Abstract
Regulated protein degradation is essential for eukaryotic cell cycle progression. The anaphase-promoting complex/cyclosome (APC/C) is responsible for the protein destruction required for the initiation of anaphase and the exit from mitosis, including the degradation of securin and B-type cyclins. We initiated a study of the APC/C in the multinucleated, filamentous ascomycete Ashbya gossypii to understand the mechanisms underlying the asynchronous mitosis observed in these cells. These experiments were motivated by previous work which demonstrated that the mitotic cyclin AgClb1/2p persists through anaphase, suggesting that the APC/C may not be required for the division cycle in A. gossypii. We have now found that the predicted APC/C components AgCdc23p and AgDoc1p and the targeting factors AgCdc20p and AgCdh1p are essential for growth and nuclear division. Mutants lacking any of these factors arrest as germlings with nuclei blocked in mitosis. A likely substrate of the APC/C is the securin homologue AgPds1p, which is present in all nuclei in hyphae except those in anaphase. The destruction box sequence of AgPds1p is required for this timed disappearance. To investigate how the APC/C may function to degrade AgPds1p in only the subset of anaphase nuclei, we localized components and targeting subunits of the APC/C. Remarkably, AgCdc23p, AgDoc1p, and AgCdc16p were found in all nuclei in all cell cycle stages, as were the APC/C targeting factors AgCdc20p and AgCdh1p. These data suggest that the AgAPC/C may be constitutively active across the cell cycle and that proteolysis in these multinucleated cells may be regulated at the level of substrates rather than by the APC/C itself.
Regulated protein degradation ensures linear order in the cell division cycle. Timely protein degradation prevents premature passage through future stages of the cycle and limits slippage back to previous stages once they are completed. A series of positive and negative feedback loops connect cyclin-dependent kinase (CDK)/cyclin complexes to the machinery that targets specific proteins for degradation by the proteosome. This web of interactions culminating in protein destruction gives accuracy to cell division (27, 32, 37).
The anaphase-promoting complex/cyclosome (APC/C) is one of several multisubunit protein complexes that add a polyubiquitin modification to specific protein substrates, which leads to their termination in the proteosome (33, 37). The components of the APC/C include the enzymatic machinery for attaching ubiquitin as well as factors for substrate docking and recognition. There are at least 13 conserved subunits to the eukaryotic APC/C which coordinate the tagging of proteins for destruction, although the exact means by which they function and assemble together as a complex is not yet known (35). In yeast cells, the APC/C is present throughout the cell cycle but its activity peaks in M through G1 to promote anaphase entry and mitotic exit and facilitate prereplication complex formation (17).
How does the APC/C recognize specific substrates destined for the proteosome? Two conserved targeting/adaptor factors, Cdc20p and Cdh1p, which contain WD40 repeats are responsible for linking the APC/C to specific substrates (42, 43, 50). In Saccharomyces cerevisiae, these two factors seem to preferentially interact with distinct sets of target proteins and function in different windows of time and are themselves controlled by different mechanisms (17). ScCdc20p functions early in mitosis by marking the S-phase cyclin ScClb5p and a fraction of the pools of ScClb2p for destruction. The anaphase inhibitor ScPds1p (securin) is another target of ScCdc20p, and ScPds1p degradation leads to the loss of sister chromatid cohesion and anaphase (11, 18, 30, 41, 44, 45, 49, 51, 56). ScPds1p degradation also helps to set off the FEAR/MEN signaling cascade that leads to the activation of ScCdh1p, which will then cover the bulk of APC/C targeting through G1 and early S phase, including marking the mitotic cyclin ScClb2p for degradation (21, 43). Cdc20p is itself degraded by the APC/C via Cdh1p in many systems, whereas Cdh1p persists across the cell cycle but its activity is limited from S to late M by inhibitory phosphorylation delivered by CDK/cyclin complexes (23, 57). An additional conserved factor called Doc1p functions to stabilize substrate-APC/C interactions by associating with substrates, but it remains associated with the APC/C throughout the cell cycle (7, 8, 16, 36).
