Abstract
The objective of the present study was to design a PCR-generated DNA probe and determine the specificity of the probe for the identification of clinical isolates of Streptococcus sanguinis. To do this, we examined over 200 arbitrarily primed PCR (AP-PCR) amplicon patterns obtained with DNA from clinical isolates of S. sanguinis. A 1.6-kb DNA amplicon that was common to all AP-PCR profiles was extracted from agarose gels and then cloned and sequenced. A search for a similar sequence in the GenBank database with the BLASTN program revealed that the 1.6-kb DNA fragment comprised an intergenic region between two housekeeping genes, uncC (proton-translocating ATPase) and murA (UDP-N-acetylglucosamine enolpyruvyl transferase). Three digoxigenin-labeled DNA probes were synthesized on the basis of the sequence of the 1.6-kb fragment: the sequence of probe SSA-1 contained the proton-translocating ATPase (uncC) and the entire intergenic region, the sequence of probe SSA-2 contained only the intergenic region, and the sequence of probe SSA-3 contained an internal region of the murA gene. Dot blot hybridization showed that the three probes displayed signals for hybridization to both S. sanguinis strain ATCC 10556 and the S. sanguinis clinical isolates. Probe SSA-1, however, hybridized to DNA from S. oralis and S. mitis. Probe SSA-3 hybridized to DNA from S. gordonii, S. mitis, S. oralis, S. parasanguinis, and S. vestibularis. The probe SSA-2-specific intergenic region appeared to be specific for S. sanguinis. The results from this study suggest that probe SSA-2 may serve as a species-specific DNA probe for the identification of clinical isolates of S. sanguinis.
The heterogeneous group of oral streptococci collectively named Streptococcus sanguinis (formerly S. sanguis) are members of the indigenous oral biota colonizing dental plaque (6, 25). S. sanguinis first colonizes an infant's oral cavity at about 9 months of age (8) and may serve a protective or antagonistic role against the cariogenic bacterium S. mutans (8, 26, 27). On the other hand, S. sanguinis may also cause life-threatening bacterial endocarditis (10) and septicemia (15).
Previous methods for identifying S. sanguinis are based primarily on physiological and biochemical characteristics. The reliability and reproducibility of the conventional phenotypic identification, however, varied among methodologies and investigators (5, 21). For example, previous studies were unable to demonstrate agreement between the genotypic and phenotypic methods for identifying clinical S. sanguinis isolates (8, 30). Accordingly, other methods are being examined, including those that combine PCR with nucleic acid probes for detection and identification of S. sanguinis and other oral bacteria (14, 17, 19, 22, 23, 29, 35, 36, 37).
In the present study, we used a PCR-based approach to develop a DNA probe for identifying S. sanguinis based on a common amplicon present on arbitrarily primed PCR (AP-PCR) profiles. The specificity of this probe was tested against a panel of previously confirmed clinical isolates of S. sanguinis (8, 30). The results of the study suggest that this species-specific probe may serve as a useful tool in the identification of S. sanguinis from clinical samples.
MATERIALS AND METHODS
Bacterial strains.
Sixteen reference strains obtained from the American Type Culture Collection (ATCC; Manassas, Va.) were included in this study (Table 1). Two other reference strains, S. pneumoniae WU2 and Escherichia coli JM109, were obtained from J. Yother at the University of Alabama at Birmingham. S. sanguinis strain ATCC 10556 was selected as a positive control. Strains representing other species served as negative controls. An additional 78 clinical isolates of S. sanguinis that were confirmed to be S. sanguinis strains by biochemical tests and 16S rRNA gene (rDNA) sequence analyses in a previous study (8, 30) were also included. The 78 clinical isolates with unique genotypes were collected from 16 individuals who visited the maternity and pediatric clinics at the Jefferson County Department of Health in Birmingham, Alabama. The details of the sample collection procedure and S. sanguinis isolation have been reported in previous studies (8, 30). Briefly, saliva samples and dental plaques were collected, dispersed, and plated onto MM10-sucrose agar (41). S. sanguinis was initially identified on the basis of its distinct colony morphology on MM10-sucrose medium (8, 25, 41), and its identity was then confirmed by biochemical tests (30). Sixteen (20%) of those clinical S. sanguinis isolates (listed in Table 1) were randomly selected and further confirmed to be S. sanguinis according to their 16S rDNA sequences.
