Abstract
The activity of myogenic regulatory factor (MRF) genes is essential for vertebrate muscle development whereas invertebrate muscle development is largely independent of MRF function. This difference indicates that myogenesis is controlled by distinct regulatory mechanisms in these two groups of animals. Here we used over expression and gene knockdown to investigate the role in embryonic myogenesis of the single MRF gene of the invertebrate chordate Ciona intestinalis (Ci-MRF). Injection of Ci-MRF mRNA into eggs resulted in increased embryonic muscle-specific gene activity and revealed the myogenic activity of Ci-MRF by inducing the expression of four muscle-marker genes, Acetylcholinesterase, Actin, Troponin I, and Myosin Light Chain in non-muscle lineages. Conversely, inhibiting Ci-MRF activity with antisense morpholinos down regulated the expression of these genes. Consistent with the effects of morpholinos on muscle gene activity, larvae resulting from morpholino injection were paralyzed and their “muscle” cells lacked myofibrils. We conclude that Ci-MRF is required for larval tail muscle development and thus that an MRF-dependent myogenic regulatory network probably existed in the ancestor of tunicates and vertebrates. This possibility raises the question of whether the earliest myogenic regulatory networks were MRF dependent or MRF independent.
Keywords: ascidian, chordate, tunicate, MyoD, myogenic regulatory network, myogenesis
INTRODUCTION
Myogenic regulatory factor (MRF) genes are a family of conserved basic-helix-loop-helix (b-hlh)-containing transcription factors that participate in muscle development in a wide variety of animals (reviewed by Baylies and Michelson, 2001; Pownall et al, 2002; Buckingham et al, 2003; Tajbakhsh, 2005; Tapscott, 2005). Vertebrates possess four MRF genes; MyoD, Myf5, myogenin, and MRF4 whose combined activity is required for muscle development. Despite their considerable functional redundancy, gene knockout experiments indicate that Myf5 and MyoD function to specify the skeletal muscle lineage (Rudnicki et al, 1993; Kablar et al, 2003), whereas myogenin plays a key role in terminal differentiation (Hasty et al, 1993; Nabeshima et al, 1993; Venuti et al, 1995). MRF4 appears to function in both muscle specification and differentiation (Kassar-Duchossoy et al, 2004). In contrast most invertebrates have a single MRF gene, which in those that have been studied (C. elegans and Drosophila) plays a much less important role in myogenesis than do the MRFs of vertebrates. These differences indicate that vertebrate and invertebrate myogenesis have significantly different requirements for MRF gene activity.
Ascidians are marine invertebrates of the subphylum Tunicata that together with the subphyla Cephalochordata and Vertebrata comprise the phylum Chordata. Recent studies indicate that tunicates are the closest living relatives of vertebrates (Ruppert, 2005; Delsuc et al, 2006), and thus, that ascidians are phylogenetically well suited for studying how differences in invertebrate and vertebrate MRF activities may have evolved. Like most invertebrates, ascidians have a single MRF gene that is equally related by sequence to the four vertebrate MRFs (Araki et al, 1994; Meedel et al, 1997; Dehal et al, 2002). In Ciona intestinalis this gene encodes two transcripts that are present in embryonic and adult body wall muscle but not in heart muscle, and whose expression is closely correlated with muscle differentiation under a variety of circumstances (Meedel et al, 1997; Meedel et al, 2002). These observations provide strong circumstantial evidence that it has a significant role(s) in myogenesis. Originally this gene was named CiMDF (Ciona intestinalis Muscle Determination Factor; Meedel et al, 1997) and has also been referred to as Ci-MyoD (e.g. Imai et al, 2004). Here we suggest the name Ci-MRF (Ciona intestinalis Myogenic Regulatory Factor) to indicate its equal relationship with the four vertebrate MRFs and that, as shown in this communication, it functions as a myogenic regulatory factor.
Muscle of the ascidian larva is confined to two bilaterally symmetric bands in the tail that flank a central notochord and a dorsal hollow nervous system (Katz, 1983; Satoh, 1994). Its organization and function most resemble vertebrate axial muscle (Bone, 1989), and it is a striated member of the troponin/tropomyosin-regulated class of muscles that includes vertebrate skeletal and cardiac muscles and ascidian body-wall muscle (Meedel and Hastings, 1993). Species of ascidians typically used for embryological studies have only 36–42 mononucleate tail muscle cells with thoroughly documented lineage histories (Conklin, 1905; Ortolani, 1955; Nishida, 1987). Muscle in the anterior and middle of the tail is designated as the primary lineage, and develops autonomously under the control of asymmetrically localized maternal determinants; muscle in the posterior is designated as the secondary lineage and develops conditionally in response to intercellular signaling (Meedel et al, 1987; Nishida, 1990). Therefore, ascidian larval muscle is a well-characterized and exceptionally simple tissue whose development can be studied at the resolution of individual cells.
In this study we present a comprehensive analysis of Ci-MRF expression and function that appeared previously in abstract form (Meedel et al, 2006). In situ hybridization experiments showed that Ci-MRF is expressed abundantly only in cells that are uniquely fated to form muscle. Gene over-expression and knockdown experiments demonstrated that Ci-MRF is a bona fide MRF whose myogenic activity is necessary for larval muscle development. Our results lead us to conclude that an MRF-dependent regulatory network is a shared feature of tunicate and vertebrate myogenesis.
MATERIALS AND METHODS
Embryo culture and injections
Adult Ciona intestinalis were obtained from the Station Biologique in Roscoff, France. Eggs and sperm were removed surgically from adults, and eggs were dechorionated chemically as described by Mita-Miyazawa et al (1985). Fertilizations were done using dilute sperm suspensions and embryos were raised on 1% agarose-coated petri dishes at 16–19°C in artificial sea water (Hudson and Lemaire, 2001) containing 50 μg/ml kanamycin. Microinjections were done as previously described (Hudson et al, 2003); each egg was injected with ~30 pl of test solution. We initially tested a variety of concentrations of RNAs or morpholinos, but in the experiments reported we injected CiMRFb mRNA at 0.5 mg/ml and morpholinos at 0.5–1.0 μM.
mRNA injection constructs and morpholinos
In order to create a full-length cDNA, genomic DNA was amplified with the primers 5’-TGACAGAATTCCACCATGACTTGCATCTCTCTAGAGG-3’ and 5’-CAACCAGACGCCATATTACTGAGC-3’ using PFU polymerase (Stratagene). This resulted in ~320bp fragment, which included the translation start site of CiMRFb RNA (shown in bold in the first primer); this fragment was digested with EcoRI and SacI and inserted together with a ~2.3 kb SacI/NotI fragment encoding most of CiMRFb RNA (Genbank accession number U80079; Meedel et al, 1997) into EcoRI/NotI digested pRN3 (Lemaire et al, 1995) to create pRN3CiMRFb. mRNA was synthesized from SfiI linearized plasmids using mMessage Machine kits (Ambion). Antisense morpholinos were purchased from Gene Tools, LLC. Morpholinos used to target Ci-MRF transcripts were ATGTCATACTACCGGCTGGATTTGC (MO1640) and CTAGAGAGATACACGTCATCGTATA (MO4468).