Most targets of the APC/C have a short N-terminal motif called the destruction box (D box) and/or a KEN box, both of which facilitate, though are not essential in all cases, identification by Cdc20/Cdh1 proteins (5, 18, 38). Additionally, the D-box motif contributes to the assembly and stability of the substrate-APC/C-Cdh1 ternary complex (6, 28). Regulated degradation of both the B-type cyclins and securin was shown to be the only essential role of the APC/C for accurate and orderly progression in budding yeast (47, 48).
In addition to uninucleated yeasts, some components of the APC/C have also been studied in several filamentous fungi and genes encoding components of the APC/C, BimA and BimE (blocked in mitosis), were identified nearly 30 years ago by genetic screens in Aspergillus nidulans (31). Subsequent work identified these gene products as negative cell cycle regulators and central components of the APC/C, with BimA homologous to Cdc27/APC3 and BimE homologous to APC1. The APC/C in A. nidulans functions both in exit from mitosis and during interphase, where it regulates the stability of NimA, a kinase that promotes mitosis (22, 34, 55). A Cdh1p homologue, Cru1, has also been characterized in Ustilago maydis, where it was shown to target the degradation of B-type cyclins and control the length of G1 (9). Interestingly, Cru1 expression is regulated by cyclic AMP and mutants lacking Cru1 are defective in pathogenicity. Little is known about how the APC/C may be spatially regulated in large, multinucleated cells. The APC/C changes localization and exists in functionally distinct subcomplexes in Drosophila melanogaster syncytial embryos; thus, there is some limited data to suggest that the APC/C can be uniquely regulated in space in multinucleated cells (20). Spatial control of the APC/C has not yet been explored in multinucleated, filamentous fungal cells.
We have begun to investigate how the APC/C, Cdc20p, and Cdh1p function in the filamentous, multinucleated ascomycete Ashbya gossypii. A. gossypii is evolutionarily related to the budding yeast S. cerevisiae but has a notably different growth and nuclear division cycle (12). Nuclei in A. gossypii cells divide asynchronously so that a mitotic nucleus divides in the same cytoplasm beside neighbors in different stages. Furthermore, the mitotic cyclin homologue AgClb1/2p appears to persist across all nuclear division cycle stages rather than be completely destroyed at the exit of mitosis, as is common in most eukaryotic systems (14). We hypothesized that in a syncytial and asynchronous system such as A. gossypii, other forms of negative cell cycle regulation may be favored over protein destruction. In these cells, proteins are continually being expressed from adjacent nuclei in different nuclear cycle stages, and we speculated that it would therefore be challenging for degradation in a single nucleus to match the influx of new proteins from multiple neighbors. Thus, in this paper, we set out to analyze how the APC/C and associated factors function within this asynchronous nuclear division cycle. Is the APC/C essential for nuclear progression? Is the APC/C spatially regulated to be active or present in nuclei of a particular cell cycle stage?
To address these questions, we have identified and characterized how the homologues of core APC/C factors, the targeting subunits, and the potential APC substrate AgPds1p function in the asynchronous A. gossypii nuclear cycle. We have found that despite the lack of AgClb1/2p oscillation in these cells, both the APC/C and the targeting factors AgCdh1p and AgCdc20p are essential in A. gossypii and mutants arrest with a clear, nearly uniform block in mitosis. The predicted APC substrate AgPds1p was also examined and found to be an essential gene, unlike in budding yeast cells. AgPds1p was absent in anaphase nuclei, and its D box was required for this disappearance. This result raised the question of how the APC/C may be locally activated to degrade AgPds1p in only a subset of nuclei in these cells. Thus, we have also evaluated the spatial and temporal appearances of the APC/C in multinucleated hyphae.
MATERIALS AND METHODS
Strains, media, plasmids, DNA manipulations, and transformation.