TABLE 1.
Bacterial species and clinical isolates tested in this study
| Bacterial species and strain | Source of isolation |
|---|---|
| Reference species | |
| S. sanguinis ATCC 10556 | Patient with bacterial endocarditis |
| S. oralis ATCC 10557 | Patient with bacterial endocarditis |
| S. oralis ATCC 35037 | Human mouth |
| S. oralis ATCC 9811 | Human mouth |
| S. gordonii ATCC 10558 | Patient with bacterial endocarditis |
| S. cristatus ATCC 49999 | Coronal dental plaque |
| S. mitis ATCC 903 (biovar 2) | Ulcerated sore throat |
| S. parasanguinis ATCC 15911 | Human throat |
| S. pneumoniaea WU2 | Normal mouse serum |
| S. mutans ATCC 25175 | Human carious dentine |
| S. salivarius ATCC 7073 | Patient with acute articular rheumatism |
| S. sobrinus ATCC 33478 | Human dental plaque |
| S. ratti ATCC 19645 | Caries lesion in rat |
| S. vestibularis ATCC 49124 | Human oral cavity |
| Lactobacillus acidophilus ATCC 4356 | Human mouth |
| Actinomyces naeslundii ATCC 12104 | Human sinus |
| E. coli JM109 | |
| S. sanguinis clinical isolatesb | |
| UASa14 | 18-mo-old boy |
| UASa22 | 24-mo-old girl |
| UASa25 | 24-mo-old boy |
| UASa30 | 24-mo-old boy |
| UASa33 | 18-mo-old boy |
| UASa38 | 24-mo-old boy |
| UASa39 | 15-mo-old boy |
| UASa42 | 18-mo-old girl |
| UASa43 | 24-mo-old boy |
| UASa44 | 18-mo-old boy |
| UASa49 | 18-mo-old girl |
| UASa51 | 24-mo-old girl |
| UASa56 | 24-mo-old boy |
| UASa57 | 24-mo-old boy |
| UASa70 | 15-mo-old girl |
| UASa88 | 24-mo-old girl |
Genomic DNA isolation.
Genomic DNA was isolated from overnight cultures grown in Todd-Hewitt broth at 37°C in an anaerobic chamber (85% N2, 10% CO2, 5% H2) with a commercially available DNA extraction kit (Wizard Genomic DNA Purification Kit; Promega Corp. Madison, Wis.), according to the instructions of the manufacturer. Purified DNA was dissolved in 10 mM Tris-HCl buffer containing 1 mM EDTA (pH 8.0); the final concentrations were adjusted spectrophotometrically to 50 μg/ml.
AP-PCR experiments.
AP-PCR was performed with all DNA samples by previously described methods (22, 23, 30). A total of 40 commercially available single-stranded 10-mer oligonucleotide primers (Kit A and Kit B; Operon Technologies, Inc., Alameda, Calif.) were screened for their suitabilities in differentiating S. sanguinis strains from non-S. sanguinis strains. Because primer OPA-02 (5′-TGCCGAGCTG-3′) was able to produce discriminative amplification patterns (data not shown), it was selected for use in this study. All amplification reactions were conducted in a thermal cycler (GeneAmp 2400; Applied Biosystems, Foster City, Calif.) with a total volume of 50 μl containing 1× PCR buffer (10 mM Tris-HCl, 50 mM KCl [pH 8.3]), 200 μM each nucleotide, 100 pmol of primer OPA-02, 2.5 U of Taq polymerase, 3.5 mM MgCl2, and 50 ng of purified template DNA. The temperature profile was 40 cycles at 94°C for 1 min, 36°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. The resulting AP-PCR amplicons were separated on a 1.5% agarose gel in TBE (Tris-borate-EDTA) buffer and stained with an ethidium bromide solution (1 μg/ml). The final images of the gels were captured with a digital camera and saved in the tagged image file format for further comparisons.
