Abstract
We analyzed the Bordetella pertussis and Bordetella parapertussis isolates circulating in Saint Petersburg that were collected between 1998 and 2000 and compared them with isolates collected 40 years ago and Russian vaccine strains. The analysis involved serotyping, pulsed-field gel electrophoresis of chromosomal DNA after digestion with XbaI and SpeI, and sequencing of the ptxS1 and prn genes, which encode the S1 subunit of the pertussis toxin and the major adhesin pertactin, respectively. The Russian isolates were classified in five of the six pulsed-field gel electrophoresis groups identified in other European countries. The B. pertussis isolates currently circulating in Saint Petersburg differed from the Russian whole-cell vaccine strains and the isolates collected in the prevaccine era. However, their repartition in the major pulsed-field gel electrophoresis groups was slightly different from that of isolates collected in countries that have had a high level of vaccine coverage for a long time, probably because the level of vaccine coverage in Saint Petersburg has increased only recently, after decreasing until the early 1990s. Most of the B. parapertussis isolates studied were similar to those circulating in France. However, some variants were observed, perhaps because B. parapertussis infections are more common in children in this area.
The introduction of general vaccination against whooping cough in developed countries led to a dramatic decrease in morbidity and mortality among infants and children from this disease, a decrease in the intensity of epidemics, and a shift from irregular episodes to regular synchronous epidemics (20). Furthermore, the transmission of the disease has changed in populations highly vaccinated with an efficient vaccine for several decades. In fact, young children, who were the major group of infected individuals in the prevaccination era, are now protected by the vaccine. However, the number of cases of whooping cough is now increasing again, with an increase in morbidity and mortality among nonvaccinated infants who contract the disease from adults (1, 3, 4). This change seems to be due mainly to the lack of vaccine and natural boosters. However, it may also be due to a decrease in the efficiency of the pertussis vaccine used, a decrease in coverage, or a change in the circulating isolates.
Bordetella pertussis and Bordetella parapertussis are the two agents that cause whooping cough. Little heterogeneity has been detected within these species so far, although the isolates that were circulating during the prevaccine era differ from those that are circulating currently (7, 9, 13, 16, 22, 23). Differences in strains can be detected by typing techniques such as pulsed-field gel electrophoresis (PFGE) and by sequencing the structural genes encoding B. pertussis virulence factors. As described in the recent review by Mattoo et al., these virulence factors include toxins and adhesins (14). The major toxins are pertussis toxin (PT), tracheal cytotoxin, and adenylate cyclase-hemolysin. The major adhesins are filamentous hemagglutinin, fimbriae 2 and 3 (Fim2 and Fim3), and pertactin. B. parapertussis expresses similar factors with the exception of PT.
Polymorphisms have been observed in the B. pertussis genes encoding pertactin and the S1 subunit of PT. However, the role of vaccination in the observed polymorphism in circulating isolates of B. pertussis is not known, mainly because only a few isolates from the prevaccine era could be analyzed. We have recently shown in France that the B. pertussis population seems to evolve continuously every 3 years in correlation with pertussis cycles (23). The heterogeneity observed between isolates is very low and is observed only by the PFGE technique (23).
In this study, we analyzed B. pertussis and B. parapertussis isolates collected in Saint Petersburg, Russia, between 1998 and 2000 and compared them to the vaccine strains currently in use and to some isolates collected during the prevaccine era.
MATERIALS AND METHODS
Isolates.
This study included 61 B. pertussis and 11 B. parapertussis isolates collected in Saint Petersburg and the four vaccine strains currently used in Russia (Table 1). For comparison, we also included the French vaccine strains (18) and the PFGE reference strains (15, 23).
TABLE 1.
