Abstract
An analysis of the expression and activity of silicon transporters (SITs) was done on synchronously growing cultures of the diatom Thalassiosira pseudonana to provide insight into the role these proteins play in cellular silicon metabolism during the cell cycle. The first SIT-specific polyclonal peptide antibody was generated and used in the immunoblot analysis of whole-cell protein lysates to monitor SIT protein levels during synchronized progression through the cell cycle. Peaks in SIT protein levels correlated with active periods of silica incorporation into cell wall substructures. Quantitative real-time PCR on each of the three distinct SIT genes (TpSIT1, TpSIT2, and TpSIT3) showed that mRNA levels for the most highly expressed SIT genes peaked during the S phase of the cell cycle, a period prior to maximal silicon uptake and during which cell wall silicification does not occur. Variations in protein and mRNA levels did not correlate, suggesting that a significant regulatory step of SITs is at the translational or posttranslational level. Surge uptake rates also did not correlate with SIT protein levels, suggesting that SIT activity is internally controlled by the rate of silica incorporation. This is the first study to characterize SIT mRNA and protein expression and cellular uptake kinetics during the course of the cell cycle and cell wall synthesis, and it provides novel insight into SIT regulation.
Silicon is an important element in biology, from bacteria to humans (7). The hydrated form of silicon, called silicic acid, is considered an important nutrient for plant growth (23, 54), and silica, the polymerized form of silicon, is used by certain plants for rigidity, fungal resistance, and defense against grazers. In animals, silicon has a wide range of systemic effects (5) in addition to being essential for proper bone and collagen formation (12, 57). Despite the importance of silicon to life on Earth, the molecular details of biological interactions with silicon and regulatory mechanisms are poorly understood. One of the largest groups of silicifying organisms is diatoms, unicellular, eukaryotic phytoplankton that use silica as a cell wall material. These organisms are found predominantly in aquatic environments but are capable of living in soils and ice. Diatoms play a dominant role in silicon biogeochemistry (49, 63), and because they are estimated to contribute 20% of global primary production (49), they play an important role in the global carbon cycle. Because most diatom species have an obligate silicon requirement for growth (19) and naturally process large amounts of silicon, they are an excellent model system for investigations into biological interactions with silicon.
The silicified diatom cell wall, or frustule, is composed of two overlapping halves, with the upper half called the epitheca and the lower half the hypotheca. Thecae consist of a valve, the species-specific structure capping each end, and girdle bands, a series of overlapping siliceous strips extending on the sides and in the region overlapping the two thecae. Vegetative cell division in certain diatom species begins with the mother cell expanding by synthesizing girdle bands (52). Cytokinesis follows, and on adjacent areas of the two daughter cell protoplasts (still contained within the mother cell), new valves are formed. Silica polymerization occurs within an organelle called the silica deposition vesicle, bounded by a membrane called the silicalemma (18, 53, 56). Once the valve is completely formed, it is exocytosed and the daughter cells separate. This intimate connection between cell wall synthesis and the cell cycle results in a tight coupling of silicon metabolism and cell division.
In diatoms, silicon is taken up from the environment predominantly as silicic acid (20). Although the average oceanic concentration of silicic acid is 70 μM, in surface waters, where diatoms are most common, levels can be less than 10 μM (63). In contrast, intracellular concentrations of silicic acid can be several hundred millimolar depending on the species (45); therefore, diatoms must posses an efficient uptake system to overcome this 1,000-fold difference. Data suggest that silicon uptake in diatoms follows Michaelis-Menten saturation kinetics with Ks values between 0.2 and 7.7 μM and the maximum rate of uptake ranging from 1.2 to 950 fmol Si cell−1 h−1 (6, 39, 40, 45, 59, 60, 64). The coupling of the diatom cell cycle and silicon metabolism (9, 13, 17, 55) has an effect on transport. Rates of silicon uptake vary during synchronized growth of cultures, suggesting that silicon uptake is cell cycle dependent (59), which has led to the understanding that uptake parameters measured for exponentially growing cultures are underestimates because cells are at different stages of the cell cycle and not necessarily utilizing maximum uptake rates (9).
Chemostat studies monitoring silicon uptake have revealed three modes of uptake: surge uptake, externally controlled uptake, and internally controlled uptake (15, 16). Surge uptake occurs upon the initial addition of silicon to silicon-starved cells, with uptake rates maximal during this time. Externally controlled uptake occurs when extracellular levels of silicon are low and the rate of uptake is controlled by the external substrate concentration. In internally controlled uptake, the rate of silica deposition into the cell wall is proposed to control the rate of uptake (15, 16). On longer time scales (hours), uptake is largely internally controlled (15, 16, 30). On shorter time scales (minutes), silicon transport is a dynamic process, involving both uptake and efflux (46, 60).
Diatom silicon transporters (SITs), first identified in the marine pennate diatom Cylindrotheca fusiformis (32, 34), are membrane-associated proteins that directly interact with and transport silicic acid (34). SITs are a novel family of transporters with no known homologs, although a silicon transporter in rice homologous to aquaporins, but with no homology to diatom SITs, was recently described (42). Five SIT genes (CfSIT1-5) in Cylindrotheca fusiformis were identified, each with a distinct level or pattern of mRNA expression during cell wall synthesis. Cylindrotheca fusiformis SITs are predicted to contain 10 transmembrane segments, an intracellular N terminus, and an intracellular C-terminal coiled-coil motif, a structure known to play a role in protein-protein interactions. Genome sequencing of the centric diatom Thalassiosira pseudonana led to the identification of three distinct SIT genes (TpSIT1-3). Similar to CfSITs, TpSITs are predicted to contain 10 transmembrane segments, with intracellular N and C termini. However, they are not predicted to contain a coiled-coil motif (61). Phylogenetic analyses of TpSIT genes suggest that TpSIT1 and TpSIT2 are related through a relatively recent lineage-specific gene duplication and that TpSIT3 is evolving at a rate significantly different from that of TpSIT1 or TpSIT2 (61).
T. pseudonana is an excellent model system for investigations into SIT regulation and function because the exact number of SIT genes is known (3, 61) and a synchronized growth procedure has recently been developed (25, 33). Synchronized growth enables the evaluation of cell cycle effects, providing a unique opportunity to perform a comprehensive investigation into the molecular details of SIT function. This is the first study to investigate molecular and biochemical aspects of SITs and relate that information to silicon uptake rates. Using this approach on synchronously growing cultures of T. pseudonana revealed distinct levels of SIT regulation.
MATERIALS AND METHODS
Culture conditions.