DNA cloning and sequencing.
DNA amplification demonstrated that all S. sanguinis strains tested contained a 1,653-bp fragment (Fig. 1). This fragment was excised from the agarose gel and eluted by using the QIAquick gel extraction kit (QIAGEN Inc., Santa Clarita, Calif.). The DNA fragment was cloned (TA cloning kit; Invitrogen, Carlsbad, Calif.) and transformed into E. coli, and the insert was sequenced in both directions. A sequence similarity search of the nonredundant GenBank database was preformed by using BLASTN program (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Md.) (2). Putative open reading frames (ORFs) were identified by using the ORF Finder program (National Center for Biotechnology Information).
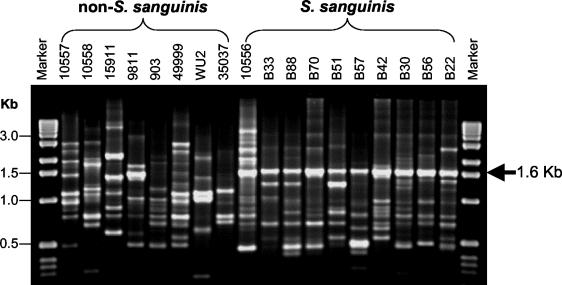
FIG. 1.
AP-PCR fingerprint profiles generated from S. sanguinis, other oral mitis group species, and S. sanguinis clinical isolates. AP-PCR results were obtained by amplification of genomic DNA with primer OPA-02. A 1.6-kb amplicon was observed for all S. sanguinis strains. Other reference strains of the mitis group showed different AP-PCR patterns and the absence of a 1.6-kb amplicon. The non-S. sanguinis strains tested were as follows: 10557, S. oralis; 10558, S. gordonii; 15911, S. parasanguinis; 9811, S. oralis; 903, S. mitis; 49999, S. cristatus; WU2, S. pneumoniae; 35037, S. oralis.
Generation of DNA-based probes and dot hybridization.
The results of the BLASTN search followed by the identification of ORFs with the ORF Finder revealed that the 1,653-bp amplicon contained three regions: portions of two housekeeping genes and an intergenic region. Accordingly, three DNA probes were designed for PCR amplification of these three regions, as illustrated in Fig. 2. The sequences and positions of the primers used to construct the probes are listed in Table 2. The three PCR products were labeled with digoxigenin, and dot hybridization was conducted according to the instructions of the manufacturer (Digoxigenin-High Prime DNA Labeling and Detection Starter Kit I; Roche Molecular Biochemicals, Indianapolis, Ind.). Briefly, heat-denatured chromosomal DNA (1 μg) was applied to a positively charged nylon membrane through the wells of Bio-Dot apparatus (Bio-Rad Laboratories, Hercules, Calif.). The filter was baked at 120°C for 30 min and then hybridized with a digoxigenin-labeled probe at 68°C (no formamide) or at 42°C (containing 50% formamide), washed at 68°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate (highly stringent conditions), and visualized by colorimetric detection with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate.
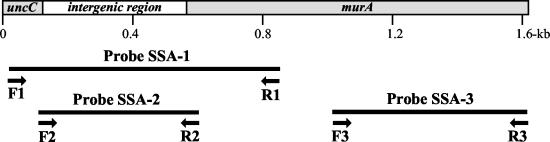
FIG. 2.
Locus and composition of DNA-based probes used for dot hybridization. The three probes were designed on the basis of the sequence of a 1,653-bp fragment from strains of S. sanguinis. The sequence of probe SSA-1 comprised a portion of the first ORF (uncC gene) and the entire intergenic region, the sequence of probe SSA-2 comprised only the intergenic region, and the sequence of probe SSA-3 comprised an internal region of the second ORF (murA gene).
TABLE 2.