In situ hybridization and acetylcholinesterase histochemistry
In situ hybridization was done using digoxigenin-labeled antisense RNA probes essentially as described by Wada et al (1995). Incubation times for color development ranged from 20 minutes to 48 hours depending on probes. Probes to detect mRNAs for Ci-Muscle Actin (Hudson and Yasuo, 2005), Ci-MRF (CiMRFa, Meedel et al 1997) and Ci-Troponin I (pcTp2; MacLean et al 1997) have been described previously; Ci-Myosin Alkali Light Chain (cilv022o11) was obtained from the Ciona gene collection plates (e.g. Satou et al., 2002). Hoechst staining was done as described by Hudson and Yasuo (2005).
For acetylcholinesterase (AChE) histochemistry, embryos were fixed for 40 minutes on ice in seawater containing 4% para-formaldehyde. AChE activity was localized according the method of Karnovsky and Roots (1964). Incubation times for color development were 2–4 hours at room temperature.
Transmission electron microscopy
Larvae were fixed for electron microscopy using the method of Crowther and Whittaker (1986). Briefly, larvae were collected when uninjected controls were actively swimming ~20 hours post fertilization), and fixed for 30 minutes at room temperature in 2.5% glutaraldehyde, 0.2M phosphate buffer pH7.2, 0.34M NaCl. After rinsing in 0.2M phosphate buffer pH7.2, specimens were post-fixed for 30 minutes at room temperature in 1% OsO4, 1.5% potassium ferrocyanide, 2.5% bicarbonate buffer pH 7.2, rinsed with water, and en-bloc stained in 2% uranyl acetate overnight at 4°C on a rotary shaker. Following dehydration through an ethanol series, larvae were embedded in Spurr resin and baked for 18 hours at 60°C. Thin sections (silver) were made with diamond knife using a Reichert OM U3 ultramicrotome and observed directly or after post staining with uranyl acetate and lead citrate, with a Hitachi H600 electron microscope at 100kV. Photographs were digitally scanned without significant image modification.
RESULTS
Zygotic expression of Ci-MRF coincides with restriction to muscle fate
We previously showed that Ci-MRF is expressed only in muscle cells of tail-formation stage embryos (Meedel et al, 1997). However, the observation that MRF transcripts exist at low concentrations in non-muscle lineage cells of many organisms at early developmental stages (e.g. Harvey, 1990; Scales et al, 1991; George-Weinstein et al, 1996; Gerhart et al, 2000; Kiefer and Hauschka, 2001), coupled with the occurrence of minute levels of the small Ci-MRF mRNA (CiMRFa) in eggs and early-cleavage stage Ciona embryos (Meedel et al, 2002) prompted us to reexamine the spatial pattern of Ci-MRF expression by in situ hybridization. Ci-MRF transcripts were first observed at the 44-cell stage in nuclei of the B7.4 blastomere pair (Fig. 1A). The nuclear localization of these transcripts indicates that Ci-MRF expression has just begun and is consistent with previous RT-PCR studies showing that zygotic transcription of Ci-MRF begins between three and four hours (32 to 64 cell stages) post fertilization (Meedel et al, 2002). Notably, the B7.4 blastomeres of the 44-cell embryo are the only muscle lineage cells whose fate is restricted to form muscle at this stage (Nishida, 1987). We did not detect Ci-MRF transcripts in eggs or earlier cleavage-stage embryos, which have been shown to contain an extremely low level of maternal CiMRFa mRNA (Meedel et al, 2002). At the 64-cell stage the B7.8 blastomere pair becomes fate restricted to form muscle (Nishida, 1987), and it was at this time that these cells first expressed Ci-MRF. With subsequent divisions of the B7.4 cells leading to the 76-cell stage and the B7.8 cells leading to the 110-cell stage three and then four cell pairs respectively expressed Ci-MRF (76 cell stage: B7.8, B8.7, B8.8; 110-cell stage: B8.7, B8.8, B8.15, B8.16). Up to and including the 110-cell stage we never detected Ci-MRF expression in the B7.5 cells (see diagram in Fig. 2), which are the other progenitors of the primary muscle lineage. The B7.5 blastomere pair is not fate restricted, and will give rise to cardiac muscle and the anterior-most larval tail muscle (Nishida 1987). Therefore, the zygotic expression of Ci-MRF in the primary muscle lineages occurs exclusively in cells whose fate is restricted to tail muscle (Fig. 1B). These results are in agreement with those of Imai et al (2004) except that they did not report on Ci-MRF expression as early as the 44-cell stage.
Fig. 1.

Localization of Ci-MRF transcripts during embryogenesis shown by in situ hybridization. (A) Vegetal views of transcript distribution in cleavage, gastrula, and neural plate stage embryos. The developmental stage is indicated in each panel; arrows in the 110-cell embryo and early gastrula embryo indicate A8.16 cells that have neural and muscle fates. Only cells on one side of the bilaterally symmetrical embryo are labeled in each panel. Labels on the neural plate stage embryo indicate muscle (A9.31) and neural (A9.32) precursors; in order to distinguish these cells, this embryo was stained with Hoechst dye. Approximate ages of embryos in hours after fertilization, when reared at 18°C are 44-cell (3.75 hr), 64-cell (4.25 hr), 76-cell (4.5 hr), 110-cell (5 hr), gastrula (6 hr), neural plate (7 hr). (B) Diagram of the primary muscle lineage; cells that are fate restricted to form muscle are shown in red. Numbers at the top of the diagram indicate the cell-stage. (C) Vegetal and lateral views of Ci-MRF transcript distribution in a single neurula-stage embryo (~8 hr post fertilization at 18°C). (D) Lateral (upper row) and dorsal (lower row) views of Ci-MRF transcript distribution during tail formation; Orientation of each embryo is anterior (top, left), posterior (bottom, right). Approximate developmental ages at 18°C are 9 hr (Early), 11 hr (Middle) and 13 hr (Late). Abbreviations are A (anterior), P (posterior), An (animal), Vg (vegetal), ep (epidermis), N (notochord). All scale bars are 50 μm.
Fig. 2.

In situ hybridization of early gastrula-stage embryos injected with CiMRFb mRNA. Embryos shown represent the two most common classes of result we obtained; the percentage of embryos in each class is indicated as are the number of embryos analyzed (N). Embryos in lateral view are oriented with the vegetal half facing left, and animal half facing right. The scale bar is 50 μm. The diagram shows an early gastrula-stage embryo in vegetal view.