T. pseudonana Hasle et Heimdale clone 3H CCMP1335, Thalassiosira weissflogii (Grunow) Fryxell et Hasle CCMP1336, Skeletonema costatum (Greville) Cleve CCAP1281, Cyclotella meneghianana CCMP338, Bacillaria paxillifer (O. F. Müller) N. I. Hendey 1951 CCAP1006/2, Cylindrotheca fusiformis Reimann et Lewin CCMP343, Ditylum brightwellii (T. West) Grunow ex van Heurck 1883 CCAP1022/1, Navicula pelliculosa (Brébisson) Hilse SAG1050-3 (a freshwater strain referred to in the text as Navicula pelliculosa FW), Navicula pelliculosa (Brébisson et Kuetzing) Hilse CCMP543 (a marine strain referred to in the text as Navicula pelliculosa M), and Phaeodactylum tricornutum Bohlin CCMP1327 were grown in batch culture under continuous illumination with cool white fluorescent lights at 150 μmol m−2 s−1 at 18 to 20°C. N. pelliculosa FW was grown in fresh water-tryptone medium (53). T. pseudonana was grown in sterile artificial seawater medium (ASW) (19) supplemented with biotin and vitamin B12, each at 1 ng liter−1, with or without Bacto tryptone added to 1 g liter−1. All other diatom species were grown in sterile ASW with 0.1% Bacto tryptone [ASW(T)] or f/2 medium made with local seawater filtered through a 0.2-μm-pore-size filter and autoclaved (27, 28).
Synchronized cell growth.
Synchronized growth of T. pseudonana cultures was carried out as previously described (25, 33). Briefly, exponential-phase cultures of T. pseudonana were harvested by centrifugation and washed with silicon-free ASW(T). Cells were resuspended in silicon-free ASW(T) at 0.8× 106 to 1.0 × 106 cells ml−1 and allowed to incubate in the light with aeration for 24 h in a polycarbonate bottle. After 24 h, a sample was harvested (representing 0 h), and sodium silicate was added to the remaining culture at a final concentration of 200 μM to initiate synchronized progression through the cell cycle. Samples (750 ml) were harvested by centrifugation at 3,000 × g for 12 min in a Composite KA-14.250 rotor every hour, washed in 3.5% (wt/vol) NaCl, and stored at −80°C. The formation of specific cell wall structures was monitored by visualization with rhodamine 123 (Sigma, St. Louis, MO) staining as described previously (25) or with 2-4-pyridyl-5-{[4-(2-dimethylaminoethylaminocarbomoyl)methoxy]-phenyl}oxazole (PDMPO; Molecular Probes, Carlsbad, CA) used at 100 nM in the culture.
mRNA extraction, cDNA synthesis, and quantitative real-time PCR.
Total RNA was isolated using TriReagent (Sigma, St. Louis, MO) as described previously (31). Aliquots of total RNA were subjected to DNase treatment using an RNeasy kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's protocol. cDNA was prepared from equivalent amounts of DNase-treated RNA by using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Quantitative real-time reverse transcription PCR (qRT-PCR) was performed using a LightCycler system and LightCycler DNA master SYBR green I (Roche Applied Science, Indianapolis, IN). Because of the high level of sequence conservation among TpSIT genes, primer sets were designed to amplify different-size fragments and PCR was used to verify that each primer set amplified only the SIT of choice. Primers used were as follows: for TpSIT1, TpSIT1 5′ (5′-AGATGGAAGGGTATTTGACAGAG) and TpSIT1 3′-2 (5′-TATCTTCACATGCATCCTTGGG); for TpSIT2, TpSIT2 5′ (5′-GTCCGTTCAACAAGGCAGAA) and TpSIT2 3′-2 (5′-CATCTTCACGTGCATTCTCGGA); and for TpSIT3, TpSIT3 5′-2 (5′-CATCCGGAGTAGTTACTCGTG) and TpSIT3 3′ (5′-GACGTCCAAGATGCGAAGAG). Standards for qRT-PCR were as previously described (25), using dilutions of T. pseudonana genomic DNA. This resulted in genome equivalent nanogram values for mRNA levels rather than an absolute amount of RNA per cell to avoid possible artifacts generated by using rRNA, whose levels change during the cell cycle (47), as a standard. Equivalent amounts of DNase-treated RNA were used as material for cDNA synthesis (11). Dilutions of cDNA were done to ensure that amplification was in the linear range. PCR amplification of RNA samples not treated with reverse transcriptase did not produce an amplification product, indicating that there was no genomic DNA contamination. qRT-PCR measurements of mRNA levels were done on two independent cultures (biological replicates). For the first culture (denoted Sync1), independent qRT-PCRs with the same set of cDNA samples were performed in triplicate for TpSIT1 and TpSIT2 while a single qRT-PCR was performed for TpSIT3. In the second culture (denoted Sync2), duplicate qRT-PCRs were performed for each SIT. To compare the patterns of mRNA changes for each SIT gene, replicates from each culture were normalized to the mean value for all time points for a given SIT. The mean of the normalized values was plotted along with the standard error. For Sync1, one of the triplicate results for 1 h was removed because it was approximately 2 standard deviations away from the mean.
Anti-TpEL4 production.
The sequence of a 15-residue region corresponding to amino acids 318 to 332 of TpSIT1 (sequence PANERGTSFFGRKTC) was provided to Sigma Genosys (The Woodlands, TX) and used to generate a peptide conjugated to keyhole limpet hemocyanin. This peptide was used as an antigen to generate polyclonal antibodies in rabbits (Sigma Genosys, The Woodlands, TX). The antibody was designated anti-TpEL4 because the peptide corresponds to a region of extracellular loop 4 (61).
SDS-polyacrylamide gel electrophoresis.
During the course of synchronized growth, 13-ml samples were harvested by centrifugation at 16,500 × g in an HB-4 rotor at 0 min, 5 min, and every 30 min thereafter for 8 h. Cell pellets were stored at −80°C until ready for use. Either equal cell numbers or equal protein amounts were analyzed. For equal cell numbers, cells were resuspended in 2× sodium dodecyl sulfate (SDS)-sample buffer (Invitrogen, Carlsbad, CA) and heated to 95°C for 5 min. Thirty microliters of each sample was separated by electrophoresis on 4 to 20% gradient gels (Bio-Rad, Hercules, CA). For experiments in which equal protein concentrations were loaded, 50 μl of 2% SDS was added to each sample and the protein concentration was measured using a DC protein assay kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Ten micrograms of total protein was added to 2× SDS-sample buffer, heated at 95°C for 5 min, and electrophoresed as described above. Protein lysates from species other than T. pseudonana were obtained using the same method described for equal-protein-concentration experiments but samples were electrophoresed on 10% polyacrylamide gels (Bio-Rad, Hercules, CA).
Peptide competition.
The TpEL4 peptide used as the antigen for antibody production was generated and provided by Sigma Genosys (The Woodlands, TX). TpEL4 was reconstituted in 0.2 M boric acid, pH 8, to a concentration of 1 mg/ml. Various concentrations of peptide were preincubated with 2.5 μl of anti-TpEL4 antiserum at 4°C overnight with mixing. This mixture was then used as the primary antibody for immunoblot analysis as described below.
Immunoblot analysis.
Electrophoretically separated proteins were transferred onto polyvinylidene difluoride membrane in blotting buffer containing 25 mM Tris and 192 mM glycine, pH 8.3. Transfer efficiency was monitored by using SeeBlue prestained protein standards (Invitrogen, Carlsbad, CA). Membranes were blocked in 1× TBST, pH 7.5 (25 mM Tris-HCl, 137 mM NaCl, 27 mM KCl, 0.2% [vol/vol] Tween 20), and 3% nonfat dried milk at 4°C overnight with shaking. The membranes were washed three times for 10 min each in 1× TBST and then incubated in blocking buffer containing preimmune or anti-TpEL4 antisera at a 1:5,000 dilution in initial experiments but at a 1:10,000 dilution in subsequent experiments for 1 h at room temperature. After three 10-min washes in 1× TBST, the membranes were incubated in blocking buffer containing horseradish peroxidase-conjugated goat anti-rabbit antibody at a 1:10,000 dilution for 1 h at room temperature. Three additional washing steps were done, and chemiluminescence detection was performed using a SuperSignal West Pico chemiluminescent substrate kit (Pierce, Rockford, IL) and following the manufacturer's protocol.