Sequences and positions of primers for PCR-based probe construction
| Probe | Primer sequence | Sequence position |
|---|---|---|
| SSA-1 | F1: 5′-GATTGACCAAGAACGCCGGGCT-3′ | 36-57 |
| R1: 5′-CCTCCTCAGACAAAGGCTGGGTGGCAT-3′ | 841-815 | |
| SSA-2 | F2a: 5′-GAAGCCATTTTGCCTAGATTGATGG-3′ | 112-136 |
| R2: 5′-CCATACCGATTCCTTACTCTAAATTT-3′ | 586-561 | |
| SSA-3 | F3: 5′-CGAAGCAAAGGCAGAGCGGCTCAAGGG-3′ | 1026-1052 |
| R3: 5′-CGCATGATATCAGAGTGCAACCC-3′ | 1640-1618 |
F2 is 2 nucleotides outside the ATPase coding sequence.
Primer design.
Three sets of primers were designed on the basis of the sequence of the 1,653-bp fragment from S. sanguinis type strain ATCC 10556. The forward primer, primer F1 (5′-GATTGACCAAGAACGCCGGGCT-3′), was derived from nucleotides (nt) 36 to 57 of the fragment. The reverse primer, primer R3 (5′-CGCATGATATCAGAGATGCAACCC-3′), was derived from nt 1618 to 1640 of the fragment. The specificities of the primers for all 20 reference and 78 clinical strains were tested. All amplification reactions were performed by a standardized PCR protocol, as described above, except that the temperature profile consisted of 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. The PCR products were separated in 1% agarose gels and stained with ethidium bromide.
Three clinical isolates (isolates UASa11, UASa22, and UASa33) representing three S. sanguinis biovars (30) were selected for sequencing after the amplification. The 1,653-bp amplicons generated by PCR were isolated with the QIAquick PCR purification kit (QIAGENE, Valencia, Calif.) and sequenced in both directions. The sequences were aligned (MacVector, version 7.1.1; Accelrys, Madison, Wis.) to compare the 1,653-bp sequences of the S. sanguinis strains of different biovars with that of S. sanguinis type strain ATCC 10556.
Nucleotide sequence accession number.
The 1,653-bp sequence from S. sanguinis ATCC 10556 is available in the GenBank database under accession number AF343003. The sequence of the intergenic region of S. sanguinis is available in the GenBank database under accession number AY277586.
RESULTS
The AP-PCR fingerprints demonstrated that a 1.6-kb amplicon could be consistently observed in the PCR profiles of type strain ATCC 10556 and all other strains of S. sanguinis tested (Fig. 1). In contrast, this fragment was not present in the AP-PCR fingerprints of other mitis group streptococcal species. Sequencing of the 1,653-bp DNA fragment revealed two partial ORFs with high degrees of similarity to other Streptococcus species; one was from nt 1 to 111 and the other was from nt 583 to 1653. A search with the BLASTN program found that the region from nt 1 to 111 bore a high degree of similarity (92%) to the distal portion of the proton-translocating ATPase uncC gene of S. sanguinis reported by Quivey et al. (34) (GenBank accession no. AF001955). The second ORF (nt 583 to 1653) closely resembled murA (UDP-N-acetylglucosamine enolpyruvyl transferase) from S. pneumoniae, S. pyogenes, and S. mutans, among others. A putative noncoding intergenic region was found between the two housekeeping genes.
On the basis of the sequence of the 1.6-kb fragment, three digoxigenin-labeled DNA probes were synthesized: the sequence of probe SSA-1 contained the proton-translocating ATPase (uncC) and the entire intergenic region, the sequence of probe SSA-2 contained only the intergenic region, and the sequence of probe SSA-3 contained an internal region of the murA gene. The stringency of hybridization was examined by dot blot hybridization with the genomic DNA of S. sanguinis and the other species listed in Table 1. The results showed that all three probes hybridized to the DNA of the type strain S. sanguinis ATCC 10556 (Fig. 3A to C). Probe SSA-1 (Fig. 3A) and probe SSA-3 (Fig. 3C) weakly hybridized to most of the Streptococcus species, including S. oralis (ATCC 10557), S. gordonii (ATCC 10558), S. oralis (ATCC 9811), S. parasanguinis (ATCC 15911), and S. vestibularis (ATCC 49124) but did not hybridize to actinomyces, lactobacilli, or E. coli.