Unlike the primary lineage, secondary muscle lineage cells sometimes expressed Ci-MRF before they were fate restricted to muscle. Expression was detected in the A8.16 cells at the 110-cell/early gastrula stages in approximately one-third of embryos analyzed (arrows in Fig. 1A); this cell pair has both neural and muscle fates before dividing to form the fate restricted A9.31 (muscle) and A9.32 (neural) cells (Nishida 1990; Nicol and Meinertzhagen 1988; Cole and Meinertzhagen 2004). Following cleavage, only the A9.31 cell continues to express Ci-MRF, and transcript levels become comparable to the primary lineage (Fig. 1A).
At none of the later stages examined was there any indication that Ci-MRF was expressed in cells other than those that give rise solely to muscle. Muscle cells in neurula stage embryos are bounded by epidermis and confined to the posterior-vegetal region of the embryo (Conklin, 1905); only these cells expressed Ci-MRF at this stage (Fig. 1C). Later, during tail formation, the muscle cells form two lateral bands in the tail, which flank the central notochord and are bounded by epidermis. Viewed laterally these cells appear as a mass that fills the tail. Viewed from the dorsal or ventral surfaces this mass of muscle cells appears “U-shaped” in early-middle tail formation stages; later it separates at the posterior end to form two bilateral bands of muscle (Conklin, 1905). Only cells corresponding to these locations expressed Ci-MRF during tail formation stages (Fig. 1D).
Ci-MRF overexpression increases muscle gene activity and leads to the formation of ectopic muscle-like cells
A hallmark of MRF family genes is their ability to direct myogenesis when they are experimentally activated in non-muscle cells (e.g. Davis et al, 1987; Weintraub et al, 1989; Choi et al, 1990; Venuti et al, 1991; Fukushige and Krause, 2005). We studied whether Ci-MRF had this potential by injecting CiMRFb mRNA into eggs, which were then fertilized and examined for muscle-specific gene activity at early gastrula or early-mid tail formation stages. We focused on CiMRFb because the protein it encodes contains all known MRF functional domains, whereas CiMRFa lacks Domain III (Rhodes and Konieczny, 1989) and thus might not be a fully functional MRF. Four muscle-specific genes were assayed in order to assess diverse aspects of the differentiated phenotype: Acetylcholinesterase served as a marker of the cholinergic system and was assayed histochemically; muscle Actin and Troponin I (TnI) served as thin filament markers and Myosin (alkali) light chain (MLC) served as a thick filament marker. All three mRNAs encoding proteins of the contractile apparatus were assayed by whole mount in situ hybridization.
Embryos reared from CiMRFb mRNA-injected eggs cleaved normally and developed into normal looking early gastrula stage embryos (Fig. 2), making it possible to identify cell lineages. As reflected in the in situ hybridization staining intensity, the muscle lineage cells of injected embryos showed dramatic increases in the levels of all three contractile apparatus mRNAs assayed (Fig. 2). In addition to enhanced activity within the primary muscle lineage itself, injected embryos also expressed these three genes in B-lineage cells that give rise to endoderm, mesenchyme and notochord (Conklin, 1905; Nishida, 1987). For Actin and MLC we observed two major phenotypes that together accounted for ≥ 90% of injected embryos (Fig. 2). Most commonly these two genes were expressed in all B-lineage cells, while less often they were primarily active in the mesenchyme lineages in addition to the primary muscle. Some A-lineage cells also expressed Actin and MLC transcripts at lower levels. TnI appeared to be particularly sensitive to CiMRFb injection, and its transcripts were detected at high levels in all the vegetal cells (i.e. both A- and B- lineages) in about 70% of injected embryos (Fig. 2). In about one-quarter of the embryos, TnI expression, while still apparent in multiple A-lineage cells, was most abundantly expressed in the B-lineage (Fig. 2). We never observed expression of any of the contractile protein genes analyzed in the animal half of the embryo as was evident when embryos were viewed laterally (Fig. 2).
Embryos reared to the tail formation stage from eggs injected with CiMRFb mRNA did not develop normally; instead they formed amorphous embryos without a distinct head or tail, but with an obvious epidermal covering (Fig. 3A). Acetylcholinesterase, Actin, and MLC expression patterns were similar in these specimens: epidermal cells were uniformly negative, and approximately three-quarters of the interior cells were strongly positive while the remaining cells were negative or weakly positive. Consistent with what we observed in early gastrula stage embryos, TnI expression was particularly sensitive to CiMRFb mRNA since all interior cells of injected embryos were strongly positive. As with the other three markers, TnI transcripts were not detected in the surrounding epidermal cells (see Fig. 3 and Legend).
Fig. 3.

Muscle gene expression in tail-formation stage embryos injected with CiMRFb mRNA. (A) Control embryos are shown in lateral views; injected embryos had no clear axial organization. Numbers of injected embryos examined (N) are indicated at the bottom of the figure. Two representatives from the pool of injected specimens are shown for each marker examined. The apparent hybridization signal in the epidermis of CiMRFb mRNA-injected embryos assayed for TnI mRNA was not observed when these embryos were examined microscopically; therefore, this “signal” is an artifact caused by the intense staining reaction seen in these embryos and by photographing them in glycerol, which renders them somewhat transparent. Scale bar is 100 μm. (B) Embryos cleavage-arrested with cytochalasin B from the 64-cell stage until early tail formation. Injected embryos had an average of 18 acetylcholinesterase positive cells; the number of TnI positive cells in injected embryos could not be determined. Number of injected embryos examined (N) is indicated at the bottom.
The widespread muscle-specific gene activity present in these embryos, together with the observation that muscle specific gene expression occurred in non-muscle lineages of CiMRFb-injected embryos at the early gastrula stage suggests that myogenesis was taking place in non muscle cells of these embryos. This was confirmed by taking advantage of the ability of embryos to differentiate when cytokinesis is prevented with cytochalasin B (Whittaker, 1973). Embryos were treated with cytochalasin B beginning at the 64-cell stage and then assayed for acetylcholinesterase activity or the presence of TnI transcripts at the early-mid tail formation stage. Under these conditions, uninjected, cleavage-arrested specimens expressed Acetylcholinesterase and TnI in an average of six primary muscle lineage blastomeres (Fig. 3B); in contrast, acetylcholinesterase activity was seen in an average of 18 cells in CiMRFb injected specimens. Most of these additional cells were B-lineage blastomeres (cells in the lower half of the embryo shown), although some corresponded to the A-lineage (cells in the upper half of the embryo shown). Consistent with previous experiments indicating that its expression is especially responsive to Ci-MRF over-expression, TnI was expressed throughout the vegetal half of cleavage-arrested embryos.
MO-treatment inhibits muscle-specific gene activity
We used antisense morpholino oligonucleotides (MO) to investigate the effect on muscle development of down regulating Ci-MRF activity. Two MOs were designed for these experiments: the first (MO1640) targeted nucleotides 16–40, which corresponds to most of the 5’ untranslated region of Ci-MRF mRNAs exclusive of the 5’ trans-spliced leader sequence (Vandenberghe et al, 2001); the second (MO4468) targeted nucleotides 44–68 which includes the translation start site at nucleotides 50–52. MOs targeting these sequences are expected to prevent the accumulation of Ci-MRF protein by blocking translation. The two MOs gave similar results, indicating that they were likely to be specifically targeting Ci-MRF mRNAs.