Three immunoblot analyses were performed on two independent cultures during synchronized cell growth. For two of the immunoblots, equal cell numbers were loaded onto the gel, while for the third analysis, equal total protein concentrations were loaded. Similar results were obtained using either method. Densitometry was done for each immunoblot whereby each blot was scanned using an AlphaImager imaging system (Alpha Innotech Corp., San Leandro, CA) and the relative densitometric units (densitometric units minus background) were determined using AlphaEaseFC (Alpha Innotech Corp., San Leandro, CA). Data were normalized to the mean intensity for all points within a given experiment, and the change relative to the mean was plotted. The normalized average for all experiments was plotted along with the standard error.
Cloning of T. pseudonana SIT genes and heterologous expression in Saccharomyces cerevisiae.
TpSIT1, TpSIT2, and TpSIT3 were amplified by PCR using cDNA as a template and the following primers: EcoRI-TpSIT1, 5′-CCGGAATTCAAAAATGTCTACCGCTGAAATCCAA, and TpSIT1-Xba, 5′-CTAGTCTAGAGGCATCCTCGGCAAGAGCATC; Bam-TpSIT2, 5′-CGCGGATCCAAAAATGTCTTCTGCCGAGGTT, and TpSIT2-Xba, 5′-CTAGTCTAGAAGCCTGCGCGTCAACAGCCTC; and Bam-TpSIT3, 5′-CGCGGATCCAAAAATGCGCGCTCAAAACGATGAA, and TpSIT3-Xba, 5′-CTAGTCTAGACTTCGCCAATCCACTTTCAAC. Products were gel purified and ligated into the yeast expression vector pYES2/CT (Invitrogen, Carlsbad, CA) by using the EcoRI/XbaI site for TpSIT1 and the BamH1/XbaI site for TpSIT2 and TpSIT3. Clones were sequenced by SeqXCel (San Diego, CA) and verified to be in frame.
Saccharomyces cerevisiae INVSc1 (Invitrogen, Carlsbad, CA) was transformed with each construct using the lithium acetate method (26). Saccharomyces cerevisiae was grown in yeast extract-peptone-dextrose medium at 30°C to a density of 2 × 107 cells ml−1 and then harvested by centrifugation at 3,000 × g for 5 min. Cells were washed in sterile water and resuspended at a density of 109 cells ml−1. An aliquot of 100 μl of competent cells was transferred into a 1.5-ml microcentrifuge tube to which 1 μg of each pYES2-TpSIT construct was added, along with 100 μg of herring sperm DNA, 36 μl of 1 M lithium acetate, and 240 μl of polyethylene glycol 3500 at 50% (wt/vol). After 30 min at 42°C, the cells were centrifuged for 30 s at maximum speed, resuspended in 1 ml of water, and plated at various dilutions onto uracil-deficient synthetic medium plates. Plates were incubated at 30°C until colonies were visible (approximately 3 days). Clones were picked and grown in liquid selection medium.
SIT protein expression was induced in yeast by growing cells in the presence of 2% galactose at 30°C. After 24 h in induction medium, cells were harvested by centrifugation at 1,500 × g for 5 min. Protein lysates were generated by resuspending cells to an optical density at 600 nm of 50 in breaking buffer (50 mM sodium phosphate [pH 7.4], 1 mM EDTA, 5% glycerol, and 1 mM phenylmethylsulfonyl fluoride), adding an equal volume of acid-washed glass beads (0.4 to 0.6 mm in size; Sigma-Aldrich, St. Louis, MO), and vortexing four times at 30-s intervals at 4°C. After centrifugation at maximum speed for 10 min, the supernatant was removed and stored at −20°C until ready for use. Proteins were separated by electrophoresis on 10% polyacrylamide gels, and immunoblot analysis was performed as described above.
Measurements of silicon in the growth medium.
Silicon disappearance from the medium was monitored during synchronized cell division to measure silicon uptake and determine the onset of valve synthesis (G2/M). The timing of cellular processes during synchronized growth was determined previously with repeated cultures by observation of rhodamine 123 incorporation coupled with flow cytometric analysis to determine cell cycle stages (33). It was found that valve synthesis correlated to a decrease of silicon in the medium (i.e., increased silicon uptake into the cell); thus, replicates can be compared by measuring silicon in the growth media and determining the timing of valve synthesis. Samples were taken at regular intervals by the centrifugation of cells at 16,500 × g for 5 min. Aliquots of supernatant were removed and stored at −20°C. Silicic acid concentrations were measured using the silicomolybdate assay (58) either as described previously (33) or modified for a 96-well-plate format as follows. Twenty-five microliters of supernatant was added to 100-μl Milli-Q water; 50-μl of molybdate reagent was added, mixed, and allowed to incubate for 10 min at room temperature; and then 75 μl of reducing agent was added and mixed. Samples were incubated for 3 h at room temperature with occasional mixing prior to the measurement of the absorbance at 810 nm on a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA). The standard was sodium hexafluorosilicate (Sigma, St. Louis, MO) used in a range from 0 to 50 μM.
Measurements of surge uptake kinetics.
Silicic acid surge uptake during the course of synchronized growth was measured using 68Ge(OH)4 as a radiotracer for silicic acid (4). We have determined (K. Thamatrakoln and M. Hildebrand, unpublished data) that short-term uptake (2 min) in silicon-replete cultures of T. pseudonana is not saturable, even at extremely high silicon concentrations (>500 μM). The lack of saturability prevents the determination of a Michaelis-Menten Vmax value; therefore, uptake was monitored at 100 μM silicate and the maximum rate of surge uptake at this concentration (V100) was determined. Cells were harvested at 3,000 × g for 5 min in an HB-4 rotor, washed, and resuspended in silicon-free ASW to a concentration of 2.5 × 105 cells ml−1. One ml of cells was added to 100 μl of 0.1 μCi 68Ge(OH)4 and 100 μM silicate (59, 60) and either immediately vacuum filtered (for 0 min; background measurement) through a Millipore Isopore RTTP 1.2-μm-pore-size membrane or incubated for 2 min (surge uptake measurement) and then filtered. Filters were washed with 5 ml of 3.5% NaCl, placed in a gamma vial, and counted using an LKB Wallac 1282 Compugamma CS universal gamma counter (Perkin-Elmer, Wellesley, MA). Triplicate samples for each condition were analyzed. Subtraction of the value for 0 min from that for 2 min gave net uptake of label, and the result was converted into fmol cell−1 h−1, taking into account counting efficiency, dilution effects, and radioactive decay.
RESULTS
Comparison of SIT sequences.