Characteristics of the B. pertussis and B. parapertussis isolates and strains analyzed in this study
| Yr of collection | Species | Origin | Fimbriae expressed | PFGE group | ptxS1 allele | prn allele | No. of isolates or strains |
|---|---|---|---|---|---|---|---|
| 2000 | B. pertussis | Isolate | 2 | V | A | 3 | 1 |
| 1999-2000 | B. pertussis | Isolate | 2 | III | NDa | ND | 4 |
| 1998 | B. pertussis | Isolate | 2 | III | ND | 1 | 2 |
| 1998-1999 | B. pertussis | Isolate | 2 | III | A | 3 | 2 |
| 1999-2000 | B. pertussis | Isolate | 2 | III | A | 1 | 7 |
| 1998-2000 | B. pertussis | Isolate | 3 | III | ND | ND | 2 |
| 1989 | B. pertussis | Isolate | 3 | III | A | 1 | 1 |
| 1999 | B. pertussis | Isolate | 3 | III | A | 3 | 1 |
| 1999-2000 | B. pertussis | Isolate | 3 | IVα | ND | ND | 10 |
| 1999-2000 | B. pertussis | Isolate | 3 | IVα | A | 1 | 4 |
| 1999-2000 | B. pertussis | Isolate | 3 | IVα | A | 2 | 5 |
| 1999-2000 | B. pertussis | Isolate | 3 | IVβ | ND | ND | 9 |
| 1999-2000 | B. pertussis | Isolate | 3 | IVβ | A | 2 | 11 |
| 1961 | B. pertussis | Isolate | 2 and 3 | III | B | 1 | 1 |
| 1965 | B. pertussis | Isolate | 2 and 3 | II | D | 1 | 1 |
| 1966 | B. pertussis | Vaccine strain | 2 and 3 | II | D | 1 | 1 |
| 1970 | B. pertussis | Vaccine strain | 2 and 3 | III | B | 1 | 1 |
| 1957 | B. pertussis | Vaccine strain | 2 | III | B | 1 | 1 |
| 1967 | B. pertussis | Vaccine strain | 3 | III | B | 1 | 1 |
| 1998 | B. parapertussis | Isolate | 1.4a-2.8e | 1 | |||
| 1960-1961 | B. parapertussis | Isolate | 1.4a-2.8e | 3 | |||
| 1999-2000 | B. parapertussis | Isolate | 1.4a-2.9b | 7 |
ND, not determined.
Growth conditions.
Bacteria were grown at 36°C for 72 h on Bordet-Gengou agar supplemented with 15% defibrinated sheep blood (BGA) and subcultured on the same medium for 24 h (15). For Western blot analysis, bacteria grown on BGA were harvested in saline, resuspended in Laemmli buffer to a final concentration of 2 × 1010 CFU/ml, and boiled for 15 min (11).
Adenylate cyclase assay.
Adenylate cyclase activity was measured as described previously (10). One unit corresponds to 1 nmol of cyclic AMP formed per min at 30°C and pH 8.
Immune serum.
Groups of 10 4-week-old female BALB/c mice were subcutaneously injected with 10 μg of purified B. pertussis filamentous hemagglutinin, purified detoxified B. pertussis PT, purified B. pertussis adenylate cyclase-hemolysin, or purified B. pertussis pertactin adsorbed onto aluminum hydroxide four times at 4-week intervals. Mice were bled 7 days after the last injection. The specificity of polyclonal antibodies was checked by Western blotting with purified antigens and a whole B. pertussis bacterial suspension.
Electrophoresis and immunoblotting methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with ready-to-use 8 to 25% polyacrylamide gels and the Pharmacia PhastSystem. After electrophoresis, the proteins were transferred onto Hybond C-Super membranes (Amersham). After blocking with buffer containing 5% low-fat milk, the membranes were incubated at 4°C overnight with polyclonal serum diluted 1:1,000. Horseradish peroxidase-labeled sheep anti-mouse immunoglobulins and an enhanced chemiluminescence system were used to reveal bound antibodies (Amersham).
Fimbria expression.
Fim2 and Fim3 production was revealed with monoclonal antibodies as described by Mooi et al. (15).
PFGE.
DNA fingerprinting was carried out by PFGE. The conditions used were as described by Mooi et al. (15). PFGE data were analyzed with the neighbor-joining clustering method on representative profiles (21).
Genotyping.
Polymorphisms have been described in the genes encoding pertactin (prn) and the S1 subunit of PT (ptxS1) (17). Genotyping was limited to these genes, and the conditions used were as described by Mooi et al. (15).
RESULTS
Description of cases and isolates.
This study included 61 B. pertussis isolates, 11 B. parapertussis isolates, and the four B. pertussis strains included in the Russian vaccine. Fifty-eight of the B. pertussis (95%) and eight of the B. parapertussis (72%) isolates were collected between 1998 and 2000 (Table 1). Most of the patients from whom these isolates were collected were children aged between 3 and 13 years. Only one of the patients was an adult, and four were aged between 3 and 10 months. All index cases were symptomatic, but some of the contact cases were asymptomatic when the culture test was carried out and found to be positive.