Embryos reared from eggs injected with either MO cleaved and developed into early gastrula stage embryos with normal morphology that were then analyzed for MLC and TnI expression (Fig. 4). Both MOs reduced muscle gene expression significantly, although we did observe some variability in the response of individual embryos to MO injection. MO injection dramatically suppressed TnI expression; with either MO no TnI transcripts were detected in > 90% of embryos. This result shows that both MOs are effective in disrupting normal myogenesis. Furthermore, MLC expression was also significantly affected with down-regulation or loss of expression observed in > 75% of the embryos injected with either MO1640 or MO4468.
Fig. 4.

MOs targeting Ci-MRF mRNA reduce muscle gene expression in early gastrula embryos. Representative staining reactions are shown for control embryos assayed for MLC and TnI expression. The majority of MO-treated embryos assayed for MLC expression were negative as shown in the panels. Remaining embryos were significantly down regulated; 21% for MO1640 and 35% for MO4468, or resembled the controls (not shown). Essentially all embryos assayed for TnI expression were negative when treated with either MO. The average number of positive cells after each treatment is indicated below the panels, and includes all cells expressing a given marker irrespective of the strength of the signal detected. The number of embryos examined (n) is shown in parentheses. Scale bar = 50 μm.
Middle and late tail-formation stage embryos raised from MO-injected eggs were also examined for muscle gene expression. Like embryos examined at the early gastrula stage, we noted some variability in the response of individual embryos. Therefore, we devised a classification system to score the extent of muscle marker gene expression as determined by histochemistry (AChE) or in situ hybridization (Actin, MLC, TnI). This system was: 3+ for expression level similar to controls, 2+ for reduced level of expression relative to control, 1+ for greatly reduced level of expression relative to control, 0+ for no detectable expression. Morphologically at least 80% of the MO-treated embryos were scored “grossly normal” meaning they had a clearly distinct head and tail. The most obvious abnormality was tail malformation that ranged from relatively minor kinks (Fig. 5) to stunted tails less than 50% of normal length (not shown). Other readily observed aspects of development appeared normal in MO-treated embryos; for example pigment formation in the otolith was comparable to controls (Fig. 5).
Fig. 5.
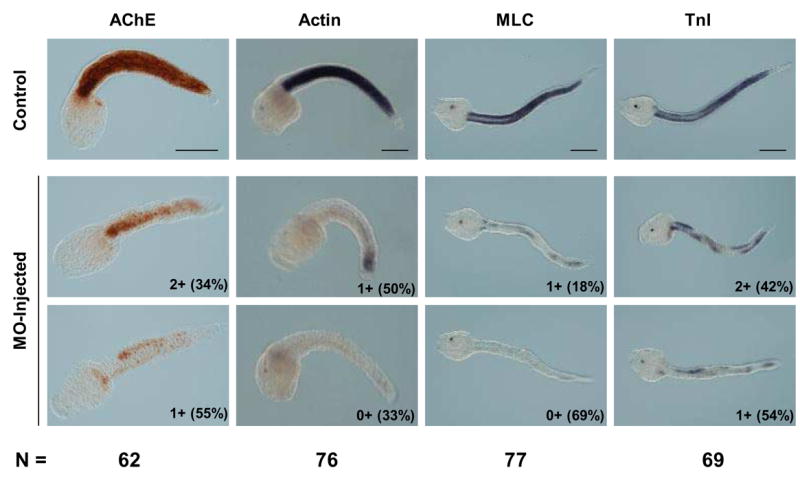
MOs targeting Ci-MRF mRNA reduce muscle gene expression in tail formation stage embryos. AChE histochemistry and Actin in situ hybridization were carried out at the mid tail formation stage; MLC and TnI in situ hybridizations were done at the late tail formation stage. Numbers outside of parentheses in the lower right corner of each MO-injected embryo refer to the stain intensity relative to the stain intensity of control embryos reacted for the same time. The system used for classifying embryos (e.g. 3+, 2+, 1+, 0+) is described in the text. Examples are shown of the two most abundant embryo classes for each marker tested and the percentage of embryos in that class is shown in parentheses. The total number of MO-injected embryos analyzed for each marker (N) is shown at the bottom of the figure. Scale bars = 100 μm.
All four markers of muscle development assayed were down regulated in MO-injected tail formation stage embryos (Fig. 5). MLC and Actin were most affected with expression of both markers either undetectable (0+) or severely down regulated (1+) in >80% of MO-injected embryos. TnI transcript levels and AChE activity were also significantly reduced by MO-injection with the majority of embryos falling into the two most reduced categories (0+ and 1+), although both markers appear less sensitive than either MLC or Actin to MO-injection.
MO-treated larvae are paralyzed and do not form myofibrils
Our results with molecular markers indicate that myogenesis is significantly perturbed following inhibition of Ci-MRF function. In order to address the consequences of blocking Ci-MRF activity on structural and functional aspects of muscle development we examined muscle ultrastructure in control and MO-treated larvae that exhibited grossly normal morphology (Fig. 6A, B). Ascidian larval tail muscle has been thoroughly characterized at the ultrastructural level (reviewed in Meedel, 1998). These cells have extremely high concentrations of centrally positioned mitochondria and, relative to vertebrate skeletal muscle, a rather modest number of sarcomeric myofibrils that are restricted to the peripheral cytoplasm near the plasma membrane. These features were apparent in sections through muscle cells of control larvae, in which the peripheral myofibrils showed the expected cross-striated pattern (Fig. 6C, E). Sections through the corresponding cells of MO-treated larvae (identified unambiguously by position in the tail and high concentration of mitochondria) occasionally showed traces of what appeared to be disorganized myofilaments, but more typically no muscle ultrastructural elements were observed (Fig. 6D, F). Consistent with the loss of organized myofibrils, MO-treated larvae rarely showed any signs of movement and those that did (<10%) exhibited only very occasional weak twitches and not the frenetic movements typical of normal larvae. These results demonstrate that Ci-MRF is required to form functional tail muscle cells.
Fig. 6.

Morphological comparison of normal (control) and morpholino-treated larvae. Differential interference contrast light micrographs of normal (A) and morpholino-injected (B) larvae. Transmission electron micrographs of the larval tail showing muscle cells in control (C, E) and morpholino-treated (D, F) specimens. Sections are slightly oblique to the longitudinal axis. The box in panel C encloses an area near the cell membrane of a muscle cell from a control larva that is rich in cross-striated myofibrils; this area is seen at higher magnification in panel E where white arrows point to myofibrils. The box in panel D encloses a corresponding area of cytoplasm near the cell membrane of a “muscle” cell of a MO-injected embryo. This area is seen at higher magnification in panel F; note the absence of myofibrils. Scale bars are 100μm (A), 1μm (C, D).