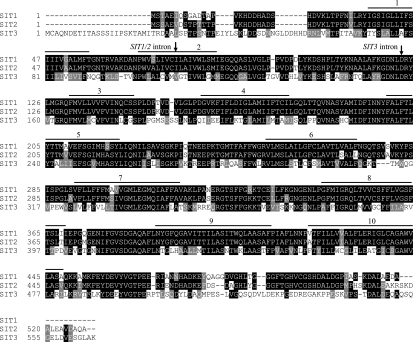
Predicted amino acid sequences of TpSIT1, TpSIT2, and TpSIT3 (61) were aligned and analyzed using CLUSTALW (14, 62) (Fig. 1). Analysis showed 44% amino acid identity and 75% similarity (identical residues plus conserved substitutions) in a comparison of all three SITs. Pairwise comparison between each SIT revealed 88% amino acid identity and 95% similarity between TpSIT1 and TpSIT2. TpSIT1 and TpSIT3 were 46% identical and 74% similar, while TpSIT2 and TpSIT3 were 46% identical and 76% similar. Each TpSIT contained a single intron ranging in size from 97 to 124 bp, located at positions corresponding to amino acid residue 108 in TpSIT1 and TpSIT2 and residue 158 in TpSIT3 (Fig. 1).
FIG. 1.
Predicted amino acid alignment of T. pseudonana SITs. Residue numbers are shown on the left. Black shading indicates identical residues, and gray shading represents conserved substitutions. Black lines and numbers above the alignment denote transmembrane segments as predicted by HMMTOP (65) with a multiple-sequence alignment of 10 SIT sequences (61). Arrows indicate the locations corresponding to introns in each SIT gene.
Characterization of TpEL4 antibody.
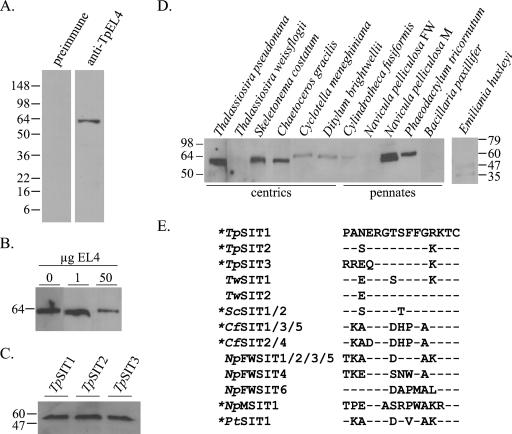
Preimmune and anti-TpEL4 antisera were tested for reactivity to whole-cell protein lysates of T. pseudonana. There was no reactivity with the preimmune serum (Fig. 2A), whereas anti-TpEL4 reacted with a single band at approximately 60 kDa (Fig. 2A), similar to the predicted molecular masses of T. pseudonana SITs. A peptide competition assay demonstrated the specificity of the antibody for this band (Fig. 2B). To determine whether anti-TpEL4 recognized all three T. pseudonana SITs, Saccharomyces cerevisiae was transformed to express individual SITs. Immunoblot analysis of whole-cell lysates showed that anti-TpEL4 recognized each T. pseudomonas SIT (Fig. 2C). To determine whether anti-TpEL4 recognized SITs from other diatom species or phytoplankton, whole-cell protein lysates were analyzed by immunoblot (Fig. 2D). Anti-TpEL4 recognized a protein of approximately 60 kDa in the centric diatoms T. pseudonana, Skeletonema costatum, Chaetoceros gracilis, Cyclotella meneghianana, and Ditylum brightwellii and the pennate diatoms Cylindrotheca fusiformis, Navicula pelliculosa M, and Phaeodactylum tricornutum but did not recognize proteins in lysates from the centric diatom Thalassiosira weissflogii or the pennate diatoms Navicula pelliculosa FW and Bacillaria paxillifer. In addition, there was no reactivity with protein lysate from the coccolithophore Emiliania huxleyi. For those species for which SIT sequences were available (61), the region corresponding to the peptide used as the antigen was compared to TpSIT1 (Fig. 2E). No distinct pattern of sequence conservation among SITs that did or did not cross-react with anti-TpEL4 was apparent. For example, SIT2 of Thalassiosira weissflogii (Fig. 2E) had only one amino acid substitution compared to TpSIT1, but there was no cross-reactivity with anti-TpEL4.
FIG. 2.
Characterization of anti-TpEL4. Molecular mass markers (kDa) are shown on the left or right side of each immunoblot derived from SDS-PAGE separations. (A) Immunoblot of T. pseudonana whole-cell protein lysates using preimmune serum and anti-TpEL4 indicated that the antibody specifically recognized a protein with a size consistent with the predicted size of SITs. (B) Immunoblot of protein lysates probed with anti-TpEL4 preincubated with 0, 1, or 50 μg of TpEL4 peptide demonstrated the specificity of the antibody for the epitope against which it was raised. (C) Immunoblot of Saccharomyces cerevisiae transformed to express TpSIT1, TpSIT2, or TpSIT3 demonstrated that the antibody recognized each of the three T. pseudonana SITs. (D) Immunoblot of 10 μg of total protein lysate from various centric and pennate diatom species and the coccolithophore Emiliania huxleyi. (E) Amino acid alignment of SITs from different diatoms in the region used to generate anti-TpEL4. The sequence of TpSIT1 is shown on the first line. SIT sequences are identified by the initials for genus and species as follows: Tp, Thalassiosira pseudonana; Tw, Thalassiosira weissflogii; Sc, Skeletonema costatum; Cf, Cylindrotheca fusiformis; NpFW, Navicula pelliculosa FW; NpM, Navicula pelliculosa M; and Pt, Phaeodactylum tricornutum. Dashes indicate residues identical to those of TpSIT1. Asterisks indicate proteins from species that cross-reacted with anti-TpEL4.
Analysis of SIT protein expression in synchronized cultures of T. pseudonana.
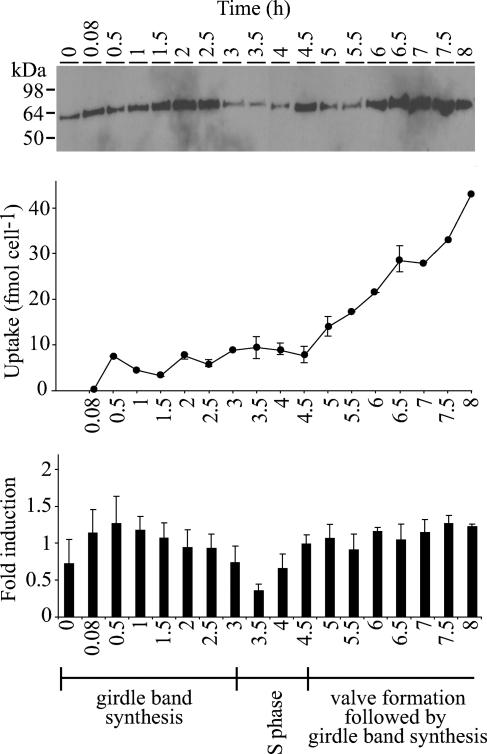
SIT protein was monitored through synchronized progression through the cell cycle by using anti-TpEL4 and immunoblot analysis (Fig. 3, top panel). In the same culture, net silicic acid uptake was determined by the measurement of silicon disappearance from the medium (Fig. 3, middle panel). Results from three immunoblot analyses performed on two independent cultures were compared by normalizing relative densitometric units (see Materials and Methods) and determining changes in protein levels (Fig. 3, bottom panel). This analysis showed that SIT protein was expressed throughout synchronized cell cycle progression but at various levels. SIT protein levels increased approximately 1.5 h after silicon replenishment and then decreased to a minimum at 3.5 h (Fig. 3). At 4.5 h, protein levels again increased and remained high until the end of the experiment. Based on measurements of silicon in the medium, valve synthesis began at 4.5 h (Fig. 3, bottom panel).