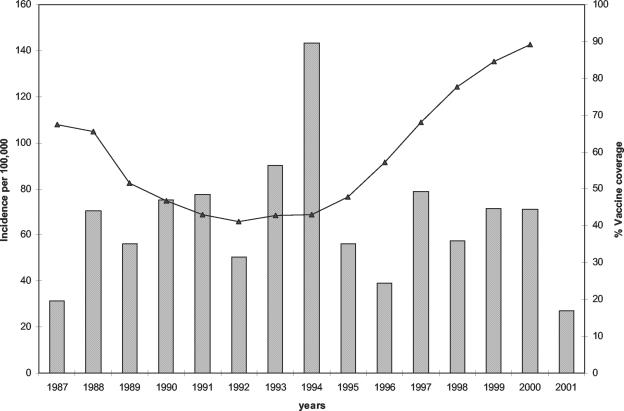
Only 11% of the 66 patients whose vaccine status was known were correctly vaccinated. All of the other patients were either not vaccinated (71%) or incompletely vaccinated or had been vaccinated after the recommended age (18%). Vaccine coverage in the Saint Petersburg region decreased by the late 1980s and started to increase again only recently (Fig. 1), but the incidence of whooping cough is still high. As in other European countries, the incidence was higher in 1993 and 1994 (23).
FIG. 1.
Incidence of whooping cough in Saint Petersburg and vaccine coverage between 1987 and 2001.
All isolates were hemolytic, indicating that they express adenylate cyclase-hemolysin, as confirmed by the detection of adenylate cyclase activity in suspensions of all isolates (data not shown). They all exhibited the bacteriological characteristics classically associated with B. pertussis and B. parapertussis. The microagglutination test was used to analyze all B. pertussis isolates for Fim2 and Fim3 expression. Of the isolates, 26% produced only Fim2, 71% produced only Fim3, and 3% produced both Fim2 and Fim3 (Table 1). The two B. pertussis isolates collected in 1961 and 1965 were the only isolates to express both Fim2 and Fim3. Two of the vaccine strains also expressed both Fim2 and Fim3, one expressed only Fim2, and the other expressed only Fim3 (Table 1).
Filamentous hemagglutinin and pertactin expression was observed in suspensions of all B. pertussis and B. parapertussis isolates by using specific antibodies (data not shown). PT expression was also detected in suspensions of the B. pertussis isolates (data not shown).
PFGE analysis of chromosomal DNA from B. pertussis and B. parapertussis.
PFGE was carried out with chromosomal DNA from the 61 B. pertussis isolates and the four vaccine strains after digestion with either SpeI or XbaI. The profiles obtained were compared to those obtained with chromosomal DNA from the PFGE reference B. pertussis strains, which represent the six major groups (15, 23). The Russian isolates were classified with the neighbor-joining method (21) in the Taxotron software package (P. A. D. Grimont, Taxotron 2000 user's manual, Institut Pasteur, Paris, France). The Russian isolates could be subdivided into PFGE groups that were similar to five of the six previously described groups (II, III, IVα, IVβ, and V) (23) (Fig. 2 and Table 1). None of the Russian isolates was similar to isolates in PFGE group I, which includes the World Health Organization reference strain 18323 and the French isolate CZ collected in 1993.
FIG. 2.
PFGE analysis of Russian isolates. Purified chromosomal DNA was cut with restriction enzymes SpeI and XbaI. Fragments were separated by electrophoresis as described previously (15). Classification was performed by the neighbor-joining clustering method with representatives from each PFGE group.
One of the Russian vaccine strains and one Russian isolate collected in 1965 were classified in PFGE group II, which includes one of the French vaccine strains, Bp1414, and the isolates that were circulating in the prevaccine era in Europe. The three other Russian vaccine strains were included in PFGE group III, as were 20 Russian isolates, 1 collected in 1961, 1 in 1989, 4 in 1998, 6 in 1999, and 8 in 2000 (Table 1). This PFGE group includes the second French vaccine strain, Bp1416, and isolates that were circulating during the pre- and postvaccine eras (23). As in France, PFGE group IV could be subdivided into two subgroups (Fig. 2 and Table 1). PFGE group IVα included 19 Russian isolates, 9 collected in 1999 and 10 collected in 2000. PFGE group IVβ included 20 Russian isolates, 3 collected in 1999 and 17 collected in 2000. PFGE group V included only one isolate (Fig. 2, and Table 1). As observed previously, isolates collected from contact cases clustered in the same PFGE groups as those isolated from index cases.