DISCUSSION
Ci-MRF expression
In recent years several genes have been found that regulate ascidian myogenesis. At the head of this regulatory hierarchy in the primary muscle lineage is the maternally expressed gene macho-1 (Nishida and Sawada, 2001). At least three early-acting zygotic genes also appear to be important regulators of myogenesis, Tbx6b, Tbx6c, and ZicL (Imai et al, 2002; Yagi et al, 2004; 2005). Our findings add Ci-MRF to this group of genes that are crucial for myogenesis and indicate that one of the critical functions of these earlier acting regulatory genes is to activate Ci-MRF expression. This view is supported by recent studies that showed Ci-MRF transcript levels were enhanced in macho-1 overexpressing embryos (Yagi et al, 2004), and down regulated in embryos treated with either Tbx6b/c/d or ZicL MOs (Imai et al, 2006).
Our results with Ci-MRF have striking parallels to earlier research on Brachyury, a gene encoding a T-box transcription factor expressed in ascidian notochord (Yasuo and Satoh, 1993; 1994; 1998). Brachyury expression is detected only in notochord lineage cells whose fates are restricted, and its initiation coincides precisely with the time of fate restriction (Yasuo and Satoh, 1994). Furthermore, Brachyury is required for the differentiation of notochord cells and is sufficient to induce notochord marker expression in certain non-notochord lineages (Takahashi et al, 1999; Hotta et al, 1999). Thus, Brachyury functions in the notochord much in the same way that our results indicate Ci-MRF functions in muscle cells. Parallels can also be drawn with other key regulatory genes such as Twist-like1 in the mesenchyme (Imai et al, 2003) and Lhx3 in the endoderm (Satou et al, 2001). Therefore, the early activation of key lineage-restricted transcription factors appears to be a common mechanism used by ascidian embryos to ensure the robust and spatially appropriate expression of terminal differentiation genes. Furthermore, while we did not investigate such a possibility in this study, Ci-MRF may also repress the activity of non-muscle genes. This would be consistent with the suggestion that vertebrate MyoD may act as a repressor through its association with histone deacetylases (Tapscott, 2005).
Ci-MRF is required for normal muscle development
Antisense MOs were used to examine the extent to which muscle development depended on Ci-MRF. MO-treated embryos expressed muscle genes at reduced levels compared with controls and developed into larvae that were almost always paralyzed and whose “muscle” cells lacked myofibrils. Notably, these specimens were quite normal in appearance indicating that MOs were not generally detrimental to embryogenesis and that the effects we observed were specific to muscle development. Therefore, our results provide strong evidence that Ci-MRF is a key regulator of muscle development. This conclusion is supported by a recent large-scale study of the early embryonic gene regulatory network of Ciona intestinalis, which also reported that a MO targeting Ci-MRF down regulated Actin transcript levels (Supplementary Material in Imai et al, 2006).
Muscle gene expression was not uniformly affected in MO-treated embryos. A clear example of this was the variable responses of TnI and MLC. Transcripts of both genes were reduced effectively at the early gastrula stage by MO-treatment, but while MLC transcripts were still dramatically down regulated at the early-mid tail formation stage TnI transcripts were less affected at this time. Because MLC expression was strongly reduced throughout development, it is unlikely that the MOs lost their efficacy at later stages. Instead, these results indicate that other regulatory mechanisms may act in parallel with Ci-MRF to control TnI expression at later stages, or that inactivation of Ci-MRF results in misregulation of other regulatory factors that lead to TnI expression.
MO-treatment usually did not eliminate muscle marker expression. This result was not entirely surprising because expression of some terminal muscle differentiation genes, such as Actin and myosin heavy chain (MHC), is initiated before Ci-MRF is expressed (Satou et al, 1995), and, thus, some Ci-MRF-independent muscle gene activity was expected. We also cannot exclude the possibility that some of the muscle gene activity we noted was Ci-MRF dependent. However, we feel that this is unlikely since injecting both MOs together gave essentially the same results as injecting MOs separately (data not shown) consistent with the view that individual MOs efficiently targeted Ci-MRF. Development of antibodies to assess Ci-MRF protein levels in MO-treated embryos would be required to resolve this issue.
Ci-MRF is myogenic
CiMRFb mRNA-injection resulted in muscle genes being expressed in non-muscle lineages. This result demonstrates that Ci-MRF is myogenic in C. intestinalis embryos, and thus that it functions as a bona fide MRF (e.g. Davis et al, 1987; Weintraub et al, 1989; Choi et al, 1990; Venuti et al, 1991; Fukushige and Krause, 2005). Not all cells were susceptible to myogenic conversion by misexpressing Ci-MRF. B-line cells were most susceptible, A-line cells were generally less susceptible, and animal-half cells appeared to be entirely refractory. These results are consistent with the view that MRF genes function together with other positive and negative regulatory factors (e.g. chromatin remodeling complexes, kinases, E-proteins. Mef2, Id, etc.) in a complex regulatory environment (Berkes and Tapscott, 2005; Tapscott, 2005). Thus, the distribution of such cofactors could account for the divergent responses of different cells to misexpression of Ci-MRF. Interestingly, mesenchyme lineage cells were among the cells most readily converted to muscle-like cells. Many of these cells originate from the B-line (Nishida, 1987), and elaboration of their fate depends on fibroblast growth factor (FGF)-dependent signals that suppress myogenesis (Kim and Nishida, 1999, 2001; Kim et al, 2000). A recent study showing that Ci-MRF is expressed in the B-line mesenchyme cells of embryos treated with a MO targeting FGF9/16/20 (Imai et al, 2006) indicates that this FGF signal blocks Ci-MRF expression, which could otherwise lead to myogenesis in mesenchyme lineages as happened in CiMRFb mRNA-injected embryos.
MRF misexpression in embryos typically activates terminal muscle genes but the ectopic, muscle-like cells formed usually do not become fully differentiated muscle (Hopwood and Gurdon 1990; Miner et al, 1992; Faerman et al, 1993; Fukushige and Krause, 2005; but see also Ludolph et al, 1994; Delfini and Duprez, 2004). In order to evaluate the extent of muscle differentiation in CiMRFb mRNA-treated embryos we analyzed four markers that represented diverse aspects of the muscle phenotype. All of these markers were expressed ectopically, indicating that Ci-MRF misexpression elicits a broad-spectrum myogenic response in some cells. We could not use structural or physiological features such as the presence of sarcomeres or contractility to further evaluate the extent of ectopic myogenesis because these features do not appear until the mid-late tail formation stage at which time the highly abnormal morphology of CiMRFb-injected embryos prevented us from distinguishing ectopic from normal muscle. We suspect that elaboration of a muscle phenotype in non-muscle lineages leads to abnormal development by interfering with cellular activities necessary for morphogenesis.