The A. gossypii reference strain used for all experiments is a derivative of the wild-type strain (ATCC 10895) in which the AgLEU2 and AgTHR4 genes were deleted (3, 30a). Growth media, culturing conditions, spore isolations, and transformation protocols are described in references 4 and 52. Transformants were selected on Ashbya full medium (AFM) containing either Geneticin-G418 (200 μg/ml) or CloNAT (50 μg/ml). All A. gossypii strains are listed in Table 1. The diploid S. cerevisiae strain DHD5 (ura3/ura3) was used for tagging and in-frame deletions of A. gossypii genes cloned in autonomously replicating centromere vectors pRS415 or pRS416 (see Table 2). The modified plasmids were verified by DNA sequencing and then used, cleaved or noncleaved, for transformation of A. gossypii. These and other plasmids are listed in Table 2. PCR was performed with Taq polymerase from Roche (Basel, Switzerland), and restriction enzymes were obtained from New England Biolabs or Roche. Oligonucleotides were synthesized at MWG (Ebersberg, Germany) or Microsynth (Balgach, Switzerland) and are listed in Table 3. Nocodazole (Sigma) was dissolved in dimethyl sulfoxide (3 mg/ml) and used at a final concentration of 15 μg/ml to block nuclear cycles.
TABLE 1.
Ashbya gossypii strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| Reference strain | Agleu2Δthr4Δ | 3 |
| NSN01 | AgCDC23 leu2Δthr4Δ/cdc23::NAT1 leu2Δthr4Δ | This study |
| NSG01 | AgCDC20 leu2Δthr4Δ/cdc20::GEN3 leu2Δthr4Δ | This study |
| NSN02 | AgDOC1 leu2Δthr4Δ/doc1::NAT1 leu2Δthr4Δ | This study |
| NSG02 | AgCDH1 leu2Δthr4Δ/cdh1::GEN3 leu2Δthr4Δ | This study |
| NSG03 | AgPDS1 leu2Δthr4Δ/pds1::GEN3 leu2Δthr4Δ | This study |
| NSG09 | AgPDS1-6HA-GEN3 leu2Δthr4Δ | This study |
| NSG10 | AgPDS1 leu2Δthr4Δ (pAgpds1Δdb-6HA-GEN3) | This study |
| NSG11 | AgDOC1-9myc-GEN3 leu2Δthr4Δ | This study |
| GVS2 | AgCDC16 leu2Δthr4Δ (pAgCDC16-13myc-GEN3) | This study |
| GVS3 | AgDOC1 leu2Δthr4Δ (pAgDOC1-13myc-GEN3) | This study |
| GVS4 | AgCDC23 leu2Δthr4Δ (pAgCDC23-13myc-GEN3) | This study |
| NSG12 | AgCDH1 leu2Δthr4Δ (pGEN3-prom-GFP-AgCDH1) | This study |
| NSG13 | AgCDC20 leu2Δthr4Δ (pGEN3-prom-GFP-AgCDC20) | This study |
All NSN, NSG, and GVS strains are derived from the reference strain. Individual nuclei are haploid, and most strains are heterokaryotic (mixture of nontransformed and transformed nuclei). The genotypes of the two different types of nuclei are separated by a slash. A plasmid name in parentheses indicates a replicating plasmid which is maintained under selection pressure.
TABLE 2.
