identify and control in hospital settings, since this bacterial species may survive on dry surfaces for a long time (2).
Acknowledgments
This work was financed by a grant (grant UPRES-EA3539) from the Ministère de l'Education Nationale et de la Recherche, Université Paris XI, Paris, France.
We thank A. Karim for the identification of the ESBL-positive A. baumannii isolate.
REFERENCES
- 1.Amyes, S. G. B., and H.-K. Young. 1996. Mechanisms of antibiotic resistance in Acinetobacter spp.—genetics of resistance, p. 185-223. In E. Bergogne-Berezin, M. L. Jolly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, Inc., New York, N.Y.
- 2.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cisneros, J. M., and J. Rodriguez-Bano. 2002. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin. Microbiol. Infect. 8:687-693. [DOI] [PubMed] [Google Scholar]
- 5.Corbella, X., A. Montero, M. Pujol, M. A. Dominguez, J. Ayats, M. J. Argerich, F. Garrigosa, J. Ariza, and F. Gudiol. 2000. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J. Clin. Microbiol. 38:4086-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Champs, C., L. Poirel, R. Bonnet, D. Sirot, C. Chanal, J. Sirot, and P. Nordmann. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob. Agents Chemother. 46:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girlich, D., T. Naas, A. Leelaporn, L. Poirel, M. Fennewald, and P. Nordmann. 2002. Nosocomial spread of the integron-located veb-1-like cassette encoding an extended-spectrum β-lactamase in Pseudomonas aeruginosa in Thailand. Clin. Infect. Dis. 34:603-611. [DOI] [PubMed] [Google Scholar]
- 8.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann. 2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum β-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouby, A., M. J. Carles-Nurit, N. Bouziges, G. Bourg, R. Mesnard, and P. J. M. Bouvet. 1992. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acineobacter baumannii. J. Clin. Microbiol. 30:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarlier, V., M.-H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 11.Koeleman, J. G., M. W. Van Der Bijl, J. Stoof, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2001. Antibiotic resistance is a major risk factor for epidemic behavior of Acinetobacter baumannii. Infect. Control Hosp. Epidemiol. 22:284-288. [DOI] [PubMed] [Google Scholar]
- 12.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mah, M. W., Z. A. Memish, G. Cunningham, and R. M. Bannatyne. 2001. Outbreak of Acinetobacter baumannii in an intensive care unit associated with tracheostomy. Am. J. Infect. Control 29:301-305. [DOI] [PubMed] [Google Scholar]
- 15.Naas, T., F. Benaoudia, S. Massuard, and P. Nordmann. 2000. Integron-located VEB-1 extended-spectrum beta-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46:703-711. [DOI] [PubMed] [Google Scholar]
- 16.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:225-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas, T., L. Poirel, A. Karim, and P. Nordmann. 1999. Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176:411-419. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Nordmann, P. 1998. Trends in β-lactam resistance among Enterobacteriaceae. Clin. Infect. Dis. 27:S100-S106. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., A. Karim, A. Mercat, I. Le Thomas, H. Vahaboglu, C. Richard, and P. Nordmann. 1999. Extended-spectrum β-lactamase-producing strain of Acinetobacter baumannii isolated from a patient in France. J. Antimicrob. Chemother. 43:157-158. [PubMed] [Google Scholar]
- 22.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 25.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Takahashi, A., S. Yomoda, I. Kobayashi, T. Okubo, M. Tsunoda, and S. Iyobe. 2000. Detection of carbapenemase-producing Acinetobacter baumannii in a hospital. J. Clin. Microbiol. 38:526-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tankovic, J., P. Legrand, G. De Gatines, V. Chemineau, C. Brun-Buisson, and J. Duval. 1994. Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J. Clin. Microbiol. 32:2677-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towner, K. J., T. Gee, and T. Boswell. 2002. An unwanted import to the UK: a carbapenem-resistant clinical isolate of Acinetobacter baumannii producing metallo-β-lactamase. J. Antimicrob. Chemother. 50:1092-1093. [DOI] [PubMed] [Google Scholar]
- 31.Vahaboglu, H., F. Coskunkan, O. Tansel, R. Ozturk, N. Sahin, I. Koksal, B. Kocazeybek, M. Tatman-Otkun, H. Leblebicioglu, M. A. Ozinel, H. Akalin, S. Kocagoz, and V. Korten. 2001. Clinical importance of extended-spectrum β-lactamase (PER-1-type)-producing Acinetobacter spp. and Pseudomonas aeruginosa strains. J. Med. Microbiol. 50:642-645. [DOI] [PubMed] [Google Scholar]
- 32.Vahaboglu, H., R. Ozturk, H. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Lecblebicioglu, I. Balik, K. Aydin, and M. Oktun. 1997. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]