All isolates included in PFGE groups IVα and IVβ expressed Fim3 (Table 1), as did the French isolates (23), whereas all isolates from PFGE group II expressed both Fim2 and Fim3. Most of the isolates in PFGE group III expressed Fim2.
Polymorphisms in pertactin gene and in S1 subunit of pertussis toxin gene.
We sequenced the ptxS1 and prn genes from selected isolates from each of the five PFGE groups described above and from the vaccine strains. Three pertussis toxin alleles, ptxS1A, ptxS1B, and ptxS1D, were identified among the Russian B. pertussis isolates, similar to those previously observed in Europe. The ptxS1D allele was expressed by one of the vaccine strains and by the isolate collected in 1965. As in other European countries, this allele was harbored by isolates circulating during the prevaccine era. All tested isolates collected between 1998 and 2000 expressed the ptxS1A allele, as already observed in Europe. The ptxS1B allele was expressed only by the three other vaccine strains and by one isolate collected in 1961.
Similarly, analysis of the Russian B. pertussis prn genes revealed alleles similar to those observed in Europe. All of the vaccine strains expressed the type 1 allele, as did the isolates collected in the 1960s and some of the isolates belonging to PFGE groups II and III. Isolates belonging to PFGE group IVα or IVβ expressed the type 2 pertactin allele, as observed before. Previous studies showed that isolates expressing the prn3 allele were always in PFGE group V (23). It was also the case in this analysis for one isolate, but the others were associated with PFGE type III (Table 1).
We also sequenced the repeated regions of the prn gene in the Russian B. parapertussis isolates. We compared the deduced amino acid sequences of the two repeated regions (I and II) with those of the equivalent regions in previously described human B. parapertussis isolates (2, 12). The sequences of 7 of the 11 Russian B. parapertussis isolates were identical to those of previously described sequences, which comprise the unique type found so far (2). This type is 1.4a/2.9b, according to the classification proposed by K. Register for B. bronchiseptica pertactin, which could be applied to B. parapertussis pertactin (19). Surprisingly, four B. parapertussis isolates harbored a prn gene with a different region II. No variation was observed in region I. According to Register's nomenclature, this new type can be designated type 1.4a/2.8e, since region II is characterized by a glycine deletion between the third and fourth Pro-Gln-Pro (PQP) repeat sequence (accession number AJ542533). Three B. parapertussis isolates harboring the new pertactin variant were collected in the 1960s and one was collected in 1998 (Table 1).
DISCUSSION
Despite the introduction of pertussis vaccination in the Saint Petersburg area in 1959, B. pertussis and B. parapertussis are still circulating. Coverage was initially high, but in the late 1980s it decreased, as in other countries, because of reactions to the vaccine. Coverage is now increasing again (Fig. 1). The recommended vaccination schedule is primary vaccination with three injections at 3, 4, and 5 months of age, followed by a booster at 16 to 18 months. If a child is not vaccinated on time, he can be vaccinated before the age of 4 years. In this case, the recommended schedule is still a primary vaccination with three injections at 1-month intervals and a booster vaccination 12 months after the third injection.
Most of the isolates collected between 1998 and 2000 were collected from unvaccinated children aged between 3 and 13 years. This is consistent with a low level of vaccine coverage in the preceding period (5). In countries with a high level of coverage, such as France, most isolates are collected from infants or adolescents, and isolates are rarely collected from children (1, 23). However, the isolates collected in Russia were quite similar to those found in France. Of the seven fully vaccinated patients, only two children, 3 and 6 years old, were index cases, and five cases were contact cases. These contact cases were carrying the bacteria but with no or only mild clinical symptoms.
The vaccine used is a whole-cell vaccine composed of four different B. pertussis strains. One of the strains is identical to one of the two French vaccine strains, and the other three are similar to the second French vaccine strain (18, 23). The four strains all express the major toxins and adhesins.