FIG. 3.
Analysis of SIT protein levels during synchronized cell growth. Time (h) during synchronized cell growth is shown. The top panel is a representative example of immunoblots of whole-cell protein lysates probed with anti-TpEL4. The middle panel shows net silicic acid uptake during synchronized growth based on results for triplicate samples, with errors bars for standard deviations. The bottom panel shows the mean induction (fold) based on relative densitometric units for three immunoblot analyses performed on two independent replicate cultures (see Materials and Methods). Error bars represent standard errors. Approximate timing of cellular events during synchronized growth is denoted at the bottom.
Analysis of SIT mRNA levels during synchronized growth.
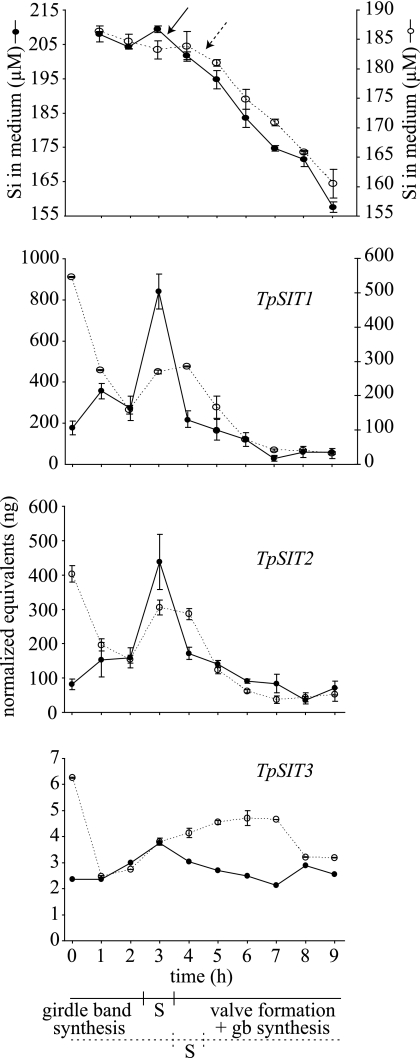
qRT-PCR was used to monitor mRNA levels for each TpSIT gene (Fig. 4) in two independent replicate cultures (designated Sync1 and Sync2). The timing of the initiation of valve synthesis differed slightly in the two replicates as determined by decreased silicate levels in the media (indicating increased cellular uptake). In Sync1, valve synthesis began between 3 and 4 h, and in Sync2, it began between 4 and 5 h (Fig. 4, top panel); from previous results (33), S phase of the cell cycle is known to occur immediately prior to these times (Fig. 4, bottom panel). For TpSIT1 andTpSIT2, mRNA levels in both replicates peaked during S phase of the cell cycle (Fig. 4). For Sync1, a distinct peak was visible at 3 h for both TpSIT1 and TpSIT2, and for Sync2, a broader peak was visible between 3 and 4 h. In both replicates, mRNA levels for TpSIT1 and TpSIT2 dropped gradually following the peak in S phase. A significant difference in the two replicates was mRNA levels at 0 h, which were relatively low in Sync1 and highly induced in Sync2. For TpSIT3 (Fig. 4, bottom panel), mRNA responses differed in the two replicates but generally varied slightly over time with the exception of the large decrease in levels at 1 h in Sync2. Comparing the means of values for all time points for each TpSIT in both replicates indicated that TpSIT1 mRNA levels were approximately twofold higher than those for TpSIT2, and TpSIT3 mRNA levels were substantially lower, approximately 40- to 80-fold lower than those for TpSIT1 and TpSIT2.
FIG. 4.
mRNA levels for TpSIT1, TpSIT2, and TpSIT3 determined by qRT-PCR on two independent synchronized cultures (designated Sync1 and Sync2). mRNA levels are plotted as normalized nanogram equivalents of standards of genomic DNA (see Materials and Methods). In each graph, data from Sync1 are represented by a solid line and closed circles and data from Sync2 are represented by a dotted line and open circles. In the top graph, the means of triplicate measurements of the silicon concentration in the medium are shown with error bars indicating standard errors. Arrows (solid line for Sync1 and dotted line for Sync2) denote a decrease in silicon in the medium, indicating the onset of valve formation, which began 1 h later in Sync2 than in Sync1. The bottom three graphs display qRT-PCR data for each TpSIT gene. Time 0 corresponds to cells harvested prior to the addition of silicate. For Sync1, the means of results for three technical replicates are shown for TpSIT1 and TpSIT2, with the exception of the value for 1 h for TpSIT1, which is the mean of results for two technical replicates (see Materials and Methods). Errors bars represent standard errors. Data from a single qRT-PCR for TpSIT3 in Sync1 are shown. For Sync2, the means of results for two technical replicates for each TpSIT gene are shown along with standard errors. Data for TpSIT1 Sync2 are plotted on the secondary y axis. Note the difference in scales of the y axis for TpSIT3 compared with TpSIT1 or TpSIT2. The approximate timing of cellular events during synchronized growth is shown in the timeline at the bottom, with solid and dotted lines representing Sync1 and Sync2, respectively.
Surge uptake rate through the cell cycle.
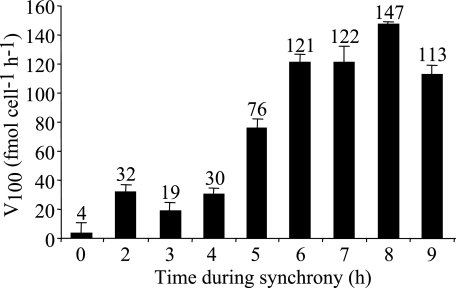
Surge uptake rates during synchronized cell cycle progression were measured using radiolabeled germanic acid as a tracer for silicic acid (Fig. 5). The rate was low at 0 h but increased and was relatively constant between 2 and 4 h, at approximately 30 fmol cell−1 h−1. At 5 h, during the period of valve synthesis, the uptake rate increased and eventually peaked at 8 h at 147 fmol cell−1 h−1. At 9 h, the rate decreased to 113 fmol cell−1 h−1.
FIG. 5.
Maximum rates of surge uptake at 100 μM silicate (V100) during synchronized cell growth. Average values (in fmol cell−1 h−1) from a sample assayed in triplicate are shown for each time point, with error bars showing standard deviations. Numbers above bars are V100 values.