Over expression of Ci-MRF by mRNA injection also led to dramatically elevated levels of terminal differentiation gene activity within the muscle lineage. This further supports our contention that Ci-MRF is a key regulator of myogenesis and is consistent with the view that it may act as a rate-limiting factor to determine the quantitative level of muscle gene expression in embryos. The presence of GC-core E-boxes in many potential target genes (Johnson et al, 2004, 2005; Kusakabe et al, 2004) indicates that it could do this directly, although its ability to regulate the expression of at least three transcription factor genes (Imai et al, 2006) indicates that some of these activities are indirect. Ci-MRF may also play a role in initiating the expression of some terminal muscle differentiation genes, consistent with properties of other MRFs (Tapscott, 2005). Of the genes studied, TnI is a good candidate to represent this class, as it was the most widely misexpressed of the genes we examined in CiMRFb-injected embryos at both the gastrula and tail formation stages. In addition, the accumulation of TnI transcripts was most negatively affected by MO-treatment when embryos were examined at the early gastrula stage, which is shortly after this gene becomes transcriptionally active (Cleto et al, 2003). Finally, E-box elements in the 5’ upstream regulatory region of TnI have been shown to be crucial for the expression of electroporated reporter constructs (Johnson et al, 2004).
Evolution of MRFs
MRF genes of invertebrates and vertebrates exhibit a number of conserved features including their sequences and expression patterns, their capacity to elicit muscle differentiation when expressed in cultured cells, and their ability to substitute for one another in functional tests (see recent reviews by Baylies and Michelson, 2001; Pownall et al, 2002; Buckingham et al, 2003; Tajbakhsh, 2005). A particularly remarkable example of this functional conservation was the ability of the Drosophila MRF, nautilus, and chicken MyoD to rescue an MRF loss of function mutant of C. elegans (Zhang et al, 1999). This high level of conservation indicates that myogenic regulatory strategies involving MRF genes are ancient and conserved. However, vertebrates have four MRF genes whereas invertebrates (with the exception of cephalochordates, which have two MRF genes; Araki et al, 1996; Schubert et al, 2003) have only one. In addition, unlike mutations of vertebrate MRFs, which have severe consequences for myogenesis (Hasty et al, 1993; Nabeshima et al, 1993; Rudnicki et al, 1993; Venuti et al, 1995; Kablar et al, 2003; Kassar-Duchossoy et al, 2004), null mutations of invertebrate MRFs, represented by C. elegans and D. melanogaster, have comparatively minor effects (Chen et al, 1994; Balagopalan, 2001). Thus, despite a high degree of conservation among MRF genes, that includes the functional capabilities of the proteins themselves, vertebrate myogenesis shows a striking reliance on MRF activity that was not seen previously in invertebrates (reviewed by Olson and Klein, 1998; Baylies and Michelson, 2001).
The results of the present study add a significant new dimension to our understanding of MRF function by demonstrating that like vertebrates, ascidians also require MRF activity for muscle development. Thus, an MRF-dependent myogenic regulatory network is not strictly a vertebrate phenomenon, and it probably existed in the ancestor of tunicates and vertebrates. Moreover, the presence of only a single MRF gene in ascidians indicates that such a network does not require multiple MRF genes. These observations raise an important question about the nature of the ancestral myogenic network: Was it MRF-dependent or MRF-independent? This question cannot be resolved based on available information because any scenarios that account for the distribution of MRF-dependent and MRF-independent myogenic networks are equally likely; i.e. gain of an MRF-dependent myogenic network in the ascidian/vertebrate ancestor, or its loss in the C. elegans/Drosophila ancestor are equally probable. This uncertainty underscores the importance of studying MRF function in a wider array of animals. For example, because gene knockdown strategies are effective in sea urchins (e.g. Davidson et al, 2002; Duboc et al, 2004) it should be possible to determine whether an MRF-dependent network predates the chordates by using MO injections to determine whether the sea urchin MRF gene, SUM-1, is required for myogenesis. Although resolving the role of the ancestral MRF in myogensis is likely to be more difficult, the discovery of JellyD1, a potential MRF homolog in the hydrozoan jellyfish Podocoryne carnea (Muller et al, 2003) indicates that this too is feasible.
Current views of the developmental basis of animal evolution focus on changes in the design of transcriptional regulatory systems (Davidson, 2001; Wilkins, 2002, Carroll et al, 2005). Evolutionary changes in the role of MRF genes without large-scale changes in the functional properties of the proteins they encode are entirely consistent with these ideas. Simply stated, functional comparison of MRF genes in invertebrates such as C. elegans and D. melanogaster, with vertebrates and the invertebrate C. intestinalis indicates that the roles of MRF genes in myogenesis depend more on the regulatory environment in which they function than on differences in the properties of the proteins they encode. The ascidian embryo is an experimental system that is well suited for identifying the components of this regulatory system and determining how they function with Ci-MRF to control muscle development.