The isolates collected in the 1960s and in 1989 are similar to isolates circulating in other European countries and the United States at that time. The isolates circulating between 1998 and 2000 were similar to those circulating in other countries with vaccination programs. However, although isolates from PFGE groups III and IVα have practically disappeared from France, some are still circulating in Saint Petersburg. However, as in France, group IVβ isolates were predominant in Saint Petersburg in 2000.
As in all countries with vaccination programs, all isolates collected between 1998 and 2000 were ptxS1 type A and prn type 2 or 3. It is noteworthy that no isolate expressing prn type 4 was found, although such strains are circulating in Finland and Poland (6, 16).
Thirteen percent of the isolates collected from children with the clinical symptoms of pertussis were B. parapertussis isolates. These data are similar to those obtained in Finland (8), where B. parapertussis infections were also detected in children. On the contrary, in France, where the pertussis whole-cell vaccine used is very efficient and the coverage is very high, only 1.4% of children with such symptoms harbor B. parapertussis (23). This raises the question of whether the incidence of B. parapertussis infections is higher in eastern European countries than in other parts of the world. If this is the case, is it due to the low pertussis vaccine coverage in this area compared to that in other countries? Probably not, since a similar incidence is also observed in Finland, where pertussis vaccine coverage was always high (8). Alternatively, it may be due to the efficiency of the pertussis whole-cell vaccine or to the genetic characteristics of the human population.
It is very difficult to answer these questions because B. parapertussis infections are not systematically recorded and the level of surveillance varies from country to country. Seven of the 11 B. parapertussis isolates recovered in Saint Petersburg between 1999 and 2000 carried a prn allele similar to that found in B. parapertussis isolates from France (2). However, the three B. parapertussis isolates collected in the prevaccination era as well as one isolate recovered recently carried a different prn allele. If the incidence of B. parapertussis infections in Russia is higher than in other countries, it is possible that polymorphism will be more common. Alternatively, the polymorphism observed could reflect an evolution of B. parapertussis populations over time, as observed for B. pertussis (23).
In conclusion, as in other vaccinated European populations, the B. pertussis isolates collected between 1998 and 2000 in Saint Petersburg are different from the Russian vaccine strains. This confirms previous results (6, 7, 13, 16, 17, 22, 23). However, these isolates are very similar to those circulating in other European countries with vaccination programs, confirming that B. pertussis does not exhibit a high degree of polymorphism. It should be noted that in Saint Petersburg, as in Finland, the relative number of B. parapertussis infections seems to be higher than in other parts of the world.
Acknowledgments
We thank the L. A. Tarassevich State Control and Standardization Institute; I. Alekseevu for providing the B. pertussis and B. parapertussis isolates collected in the 1960s and vaccine strains; N. N. Zveriakinu, V. V. Suslovu, V. P. Liaminu, and V. I. Vassilievu for providing the B. pertussis and B. parapertussis isolates collected between 1998 and 2000; GlaxoSmithKline laboratories for providing purified PT, filamentous hemagglutinin, and pertactin; and the Centre for Sanitary and Epidemiological Inspection from the Viborgskii district of Saint Petersburg for providing the pertussis vaccine. We also thank C. Boursaux-Eude for help at the beginning of this study and F. Rimlinger and G. Soubigou for technical assistance.
This work was supported by grants from the Institut Pasteur Fondation in Paris and the Pasteur Institute of Epidemiology and Microbiology in Saint Petersburg.
N.K. and V.C. contributed equally to this work and share first-author status.