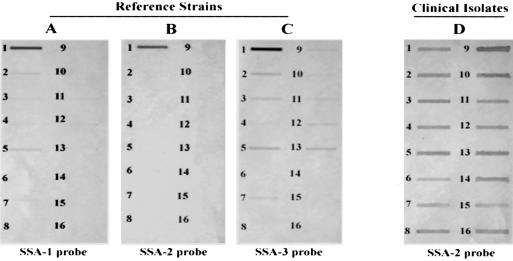
FIG. 3.
Dot blot hybridization shows the specificities of the three probes hybridized with different Streptococcus reference strains as well as with S. sanguinis clinical isolates. Probe SSA-2 was specific for type strain ATCC 10556 (B) and all S. sanguinis clinical isolates (D). SSA-1 and SSA-3 showed different degrees of hybridization to other species (A and C). The bacterial strains tested were as follows: 1, S. sanguinis; 2, S. oralis; 3, S. gordonii; 4, S. cristatus; 5, S. oralis; 6, S. mitis; 7, S. parasanguinis; 8, S. pneumoniae; 9, S. mutans; 10, S. salivarius; 11, S. sobrinus; 12, S. ratti; 13, S. vestibularis; 14, A. naeslundii; 15, L. acidophilus; 16, E. coli JM109.
Probe SSA-2, whose sequence spans the intergenic region, hybridized to the genomic DNA of S. sanguinis ATCC 10556 (Fig. 3B) as well as to the genomic DNA of all clinical S. sanguinis isolates (Fig. 3D). No cross-hybridization was observed with other representative isolates of common oral streptococci, indicating a high degree of specificity of probe SSA-2 to S. sanguinis. An extensive search of the GenBank database with the probe SSA-2 sequence failed to reveal similarities to any DNA, with E value greater than 0.2. The ORF Finder suggested the presence of a putative ORF within the intergenic region (nt 285 to 515) which resembled a histidine kinase from S. pneumoniae and S. pyogenes and a hypothetical protein from S. pyogenes, S. mutans, and S. agalactiae (E value less than 2e-05). Nonetheless, the identity between the theoretical protein sequences of the probe and those predicted from genome sequences did not exceed 31%, and the nucleotide match was even less. Indeed, the nucleotide sequence of the intergenic region bore little or no similarity to sequences in the GenBank and failed to hybridize to probe SSA-2, whose sequence contains the entire intergenic region.
A set of primers (primers F1 and R3) was designed to test whether the presence of the 1.6-kb fragment could be used to identify strains of S. sanguinis. The results showed that only S. sanguinis isolates (78 strains) exhibited the 1,653-bp fragment on 1% agarose gels, as predicted, and no PCR products were detected among the 20 other species strains tested (data not shown). Furthermore, a specific set of PCR primers (primers F2 and R2) was designed to amplify the 475-bp intergenic region, as shown in Fig. 4. DNA from three biovars of S. sanguinis clinical isolates (30) was amplified by PCR with these primers and then sequenced. Although identical in size, the sequences of the three PCR amplicons showed the presence of 12 to 18 polymorphic sites compared with the sequence of ATCC 10556, with a 4% variation in nucleotide sequence found among the three biovars of S. sanguinis isolates (data not shown).
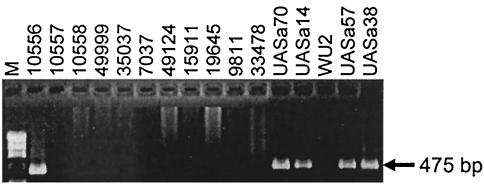
FIG. 4.
Specific amplification of the 475-bp intergenic region from S. sanguinis with primers F2 and R2. The agarose gel shows the presence of the PCR amplicon for ATCC 10556 and the S. sanguinis clinical isolates and the absence of the PCR amplicon for the other non-S. sanguinis strains tested.