Acknowledgments
We thank C. Hudson for carrying out the Hoechst staining showed in Fig. 1A and for help identifying Ci-MRF expression in the A9.31 blastomere. We also thank K. Hastings and C. Hudson for their many helpful comments on this manuscript. This work was supported by awards from the National Institutes of Health (1 R15 HD47357-01), the Rhode Island College (RIC) Faculty Research Committee, and the RIC Faculty Development Fund to THM, and from the Centre National de la Recherche Scientifique (CNRS), the Université Paris VI, The Association Française contre les Myopathies (AFM), and The Agence Nationale de la Recherche (ANR) to HY. HY and PC are CNRS staff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki I, Saiga H, Makabe KW, Satoh N. Expression of AMD1, a gene for a MyoD1-related factor in the ascidian Halocynthia roretzi. Roux's Archives of Developmental Biology. 1994;203:320–327. doi: 10.1007/BF00457803. [DOI] [PubMed] [Google Scholar]
- Araki I, Terazawa K, Satoh N. Duplication of an amphioxus myogenic bHLH gene is independent of vertebrate myogenic bHLH gene duplication. Gene. 1996;171:231–236. doi: 10.1016/0378-1119(96)00174-6. [DOI] [PubMed] [Google Scholar]
- Balagopalan L, Keller CA, Abmayr SM. Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Dev Biol. 2001;231:374–382. doi: 10.1006/dbio.2001.0162. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11:431–439. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bone Q. Evolutionary patterns of axial muscle systems in some invertebrates and fish. American Zoologist. 1989;29:5–18. [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design; Blackwell Publishing: 2005. [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87:7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleto CL, Vandenberghe AE, MacLean DW, Pannunzio P, Tortorelli C, Meedel TH, Satou Y, Satoh N, Hastings KE. Ascidian larva reveals ancient origin of vertebrate-skeletal-muscle troponin I characteristics in chordate locomotory muscle. Mol Biol Evol. 2003;20:2113–2122. doi: 10.1093/molbev/msg227. [DOI] [PubMed] [Google Scholar]
- Cole AG, Meinertzhagen IA. The central nervous system of the ascidian larva: mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev Biol. 2004;271:239–262. doi: 10.1016/j.ydbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Conklin EG. The organization and cell-lineage of the ascidian egg. Philadelphia: Academy of Natural Sciences; 1905. [Google Scholar]
- Crowther RJ, Whittaker JR. Differentiation without cleavage: multiple cytospecific ultrastructural expressions in individual one-celled ascidian embryos. Dev Biol. 1986;117:114–126. doi: 10.1016/0012-1606(86)90354-4. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic Regulatory Systems. Academic Press; 2001. [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Armore G, Hinman V, Arenas-Mena C, et al. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev Biol. 2002;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delfini MC, Duprez D. Ectopic Myf5 or MyoD prevents the neuronal differentiation program in addition to inducing skeletal muscle differentiation, in the chick neural tube. Development. 2004;131:713–723. doi: 10.1242/dev.00967. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Duboc V, Rottinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6:397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Faerman A, Pearson-White S, Emerson C, Shani M. Ectopic expression of MyoD1 in mice causes prenatal lethalities. Dev Dyn. 1993;196:165–173. doi: 10.1002/aja.1001960303. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals widespread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- George-Weinstein M, Gerhart J, Reed R, Flynn J, Callihan B, Mattiacci M, Miehle C, Foti G, Lash JW, Weintraub H. Skeletal myogenesis: the preferred pathway of chick embryo epiblast cells in vitro. Dev Biol. 1996;173:279–291. doi: 10.1006/dbio.1996.0023. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Baytion M, DeLuca S, Getts R, Lopez C, Niewenhuis R, Nilsen T, Olex S, Weintraub H, George-Weinstein M. DNA dendrimers localize MyoD mRNA in presomitic tissues of the chick embryo. J Cell Biol. 2000;149:825–834. doi: 10.1083/jcb.149.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. The Xenopus MyoD gene: an unlocalised maternal mRNA predates lineage-restricted expression in the early embryo. Development. 1990;108:669–680. doi: 10.1242/dev.108.4.669. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hopwood ND, Gurdon JB. Activation of muscle genes without myogenesis by ectopic expression of MyoD in frog embryo cells. Nature. 1990;347:197–200. doi: 10.1038/347197a0. [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Erives A, Levine M, Satoh N. Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev Growth Differ. 1999;41:657–664. doi: 10.1046/j.1440-169x.1999.00467.x. [DOI] [PubMed] [Google Scholar]
- Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100:189–203. doi: 10.1016/s0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- Hudson C, Yasuo H. Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development. 2005;132:1199–210. doi: 10.1242/dev.01688. [DOI] [PubMed] [Google Scholar]
- Hudson C, Darras S, Caillol D, Yasuo H, Lemaire P. A conserved role for the MEK signaling pathway in neural tissue specification and posteriorization in the invertebrate chordate, the ascidian Ciona intestinalis. Development. 2003;130:147–159. doi: 10.1242/dev.00200. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129:2723–32. doi: 10.1242/dev.129.11.2723. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. A Twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development. 2003;130:4461–4472. doi: 10.1242/dev.00652. [DOI] [PubMed] [Google Scholar]
- Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Zhou Q, Yagi K, Satoh N, Wong W, Sidow A. De novo discovery of a tissue-specific gene regulatory module in a chordate. Genome Res. 2005;15:1315–1324. doi: 10.1101/gr.4062605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Tajbakhsh S, Rudnicki MA. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol. 2003;258:307–318. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Karnovsky MJ, Roots L. A 'direct-coloring' thiocholine method for cholinesterase. J Histochem Cytochem. 1964;12:219–221. doi: 10.1177/12.3.219. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Katz MJ. Comparative anatomy of the tunicate tadpole, Ciona intestinalis. Biol Bull. 1983;164:1–27. [Google Scholar]
- Kiefer JC, Hauschka SD. Myf-5 is transiently expressed in nonmuscle mesoderm and exhibits dynamic regional changes within the presegmented mesoderm and somites I–IV. Dev Biol. 2001;232:77–90. doi: 10.1006/dbio.2000.0114. [DOI] [PubMed] [Google Scholar]
- Kim GJ, Nishida H. Suppression of muscle fate by cellular interaction is required for mesenchyme formation during ascidian embryogenesis. Dev Biol. 1999;214:9–22. doi: 10.1006/dbio.1999.9402. [DOI] [PubMed] [Google Scholar]
- Kim GJ, Nishida H. Role of the FGF and MEK signaling pathway in the ascidian embryo. Dev Growth Differ. 2001;43:521–533. doi: 10.1046/j.1440-169x.2001.00594.x. [DOI] [PubMed] [Google Scholar]
- Kim GJ, Yamada A, Nishida H. An FGF signal from endoderm and localized factors in the posterior-vegetal egg cytoplasm pattern the mesodermal tissues in the ascidian embryo. Development. 2000;127:2853–2862. doi: 10.1242/dev.127.13.2853. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Ikeda Y, Tsuda M. Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Dev Biol. 2004;276:563–80. doi: 10.1016/j.ydbio.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- Ludolph DC, Neff AW, Mescher AL, Malacinski GM, Parker MA, Smith RC. Overexpression of XMyoD or XMyf5 in Xenopus embryos induces the formation of enlarged myotomes through recruitment of cells of nonsomitic lineage. Dev Biol. 1994;166:18–33. doi: 10.1006/dbio.1994.1294. [DOI] [PubMed] [Google Scholar]