DISCUSSION
SITs were first identified in the late 1990s (32, 34), but until now, tools for an in-depth characterization of protein, mRNA, and uptake activities during the cell cycle were not available. In this study, biochemical, molecular, and physiological tools were used to characterize the patterns of SIT protein and mRNA expression and relate that information to silicon uptake activity. Because of the tight coupling between silicon uptake and the cell cycle, it is only with the ability to synchronize the growth of T. pseudonana cultures that these analyses were possible. Interpretation of the data requires discussion about events occurring during synchronized growth of T. pseudonana, details of which are published elsewhere (33) but are summarized here. T. pseudonana cells starved for silicon for 24 h arrest predominantly in the G1 stage of the cell cycle. Upon silicate replenishment, cells progress through G1 for approximately 3 h while girdle band synthesis occurs. As the cells enter S phase (3 to 4.5 h), girdle band production ceases (33). After S phase, valve synthesis is observed between 4 and 6 h, with the exact timing varying between synchronized cultures. Data from different synchronized cultures can be compared because the timing of valve synthesis can be monitored either by direct visualization by fluorescence microscopy with rhodamine 123- or PDMPO-stained cells or by measurement of a characteristic fivefold increase in net silicic acid uptake over that occurring during girdle band synthesis (33). Upon the completion of valve synthesis, cells separate and additional girdle bands are synthesized. Similar to that of other diatom species (10), synchronized growth of T. pseudonana cultures does not result in all of the cells entrained at the same stage of the cell cycle; typically, 80% are arrested under silicate starvation in G1, with the remaining 20% in G2/M or S phase (33).
In general, the pattern and levels of SIT protein expression correlated with active periods of silica incorporation into cell wall substructures (Fig. 3). Upon silicate replenishment, SIT protein levels increased coincident with girdle band synthesis until 3 h (Fig. 3). During S phase (3 to 4.5 h), when cells are not making silica structures (33), SIT levels were minimal (Fig. 3). Considering that 20% of the cells may not be in S phase at that time, it is possible that SIT protein levels are even lower or undetectable in S-phase cells. During valve and subsequent girdle band synthesis, when silicon demands increase, SIT protein levels increased again (Fig. 3).
SIT protein levels and silicon uptake were not strictly correlated. SIT protein levels were not substantially different between 0.5 to 2.5 h and 5 to 8 h, yet net silicic acid uptake, as measured by the disappearance of silicic acid from the medium (Fig. 3, middle panel), and surge uptake, as measured by short-term uptake of radiolabeled 68Ge(OH)4 (Fig. 5), increased fivefold in the later time period. These observations are consistent with the idea that the abundance of SIT protein is not a primary determinant of net uptake, suggesting that the cell may rely more on the regulation of the activity of SITs than the level of SIT protein. This possibility is consistent with an internally controlled uptake mechanism (15) in which the regulation of uptake is largely dictated by the rate of cell wall silica incorporation. Surge uptake data (Fig. 5) indicated that the cells were capable of taking up 7.5-fold more silicon than they did on longer time scales, also consistent with an internally controlled SIT regulatory mechanism. An alternative explanation for the lack of correlation between SIT protein levels and uptake rates could be because immunoblot analysis measures total SIT protein, it is possible that some SITs measured may not be localized to the plasma membrane. If, as has been demonstrated for glucose transporters (8, 22), SITs cycle between the plasma membrane and intracellular vesicles as part of a regulatory mechanism, changes in uptake rates without a concomitant change in total protein could result.
There was no direct correlation between mRNA and protein levels. mRNA levels for TpSIT1 and TpSIT2 in both replicates peaked during S phase of the cell cycle (Fig. 4), at a time when no silicification occurs (33) and when SIT protein levels were at a minimum (Fig. 3). This indicates that although SIT transcript levels increased, the transcripts were not translated into protein. The accumulation of nontranslated mRNA occurs in other organisms (2) and may enable a cell to respond more rapidly, or target mRNAs more specifically (37), in response to changes in cell cycle events or environmental conditions. In contrast, when SIT protein levels peaked during valve synthesis, mRNA levels decreased, suggesting either increased translational efficiency or an inhibition of protein degradation. These data are consistent with a translational or posttranslational regulatory mechanism over SIT expression.
Interestingly, mRNA levels at 0 h were consistently higher for each TpSIT in Sync2 than for those in Sync1, suggesting that the cells had different responses to silicon starvation. It is common for cells to induce nutrient transporter mRNA levels when starved for a particular nutrient (29, 31). Because diatoms can accumulate intracellular pools of silicon (45), perhaps these pools in the cells in Sync2 were more depleted than those in Sync1, giving rise to higher mRNA levels. More rigorous experiments are needed to test this hypothesis.
mRNA levels for each TpSIT were also measured in a single experiment in which the cells were grown in enough silicate to allow one complete cell division to occur before the cells became silicon starved (data not shown). These conditions may be more representative of natural environmental conditions where cells may experience periods of gradual silicon starvation. The major conclusion from this experiment was that SIT mRNA levels were not strictly controlled by extracellular silicon concentrations, as had been observed in synchronized cultures (Fig. 4). In addition, TpSIT3 mRNA levels were, on average, 30-fold lower than those of TpSIT1 or TpSIT2.
The low levels of TpSIT3 mRNA compared to those of TpSIT1 and TpSIT2 mRNA seen in both synchronized cultures (Fig. 4) and a culture gradually starved for silicon (data not shown) lead us to speculate that TpSIT3 may serve as a silicon sensor in T. pseudonana. In addition to being able to transport silicon, diatoms could also have a mechanism for sensing silicon; for example, to evaluate whether enough extracellular silicon is present to complete a round of valve and girdle band synthesis. Specific members of other transporter families have been shown to function as substrate sensors that regulate the expression of other members in the family through a signal transduction cascade (35, 51). Some known substrate sensors have distinct characteristics that differentiate them from the functional transporters (24, 66). One of these features is the lack of sequence conservation between sensors and other members of the transporter family. In the yeast ammonium transporter family, Mep1 and Mep3 share 80% amino acid identity but are only 39 to 41% identical to the ammonium sensor Mep2 (21, 41, 43, 44). Similarly, the glucose sensors Snf3p and Rgt2p are only 26 to 30% identical to other members of the family (38), while the other members share 64 to 87% identity (36, 38). Structural differences in substrate sensors have also been identified. Snf3p and Rgt2p have an extended C terminus compared to those of the other glucose transporters (51), and the amino acid permease sensor Ssy1p has an N terminus that is 140 amino acids longer and an extra 25- to 35-amino-acid region in a hydrophilic loop (35). Genes encoding Snf3p and Rgt2p are also weakly expressed compared to other family members, with expression levels being approximately 100 to 300-fold lower (48, 51).