DISCUSSION
The accurate identification of S. sanguinis has been problematic. Conventional identification protocols are usually time-consuming and exhibit ambiguities when they are used to differentiate among the members of the mitis group (21, 32). Several new PCR-based methods show promise in identifying isolates to the species level (1, 11, 12, 20, 35, 43). For example, Garnier and coworkers (11) described PCR primers based on internal fragments of the genes encoding d-alanine-d-ligases for the identification of clinically relevant viridans group streptococcal species. Recently, Rudney and Larson (35) developed AP-PCR protocols for the identification of members of the mitis group, including strains of S. sanguinis. However, the use of only the PCR-based typing method proved ineffective for differentiation of the members of the mitis group. Thus, the combination of a PCR-based typing method and DNA-based probe hybridization might increase the overall accuracy of bacterial species delineation (35).
The 16S rDNA locus in bacteria has been widely used as a target for PCR primers or probes for the identification of numerous microorganisms in cerebrospinal fluid, the gastrointestinal tract, and other sources (12, 43). Several studies have included the rDNA locus to identify S. sanguinis from clinical specimens (8, 20, 30, 33, 35). The probes that were used were usually based on variable regions within the 16S rDNA (3), but such regions often differ by only a few base pairs, especially among the members of the mitis group (20). In fact, Jacobs et al. (18) reported that the use of oligonucleotide probes specific for the 16S rRNA sequences of S. anginosus, S. constellatus, and S. intermedius resulted in the reaction of a large number of strains with both the S. constellatus- and the S. intermedius-specific probes. Among the members of the mitis group, the degrees of similarity, based on comparison of 16S rDNA sequences, between the species ranged from 96 to 99% (20), indicating an increased risk of false-positive results in hybridization reactions. Therefore, the development of a highly sensitive and specific DNA-based probe assay for the identification of S. sanguinis adds a potentially valuable new tool for the early detection and identification of S. sanguinis infection and colonization. Similar approaches have previously been reported by other investigators and successfully applied to the species identification of several microorganisms, such as Prevotella, Porphyromonas, Legionella, Candida, and Bacteroides (13, 14, 28, 40, 44).
In our study, we demonstrated that a specific PCR-generated probe and a set of primers could identify S. sanguinis from a subset of clinical isolates previously suggested to be members of the S. sanguinis complex. Although our findings show that the specifically designed probe SSA-2 possessed accuracy in discriminating S. sanguinis from the mitis group, further research is needed to determine whether the S. sanguinis-specific PCR probe or primer set described here could yield both a high degree of sensitivity and a high degree of specificity in detecting S. sanguinis from whole plaque samples containing hundreds of different phylotypes or species. One strategy would be to examine plaque samples from pre- and postdentate infants for S. sanguinis, as S. sanguinis has been shown to colonize the oral cavity before the emergence of teeth (6, 8, 24, 38), and then to monitor the oral cavities of infants for colonization with S. sanguinis. This, in turn, might then be used to predict the time to colonization with the mutans group of streptococci (8) and, perhaps, the risk for dental caries (7, 9, 24). Incorporation of a signature sequence unique to S. sanguinis into a microarray-type or reverse checkerboard assay (4, 31, 42) might be a convenient way to assay plaque samples for the presence or absence of S. sanguinis. Also noteworthy is the fact that the presence of S. sanguinis has been shown to be associated with health in periodontal diseases (16), suggesting that its activity is antagonistic against periodontopathogens, similar to that against mutans group streptococci shown previously (8, 16, 39).
In conclusion, because the PCR-generated DNA probes can precisely identify S. sanguinis at the species level, application of these species-specific DNA markers will provide valuable tools for the early detection of S. sanguinis colonization and facilitate epidemiological study of its interaction with other oral microbes associated with caries progression and periodontal diseases.