- MacLean DW, Meedel TH, Hastings KE. Tissue-specific alternative splicing of ascidian troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J Biol Chem. 1997;272:32115–32120. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- Meedel TH. Development of ascidian muscles & their evolutionary relationship to other chordate muscle types. In: Collier JR, Adiyodi KG, Adiyodi RG, editors. Reproductive Biology of Invertebrates: Progress in Developmental Biology. VIII. New York: John Wiley and Sons; 1998. pp. 305–330. [Google Scholar]
- Meedel TH, Hastings KE. Striated muscle-type tropomyosin in a chordate smooth muscle, ascidian body-wall muscle. J Biol Chem. 1993;268:6755–6764. [PubMed] [Google Scholar]
- Meedel TH, Crowther RJ, Whittaker JR. Determinative properties of muscle lineages in ascidian embryos. Development. 1987;100:245–260. doi: 10.1242/dev.100.2.245. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Farmer SC, Lee JJ. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: implications for the evolution of chordate muscle gene regulation. Development. 1997;124:1711–1721. doi: 10.1242/dev.124.9.1711. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Lee JJ, Whittaker JR. Muscle development and lineage-specific expression of CiMDF, the MyoD-family gene of Ciona intestinalis. Dev Biol. 2002;241:238–246. doi: 10.1006/dbio.2001.0511. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Yasuo H, Chang P. The Ciona intestinalis MyoD homolog is essential for myogenesis. Dev Biol. 2006;295:412. [Google Scholar]
- Miner JH, Miller JB, Wold BJ. Skeletal muscle phenotypes initiated by ectopic MyoD in transgenic mouse heart. Development. 1992;114:853–860. doi: 10.1242/dev.114.4.853. [DOI] [PubMed] [Google Scholar]
- Mita-Miyazawa I, Ikegami S, Satoh N. Histospecific acetylcholinesterase development in the presumptive muscle cells isolated from 16-cell-stage ascidian embryos with respect to the number of DNA replications. J Embryol Exp Morphol. 1985;87:1–12. [PubMed] [Google Scholar]
- Muller P, Seipel K, Yanze N, Reber-Muller S, Streitwolf-Engel R, Stierwald M, Spring J, Schmid V. Evolutionary aspects of developmentally regulated helix-loop-helix transcription factors in striated muscle of jellyfish. Dev Biol. 2003;255:216–229. doi: 10.1016/s0012-1606(02)00091-x. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev Biol. 1988;130:737–766. doi: 10.1016/0012-1606(88)90364-8. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Dev Biol. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Nishida H. Determinative mechanisms in secondary muscle lineages of ascidian embryos: development of muscle-specific features in isolated muscle progenitor cells. Development. 1990;108:559–568. doi: 10.1242/dev.108.4.559. [DOI] [PubMed] [Google Scholar]
- Nishida H, Sawada K. macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature. 2001;409:724–729. doi: 10.1038/35055568. [DOI] [PubMed] [Google Scholar]
- Olson EN, Klein WH. Muscle minus MyoD. Dev Biol. 1998;202:153–6. doi: 10.1006/dbio.1998.9020. [DOI] [PubMed] [Google Scholar]
- Ortolani G. The presumptive territory of the mesoderm in the ascidian germ. Experientia. 1955;11:445–446. [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Rhodes SJ, Konieczny SF. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Ruppert EE. Key characters uniting hemichordates and chordates: homologies or homoplasies. Can J Zool. 2005;83:8–23. [Google Scholar]
- Satoh N. Developmental Biology of Ascidians. Cambridge University Press; 1994. [Google Scholar]
- Satou Y, Kusakabe T, Araki I, Satoh N. Timing of initiation of muscle-specific gene expression in the ascidian embryo precedes that of developmental fate restriction in lineage cells. Development Growth and Differentiation. 1995;37:319–327. doi: 10.1046/j.1440-169X.1995.t01-2-00010.x. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Early embryonic expression of a LIM-homeobox gene Cs-lhx3 is downstream of β-catenin and responsible for the endoderm differentiation in Ciona savignyi embryos. Development. 2001;128:3559–3570. doi: 10.1242/dev.128.18.3559. [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, Fujiwara S, Nishikata T, Saiga H, Kusakabe T, Shin-i T, Kohara Y, Satoh N. ). Ciona intestinalis cDNA projects: expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene. 2002;287:83–96. doi: 10.1016/s0378-1119(01)00826-5. [DOI] [PubMed] [Google Scholar]
- Scales JB, Olson EN, Perry M. Differential expression of two distinct MyoD genes in Xenopus. Cell Growth Differ. 1991;2:619–629. [PubMed] [Google Scholar]
- Schubert M, Meulemans D, Bronner-Fraser M, Holland LZ, Holland ND. Differential mesodermal expression of two amphioxus MyoD family members (AmphiMRF1 and AmphiMRF2) Gene Expr Patterns. 2003;3:199–202. doi: 10.1016/s1567-133x(02)00099-6. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem and progenitor cells: reconciling genetics and lineage. Exp Cell Res. 2005;306:364–372. doi: 10.1016/j.yexcr.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller RW, Levine M, Satoh N. Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 1999;13:1519–1523. doi: 10.1101/gad.13.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Vandenberghe AE, Meedel TH, Hastings KE. mRNA 5'-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuti JM, Goldberg L, Chakraborty T, Olson EN, Klein WH. A myogenic factor from sea urchin embryos capable of programming muscle differentiation in mammalian cells. Proc Natl Acad Sci U S A. 1991;88:6219–23. doi: 10.1073/pnas.88.14.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuti JM, Gan L, Kozlowski MT, Klein WH. Developmental potential of muscle cell progenitors and the myogenic factor SUM-1 in the sea urchin embryo. Mech Dev. 1993;41:3–14. doi: 10.1016/0925-4773(93)90051-x. [DOI] [PubMed] [Google Scholar]
- Venuti JM, Morris JH, Vivian JL, Olson EN, Klein WH. Myogenin is required for late but not early aspects of myogenesis during mouse development. J Cell Biol. 1995;128:563–576. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Katsuyama Y, Yasugi S, Saiga H. Spatially and temporally regulated expression of the LIM class homeobox gene Hrlim suggests multiple distinct functions in development of the ascidian, Halocynthia roretzi. Mech Dev. 1995;51:115–126. doi: 10.1016/0925-4773(95)00359-9. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker JR. Segregation during ascidian embryogenesis of egg cytoplasmic information for tissue-specific enzyme development. Proc Natl Acad Sci U S A. 1973;70:2096–2100. doi: 10.1073/pnas.70.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AS. The Evolution of Developmental Pathways. Sunderland, MA: Sinauer Associates, Inc; 2002. [Google Scholar]
- Yagi K, Satoh N, Satou Y. Identification of downstream genes of the ascidian muscle determinant gene Ci-macho1. Dev Biol. 2004;274:478–489. doi: 10.1016/j.ydbio.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Yagi K, Takatori N, Satou Y, Satoh N. Ci-Tbx6b and Ci-Tbx6c are key mediators of the maternal effect gene Ci-macho1 in muscle cell differentiation in Ciona intestinalis embryos. Dev Biol. 2005;282:535–549. doi: 10.1016/j.ydbio.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N. Function of vertebrate T gene. Nature. 1993;364:582–583. doi: 10.1038/364582b0. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N. An ascidian homolog of the mouse Brachyury (T) gene is expressed exclusively in notochord cells at the fate restricted stage. Development Growth and Differentiation. 1994;36:9–18. doi: 10.1111/j.1440-169X.1994.00009.x. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N. Conservation of the developmental role of Brachyury in notochord formation in a urochordate, the ascidian Halocynthia roretzi. Dev Biol. 1998;200:158–170. doi: 10.1006/dbio.1998.8958. [DOI] [PubMed] [Google Scholar]
- Zhang J-M, Chen L, Krause M, Fire A, Paterson BM. Evolutionary conservation of MyoD function and differential utilization of E proteins. Dev Biol. 1999;208:465–472. doi: 10.1006/dbio.1999.9218. [DOI] [PubMed] [Google Scholar]