TpSIT1 and TpSIT2 shared 88% amino acid identity, but TpSIT3 was only 46% identical (Fig. 1). In addition, the predicted amino acid sequence based on genome data for TpSIT3 suggests a 34-amino-acid extension at the N terminus compared to TpSIT1 and TpSIT2 (Fig. 1). A BLAST search (1) using this sequence did not reveal significant similarity to known proteins, nor were any conserved functional domains identified. SignalP (50) did not predict the presence of a signal peptide cleavage site, suggesting that TpSIT3 may not necessarily be differentially targeted. qRT-PCR data showed that the level of TpSIT3 mRNA was 40- to 80-fold lower than that of TpSIT1 and TpSIT2 during synchronized growth (Fig. 4). These points are consistent with the possibility of TpSIT3 being a silicon sensor, although one alternative hypothesis could be that TpSIT3 plays a specialized role in uptake (e.g., specific intracellular targeting). High levels of expression of TpSIT1 and TpSIT2 suggest that these proteins are active transporters, but amino acid differences between them could result in different affinities or capacities for silicic acid. For example, one could be a high-affinity, low-capacity transporter and the other a low-affinity, high-capacity transporter, allowing the cell to take advantage of a wider range of silicic acid concentrations. Definitively determining whether TpSIT3 is a silicon sensor and whether TpSIT1 and TpSIT2 have different affinities or capacities for silicic acid will require additional investigation.
In conclusion, data obtained in this study indicate that a major regulatory step for SIT expression is at the translational or posttranslational level and that SIT activity is controlled largely by intracellular processes and not by protein levels. Circumstantial data suggest that TpSIT3 may be a silicon sensor, and future work focusing on determining the transport capacities and silicon affinities of SITs should provide further insight into the roles each SIT plays in meeting the overall transport needs of the cell. In addition, the first SIT-specific antibody is now available, which will be integral in future investigations into SIT function and regulation.
Acknowledgments
We thank Luciano Frigeri and Sandra Hazelaar for technical assistance, Brian Palenik for the use of an epifluorescence microscope, and Dori Landry for kindly providing a frozen Emiliania huxleyi cell pellet. We would also like to thank Michael Latz for use of lab space and equipment. Navicula pelliculosa (Brébisson et Kuetzing) Hilse CCMP543 was provided as part of a collaborative project with Sandra Hazelaar and Winfried W. C. Gieskes.
This work was supported by Air Force Office of Scientific Research Multidisciplinary University Research Initiative grant RF00965521.
Instrumentation was purchased with support from a National Science Foundation grant (0115801).
Footnotes
Published ahead of print on 15 December 2006.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 3.Armbrust, E. V., J. A. Berges, C. Bowler, B. R. Green, D. Martinez, N. H. Putnam, S. Zhou, A. E. Allen, K. E. Apt, M. Bechner, M. A. Brzezinski, B. K. Chaal, A. Chiovitti, A. K. Davis, M. S. Demarest, J. C. Detter, T. Glavina, D. Goodstein, M. Z. Hadi, U. Hellsten, M. Hildebrand, B. D. Jenkins, J. Jurka, V. V. Kapitonov, N. Kröger, W. W. Y. Lau, T. W. Lane, F. W. Larimer, J. C. Lippmeier, S. Lucas, M. Medina, A. Montsant, M. Obornik, M. S. Parker, B. Palenik, G. J. Pazour, P. M. Richardson, T. A. Rynearson, M. A. Saito, D. C. Schwartz, K. Thamatrakoln, K. Valentin, A. Vardi, F. P. Wilkerson, and D. S. Rokhsar. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79-86. [DOI] [PubMed] [Google Scholar]
- 4.Azam, F. 1974. Silicic acid uptake in diatoms studied with [68Ge]germanic acid as a tracer. Planta 121:205-212. [DOI] [PubMed] [Google Scholar]
- 5.Bendz, G., and I. Lindqvist (ed.). 1978. The biochemistry of silicon and related problems. Plenum Press, New York, NY.
- 6.Bhattacharyya, P., and B. E. Volcani. 1980. Sodium dependent silicate transport in the apochlorotic marine diatom Nitzschia alba. Proc. Natl. Acad. Sci. USA 77:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birchall, J. D. 1995. The essentiality of silicon in biology. Chem. Soc. Rev. 24:351-357. [Google Scholar]
- 8.Bryant, N., R. Govers, and D. James. 2002. Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3:267-277. [DOI] [PubMed] [Google Scholar]
- 9.Brzezinski, M. A. 1992. Cell-cycle effects on the kinetics of silicic acid uptake and resource competition among diatoms. J. Plankton Res. 14:1511-1539. [Google Scholar]
- 10.Brzezinski, M. A., R. J. Olson, and S. W. Chisholm. 1990. Silicon availability and cell cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 67:83-96. [Google Scholar]
- 11.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 12.Carlisle, E. M. 1981. Silicon in bone formation, p. 69-94. In T. L. Simpson and B. E. Volcani (ed.), Silicon and siliceous structures in biological systems. Springer-Verlag, New York, NY.
- 13.Claquin, P., V. Martin-Jézéquel, J. C. Kromkamp, M. J. Veldhuis, and G. W. Kraay. 2002. Uncoupling of silicon compared with carbon and nitrogen metabolisms and the role of the cell cycle in continuous cultures of Thalassiosira pseudonana (Bacillariophyceae) under light, nitrogen, and phosphorous control. J. Phycol. 38:922-930. [Google Scholar]
- 14.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 15.Conway, H. L., and P. J. Harrison. 1977. Marine diatoms grown in chemostats under silicate or ammonium limitations. IV. Transient response of Chaetoceros debilis, Skeletonema costatum, and Thalassiosira gravida to a single addition of the limiting nutrient. Mar. Biol. 43:33-43. [Google Scholar]
- 16.Conway, H. L., P. J. Harrison, and C. O. Davis. 1976. Marine diatoms grown in chemostats under silicate or ammonium limitation. II. Transient response of Skeletonema costatum to a single addition of the limiting nutrient. Mar. Biol. 35:187-199. [Google Scholar]
- 17.Crawford, R. M. 1981. The siliceous components of the diatom cell wall and their morphological variation, p. 129-156. In T. L. Simpson and B. E. Volcani (ed.), Silicon and siliceous structures in biological systems. Springer-Verlag, New York, NY.
- 18.Crawford, R. M., and A. M. M. Schmid. 1986. Ultrastructure of silica deposition in diatoms, p. 291-314. In B. S. Leadbeater and R. Riding (ed.), Biomineralization in lower plants and animals, vol. 30. The Systematics Society, London, United Kingdom. [Google Scholar]
- 19.Darley, W. M., and B. E. Volcani. 1969. Role of silicon in diatom metabolism: a silicon requirement for deoxyribonucleic acid synthesis in the diatom Cylindrotheca fusiformis Reimann and Lewin. Exp. Cell Res. 58:334-342. [DOI] [PubMed] [Google Scholar]
- 20.Del Amo, Y., and M. A. Brzezinski. 1999. The chemical form of dissolved Si taken up by marine diatoms. J. Phycol. 35:1162-1170. [Google Scholar]
- 21.Dubois, E., and M. Grenson. 1979. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol. Gen. Genet. 175:67-76. [DOI] [PubMed] [Google Scholar]
- 22.Dugani, C. B., and A. Klip. 2005. Glucose transporter 4: cycling, compartments and controversies. EMBO Rep. 12:1137-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epstein, E. 1994. The anomaly of silicon in plant biology. Proc. Natl. Acad. Sci. USA 91:11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 25.Frigeri, L. G., T. R. Radabaugh, P. A. Haynes, and M. Hildebrand. 2006. Identification of proteins from a cell wall fraction of the diatom Thalassiosira pseudonana: insights into silica structure formation. Mol. Cell. Proteomics 5:182-193. [DOI] [PubMed] [Google Scholar]
- 26.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 27.Guillard, R. R., and J. H. Ryther. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 28.Guillard, R. R. L. 1975. Culture of phytoplankton for feed marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY.