Acknowledgments
This study was supported by research grants DE-11147 and DE-09082 from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
REFERENCES
- 1.Alam, S., S. R. Brailsford, R. A. Whiley, and D. Beighton. 1999. PCR-based methods for genotyping viridans group streptococci. J. Clin. Microbiol. 37:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beighton, D., J. M. Hardie, and R. A. Whiley. 1991. A scheme for the identification of viridans streptococci. J. Med. Microbiol. 35:367-372. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson, J., H. Grahnen, G. Jonsson, and S. Wikner. 1970. Establishment of Streptococcus sanguis in the mouths of infants. Arch. Oral Biol. 15:1143-1148. [DOI] [PubMed] [Google Scholar]
- 7.Caufield, P. W., G. R. Cutter, and A. P. Dasanayake. 1993. Initial acquisition of mutans streptococci by infants: evidence for a discrete window of infectivity. J. Dent. Res. 72:37-45. [DOI] [PubMed] [Google Scholar]
- 8.Caufield, P. W., A. P. Dasanayake, Y. Li, Y. Pan, J. Hsu, and J. M. Hardin. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Stoppelaar, J. D., J. Van Houte, and O. Backer Dirks. 1969. The relationship between extracellular polysaccharide-producing streptococci and smooth surface caries in 13-year-old children. Caries Res. 3:190-199. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, C. W., J. Heath, K. K. Hampton, and F. E. Preston. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179-182. [DOI] [PubMed] [Google Scholar]
- 11.Garnier, F., G. Gerbaud, P. Courvalin, and M. Galimand. 1997. Identification of clinically relevant viridans group streptococci to the species level by PCR. J. Clin. Microbiol. 35:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillot, E., and C. Mouton. 1996. A PCR-DNA probe assay specific for Bacteroides forsythus. Mol. Cell. Probes 10:413-421. [DOI] [PubMed] [Google Scholar]
- 14.Guillot, E., and C. Mouton. 1997. PCR-DNA probe assays for identification and detection of Prevotella intermedia sensu stricto and Prevotella nigrescens. J. Clin. Microbiol. 35:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herzberg, M. C., K. Gong, G. D. MacFarlane, P. R. Erickson, A. H. Soberay, P. H. Krebsbach, G. Manjula, K. Schilling, and W. H. Bowen. 1990. Phenotypic characterization of Streptococcus sanguis virulence factors associated with bacterial endocarditis. Infect. Immun. 58:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillman, J. D., S. S. Socransky, and M. Shivers. 1985. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch. Oral Biol. 30:791-795. [DOI] [PubMed] [Google Scholar]
- 17.Ida, H., T. Igarashi, A. Yamamoto, N. Goto, and R. Sasa. 1999. A DNA probe specific to Streptococcus sobrinus. Oral. Microbiol. Immunol. 14:233-237. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, J. A., C. S. Schot, A. E. Bunschoten, and L. M. Schouls. 1996. Rapid species identification of “Streptococcus milleri” strains by line blot hybridization: identification of a distinct 16S rRNA population closely related to Streptococcus constellatus. J. Clin. Microbiol. 34:1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-25. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 21.Kilian, M., L. Mikkelsen, and J. Henrichsen. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended descriptions of Streptococcus sanguis, Streptococcus oralis, and Streptococcus mitis. Int. J. Syst. Bacteriol. 39:471-484. [Google Scholar]
- 22.Li, Y., and P. W. Caufield. 1998. Arbitrarily primed polymerase chain reaction fingerprinting for the genotypic identification of mutans streptococci from humans. Oral Microbiol. Immunol. 13:17-22. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., P. W. Caufield, I. R. Emanuelsson, and E. Thornqvist. 2001. Differentiation of Streptococcus mutans and Streptococcus sobrinus via genotypic and phenotypic profiles from three different populations. Oral Microbiol. Immunol. 16:16-23. [DOI] [PubMed] [Google Scholar]