REFERENCES
- 1.Baron, S., E. Njamkepo, E. Grimprel, P. Begue, J. C. Desenclos, J. Drucker, and N. Guiso. 1998. Epidemiology of pertussis in French hospitals in 1993 and 1994: thirty years after a routine use of vaccination. Pediatr. Infect. Dis. J. 17:412-418. [DOI] [PubMed] [Google Scholar]
- 2.Boursaux-Eude, C., and N. Guiso. 2000. Polymorphism of the repeated regions of pertactin in Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Infect. Immun. 68:4815-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowcroft, N. S., N. Andrews, C. Rooney, M. Brisson, and E. Miller. 2002. Deaths from pertussis are underestimated in England. Arch. Dis. Child. 86:336-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floret, D., Groupe de Pathologie Infectieuse Pédiatrique, and Groupe Francophone de Réanimation et d'Urgence Pédiatrique. 2001. Les décès par infection bactérienne communautaire. Enquête dans les services de réanimation pédiatrique français. Arch. Pédiatr. 8:705-711. [DOI] [PubMed] [Google Scholar]
- 5.Grimprel, E., S. Baron, D. Levy-Bruhl, J. M. Garnier, E. Njamkepo, N. Guiso, and P. Bégué. 1999. Influence of vaccination coverage on pertussis transmission in France. Lancet 354:1699-1700. [DOI] [PubMed] [Google Scholar]
- 6.Gzyl, A., E. Augustynowicz, I. Van Loo, and J. Slusarczyk. 2002. Temporal nucleotide changes in pertactin and pertussis toxin genes in Bordetella pertussis strains isolated from clinical cases in Poland. Vaccine 20:299-303. [DOI] [PubMed] [Google Scholar]
- 7.Hardwick, T. H., P. Cassiday, R. S. Weyant, K. M. Bisgard, and G. N. Sanden. 2002. Changes in predominance and diversity of genomic subtypes of Bordetella pertussis isolated in the United States, 1935 to 1999. Emerg. Infect. Dis. 8:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, Q. S., M. K. Viljanen, H. Arvilommi, B. Aittanen, and J. Mertsola. 1998. Whooping cough caused by Bordetella pertussis and Bordetella parapertussis in an immunized population. JAMA. 280:635-637. [DOI] [PubMed] [Google Scholar]
- 9.Khattak, M. N., R. C. Matthews, and J. P. Burnie. 1992. Is Bordetella pertussis clonal? Br. Med. J. 304:813-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ladant, D., C. Brezin, J.-M. Alonso, I. Crenon, and N. Guiso. 1986. Bordetella pertussis adenylate cyclase. Purification, characterization, and radioimmunoassay. J. Biol. Chem. 261:16264-16269. [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Li, L. J., G. Dougan, P. Novotny, and I. G. Charles. 1991. P. 70 pertactin, an outer-membrane protein from Bordetella parapertussis: cloning, nucleotide sequence and surface expression in Escherichia coli. Mol. Microbiol. 5:409-417. [DOI] [PubMed] [Google Scholar]
- 13.Mastrantonio, P., P. Spigaglia, H. van Oirschot, H. G. J. van der Heide, K. Heuvelman, P. Stefanelli, and F. R. Mooi. 1999. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology 145:2069-2075. [DOI] [PubMed] [Google Scholar]
- 14.Mattoo, S., A. K. Foreman-Wykert, P. A. Cotter, and J. F. Miller. 2001. Mechanisms of Bordetella pathogenesis. Front. Biosci. 6:1. [DOI] [PubMed] [Google Scholar]
- 15.Mooi, F. R., H. Hallander, C. H. Wirsing von Köning, B. Hoet, and N. Guiso. 2000. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur. Clin. Infect. Dis. J. 19:174-181. [DOI] [PubMed] [Google Scholar]
- 16.Mooi, F. R., H. Qiushui, H. van Oirschot, and J. Mertsola. 1999. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect. Immun. 67:3133-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. J. van der Heide, W. Gaastra, and R. J. L. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Njamkepo, E., F. Rimlinger, S. Thiberge, and N. Guiso. 2002. Thirty-five years' experience with the whole-cell pertussis vaccine in France: vaccine strains analysis and immunogenicity. Vaccine 20:1290-1294. [DOI] [PubMed] [Google Scholar]
- 19.Register, K. B. 2001. Novel genetic and phenotypic heterogeneity in Bordetella bronchiseptica pertactin. Infect. Immun. 69:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohani, P., D. J. D. Earn, and B. T. Grenfell. 1999. Opposite patterns of synchrony in sympatric disease metapopulations. Science 286:968-971. [DOI] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. A new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.van Loo, I. H. M., H. G. J. van der Heide, N. J. D. Nagelkerke, J. Verhoef, and F. R. Mooi. 1999. Temporal trends in the population structure of Bordetella pertussis during 1949-1996 in a highly vaccinated population. J. Infect. Dis. 179:915-923. [DOI] [PubMed] [Google Scholar]
- 23.Weber, C., C. Boursaux-Eude, G. Coralie, V. Caro, and N. Guiso. 2001. Polymorphism of Bordetella pertussis isolates circulating in the last ten years in France, where a single effective whole-cell vaccine has been used for more than thirty years. J. Clin. Microbiol. 39:4396-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]