- 29.Hildebrand, M. 2005. Cloning and functional characterization of ammonium transporters from the marine diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 41:105-113. [Google Scholar]
- 30.Hildebrand, M. 2000. Silicic acid transport and its control during cell wall silicification in diatoms, p. 171-188. In E. Bäuerlein (ed.), Biomineralization: from biology to biotechnology and medical applications. Wiley-VCH, Weinheim, Germany.
- 31.Hildebrand, M., and K. Dahlin. 2000. Cloning and characterization of nitrate transporter genes from the diatom Cylindrotheca fusiformis. J. Phycol. 36:30. [DOI] [PubMed] [Google Scholar]
- 32.Hildebrand, M., K. Dahlin, and B. E. Volcani. 1998. Characterization of a silicon transporter gene family in Cylindrotheca fusiformis: sequences, expression analysis, and identification of homologs in other diatoms. Mol. Gen. Genet. 260:480-486. [DOI] [PubMed] [Google Scholar]
- 33.Hildebrand, M., L. G. Frigeri, and A. K. Davis. Synchronized growth of Thalassiosira pseudonana (Bacillariophyceae) provides novel insights into cell wall synthesis processes in relation to the cell cycle. J. Phycol., in press.
- 34.Hildebrand, M., B. E. Volcani, W. Gassmann, and J. I. Schroeder. 1997. A gene family of silicon transporters. Nature 385:688-689. [DOI] [PubMed] [Google Scholar]
- 35.Iraqui, I., S. Vissers, F. Bernard, J.-O. de Craene, E. Boles, A. Urrestarazu, and B. Andre. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasahara, T., and M. Kasahara. 2003. Transmembrane segments 1, 5, 7, and 8 are required for high-affinity glucose transport by Saccharomyces cerevisiae Hxt2 transporter. Biochem. J. 372:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kindler, S., H. Wang, D. Richter, and H. Tiedge. 2005. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21:223-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ko, C. H., H. Liang, and R. F. Gaber. 1993. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:638-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewin, J. C. 1954. Silicon metabolism in diatoms. I. Evidence for the role of reduced sulfur compounds in silicon utilization. J. Gen. Physiol. 37:589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewin, J. C. 1955. Silicon metabolism in diatoms. III. Respiration and silicon uptake in Navicula pelliculosa. Can. J. Microbiol. 3:427-433. [DOI] [PubMed] [Google Scholar]
- 41.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, J. F., K. Tamai, N. Yamaji, N. Mitani, S. Konishi, M. Katsuhara, M. Ishiguro, Y. Murata, and M. Yano. 2006. A silicon transporter in rice. Nature 440:688-691. [DOI] [PubMed] [Google Scholar]
- 43.Marini, A.-M., S. Soussi-Boudekou, S. Vissers, and B. André. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marini, A.-M., S. Vissers, A. Urrestarazu, and B. André. 1994. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 13:3456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Jézéquel, V., M. Hildebrand, and M. A. Brzezinski. 2000. Silicon metabolism in diatoms: implications for growth. J. Phycol. 36:821-840. [Google Scholar]
- 46.Milligan, A., D. E. Varela, M. A. Brzezinski, and F. M. M. Morel. 2004. Dynamics of silicon metabolism and silicon isotopic discrimination in a marine diatom as a function of pCO2. Limnol. Oceanogr. 49:322-329. [Google Scholar]
- 47.Mitchison, J. 2003. Growth during the cell cycle. Int. Rev. Cytol. 226:165-258. [DOI] [PubMed] [Google Scholar]
- 48.Neigeborn, L., P. Schwartzberg, R. Reid, and M. Carlson. 1986. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol. Cell. Biol. 6:3569-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson, D. M., P. Tréguer, M. A. Brzezinski, and A. Leynaert. 1995. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 9:359-372. [Google Scholar]
- 50.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 51.Özcan, S., J. Dover, A. G. Rosenwald, S. J. Wölfl, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickett-Heaps, J., A. M. M. Schmid, and L. A. Edgar. 1990. The cell biology of diatom valve formation., p. 1-168. In F. E. Round and D. J. Chapman (ed.), Progress in phycological research, vol. 7. Biopress Ltd., Bristol, United Kingdom. [Google Scholar]
- 53.Reimann, B. E. F., J. C. Lewin, and B. E. Volcani. 1966. Studies on the biochemistry and fine structure of silica shell formation in diatoms. II. The structure of the cell wall of Navicula pelliculosa (Breb.) Hilse. J. Phycol. 2:74-84. [DOI] [PubMed] [Google Scholar]
- 54.Richmond, K. E., and M. Sussman. 2003. Got silicon? The non-essential beneficial plant nutrient. Curr. Opin. Plant Biol. 6:268-272. [DOI] [PubMed] [Google Scholar]
- 55.Schmid, A. M. M. 1994. Aspects of morphogenesis and function of diatom cell walls with implication for taxonomy. Protoplasma 181:43-60. [Google Scholar]
- 56.Schmid, A. M. M., M. A. Borowitzka, and B. E. Volcani. 1981. Morphogenesis and biochemistry of diatom cell walls, p. 63-97. In O. Kiermayer (ed.), Cytomorphogenesis in plants, vol. 8. Springer-Verlag, New York, NY. [Google Scholar]
- 57.Schwarz, K., and D. B. Milne. 1972. Growth-promoting effects of silicon in rats. Nature 239:333-334. [DOI] [PubMed] [Google Scholar]
- 58.Strickland, J. D. H., and T. R. Parsons. 1968. A practical handbook of sea water analysis. Bull. Fish. Res. Board Canada 167:1-311. [Google Scholar]
- 59.Sullivan, C. W. 1976. Diatom mineralization of silicic-acid. I. Si(OH)4 transport characteristics in Navicula pelliculosa. J. Phycol. 12:390-396. [Google Scholar]
- 60.Sullivan, C. W. 1977. Diatom mineralization of silicic acid. II. Regulation of Si(OH)4 transport rates during the cell cycle of Navicula pelliculosa. J. Phycol. 13:86-91. [Google Scholar]
- 61.Thamatrakoln, K., A. J. Alverson, and M. Hildebrand. 2006. Comparative sequence analysis of diatom silicon transporters: towards a mechanistic model for silicon transport. J. Phycol. 42:822-834. [Google Scholar]
- 62.Thompson, J. D., D. R. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tréguer, P., D. M. Nelson, A. J. Van Bennekom, D. J. DeMaster, A. Leynaert, and B. Queguiner. 1995. The silica balance in the world ocean: a reestimate. Science 268:375-379. [DOI] [PubMed] [Google Scholar]
- 64.Tréguer, P., and P. Pondaven. 2000. Silica control of carbon dioxide. Nature 406:358-359. [DOI] [PubMed] [Google Scholar]
- 65.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 66.Van Belle, D., and B. André. 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13:389-398. [DOI] [PubMed] [Google Scholar]