- 24.Loesche, W. J., S. Eklund, R. Earnest, and B. Burt. 1984. Longitudinal investigation of bacteriology of human fissure decay: epidemiological studies in molars shortly after eruption. Infect. Immun. 46:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loesche, W. J., J. Rowan, L. H. Straffon, and P. J. Loos. 1975. Association of Streptococcus mutans with human dental decay. Infect. Immun. 11:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7:201-216. [DOI] [PubMed] [Google Scholar]
- 27.Loesche, W. J., A. Walenga, and P. Loos. 1973. Recovery of Streptococcus mutans and Streptococcus sanguis from a dental explorer after clinical examinations of single human teeth. Arch. Oral Biol. 18:571-575. [DOI] [PubMed] [Google Scholar]
- 28.Lo Presti, F., S. Riffard, F. Vandenesch, and J. Etienne. 1998. Identification of Legionella species by random amplified polymorphic DNA profiles. J. Clin. Microbiol. 36:3193-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menard, C., P. Gosselin, J. F. Duhaime, and C. Mouton. 1994. Polymerase chain reaction using arbitrary primer for the design and construction of a DNA probe specific for Porphyromonas gingivalis. Res. Microbiol. 145:595-602. [DOI] [PubMed] [Google Scholar]
- 30.Pan, Y. P., Y. Li, and P. W. Caufield. 2001. Phenotypic and genotypic diversity of Streptococcus sanguis in infants. Oral Microbiol. Immunol. 16:235-242. [DOI] [PubMed] [Google Scholar]
- 31.Papapanou, P. N., P. N. Madianos, G. Dahlen, and J. Sandros. 1997. “Checkerboard” versus culture: a comparison between two methods for identification of subgingival microbiota. Eur. J. Oral Sci. 105:389-396. [DOI] [PubMed] [Google Scholar]
- 32.Pearce, C., G. H. Bowden, M. Evans, S. P. Fitzsimmons, J. Johnson, M. J. Sheridan, R. Wientzen, and M. F. Cole. 1995. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J. Med. Microbiol. 42:67-72. [DOI] [PubMed] [Google Scholar]
- 33.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quivey, R. G., Jr., R. C. Faustoferri, W. A. Belli, and J. S. Flores. 1991. Polymerase chain reaction amplification, cloning, sequence determination and homologies of streptococcal ATPase-encoding DNAs. Gene 97:63-68. [DOI] [PubMed] [Google Scholar]
- 35.Rudney, J. D., and C. J. Larson. 1999. Identification of oral mitis group streptococci by arbitrarily primed polymerase chain reaction. Oral Microbiol. Immunol. 14:33-42. [DOI] [PubMed] [Google Scholar]
- 36.Rudney, J. D., E. K. Neuvar, and A. H. Soberay. 1992. Restriction endonuclease-fragment polymorphisms of oral viridans streptococci, compared by conventional and field-inversion gel electrophoresis. J. Dent. Res. 71:1182-1188. [DOI] [PubMed] [Google Scholar]
- 37.Schmidhuber, S., W. Ludwig, and K. H. Schleifer. 1988. Construction of a DNA probe for the specific identification of Streptococcus oralis. J. Clin. Microbiol. 26:1042-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, D. J., J. M. Anderson, W. F. King, J. van Houte, and M. A. Taubman. 1993. Oral streptococcal colonization of infants. Oral Microbiol. Immunol. 8:1-4. [DOI] [PubMed] [Google Scholar]
- 39.Socransky, S. S., A. D. Haffajee, C. Smith, and S. Dibart. 1991. Relation of counts of microbial species to clinical status at the sampled site. J. Clin. Periodontol. 18:766-775. [DOI] [PubMed] [Google Scholar]
- 40.Steffan, P., J. A. Vazquez, D. Boikov, C. Xu, J. D. Sobel, and R. A. Akins. 1997. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J. Clin. Microbiol. 35:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syed, S. A., and W. J. Loesche. 1973. Efficiency of various growth media in recovering oral bacterial flora from human dental plaque. Appl. Microbiol. 26:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner, A. C., P. M. Milgrom, R. Kent, Jr., S. A. Mokeem, R. C. Page, C. A. Riedy, P. Weinstein, and J. Bruss. 2002. The microbiota of young children from tooth and tongue samples. J. Dent. Res. 81:53-57. [DOI] [PubMed] [Google Scholar]
- 43.Tannock, G. W., A. Tilsala-Timisjarvi, S. Rodtong, J. Ng, K. Munro, and T. Alatossava. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl. Environ. Microbiol. 65:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng, L. J., P. R. Hsueh, J. C. Tsai, F. L. Chiang, C. Y. Chen, S. W. Ho, and K. T. Luh. 2000. PCR assay for species-specific identification of Bacteroides thetaiotaomicron. J. Clin. Microbiol. 38:1672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]