Plasmids used in this study
| Plasmid | Vector | Important sequence features | Source or reference |
|---|---|---|---|
| pGEN3 | GEN3 | 52 | |
| pRS416 | pUC19 | ScURA3 CEN ARS | 46 |
| pRS415 | pUC19 | ScLEU2 CEN ARS | 46 |
| pAGT146 | pUC19 | 13myc-ScURA3term-GEN3 | A. Kaufmann |
| pG3-9Myc | pUC19 | 9myc-KlIPP1term-GEN3 | A. Kaufmann |
| pK16Bni-GFP | pUC19 | GEN3-promAgBNI1-GFP | M. Köhli |
| pUC19NATP | pUC19 | CloNAT | D. Hoepfner |
| pAGT145 | pUC19 | GEN3-6HA | A. Kaufmann |
| pAG7019 | pRS416 | AgCDH1 | 12 |
| pAG14820 | pRS416 | AgPDS1 | 12 |
| pAG6955 | pRS416 | AgCDC23 | 12 |
| pAG1393 | pRS416 | AgDOC1 | 12 |
| pNS01 | pRS416 | AgCDC20 | This study |
| pGVS1 | pRS415 | AgCDC16 | This study |
| pGVS2 | pRS415 | AgCDC16-13myc-GEN3 | This study |
| pGVS3 | pAG1393 | AgDOC1-13myc-GEN3 | This study |
| pGVS4 | pAG6955 | AgCDC23-13myc-GEN3 | This study |
| pNS07 | pAG14820 | Agpds1Δdb-6HA-GEN3 | This study |
| pNS09 | pAG14820 | Agpds1Δdb | This study |
| pNS015 | pRS416 | GEN3-prom-GFP-AgCDH1 | This study |
| pNS016 | pRS416 | GEN3-prom-GFP-AgCDC20 | This study |
TABLE 3.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequencea | Oligonucleotide | Sequencea | |
|---|---|---|---|---|
| CDH1-S1 | TTGAAGTGTGCTTCGGGGCTAGGTGAATAGAAAGTCAG | URA3term-rev | CGAGATTCCCGGGTAATAAC | |
| GAGGAACTgctagggataacagggtaat | M13 for | CCAGGGTTTTCCCAGTCACGA | ||
| CDH1-S2 | TAACCAATGTATGCCGGCTGGGTTATAGTTAGGAGTATA | CDC16-rev1 (I2) | ATCACTCGACGCTGCTCCTG | |
| TTATATAaggcatgcaagcttagatct | CDC23-NS1 | CATCGCATTGCGGTAGTTCTCGTCCAGCTGCCC | ||
| CDH1-G1 | TGGACCGCCATTGTACATAGC | CTGCTGCTCGCTCAGCAGGTCCccggggatcctctag | ||
| CDH1-G4 | GTTACATACCGCCTGCAAACTG | agtcg | ||
| CDH1-I | GGTCGCCAAACAGCTCATTC | CDC23-NS2 | TGAGTAACCACTAGTATTCATCCTAAACTCTTTC | |
| NtS1GFP-CDH1 | CGTATTATCCATAAATGGGCTCCCAGTACCCTCggatcctctag | TATTTACTTTTAAAGCTACCTctgcagccaaacagtgttcc | ||
| tgtttaaacc | CDC23-G1 | TGCAGCTTGCTCTTCGCCACGTCT | ||
| NtS2GFP-CDH1 | CATGTATATTTTGATCAGTGACATTTGAAGTGTGCTTCGa | CDC23-G4 | TAGCCAGAATATGTGCGGTG | |
| ccatgattacgccaagcttgc | CDC23-I3 | CGCCGTGCGGTAGATATCAA | ||
| Fw-CDC20-cl | CGTTCACTAGAGCCGGGAGTTTCACCCTGGCACAA | F1-13myc CDC23 | GCAATAGTTATTGCCAGGGAATGCAGGAAACG | |
| Rev-CDC20-cl | CGTTCACTAGACGTGCCGCAGCCATTGCACTAC | AATGGAATATCAAaaaacgacggccagtgaattcg | ||
| CDC20-S1 | GGGCATCTTCGCTGTGAAAACGCACTGTGGAGCGGTCTG | F2-13myc CDC23 | TATATAGTTAGTAAATGAGTAACCACTAGTATT | |
| GCACAGAGGCGAGGATCgctagggataacagggtaat | CATCCTAAACTCcatgattacgccaagcttgc | |||
| CDC20-S2 | GCACGTTATTAAATGTCGTTAGATACTGCTGAGAGTTTTT | CDC23-fw1 (I3) | GTATGAGCGAGTCCAGGATG | |
| AGTTTATATGCGTaggcatgcaagcttagatct | G3.3 (GEN3) | ATGTTGGACGAGTCGGAATC | ||
| CDC20-G1 | GCTGGAGCGTCGCATCTTAC | M13 rev | TCACACAGGAAACAGCTATGA | |
| CDC20-G4 | ATGGCCCTCCCTCTTACAGG | DOC1-NS1 | TGAAAAGAGATAAGTAGACCGCGCGCGCAGCC | |
| CDC20-I | CTTCACCGCCTCCTCCTTG | CAGCCGTCCTCGCCCACGCAGCAccggggatcctct | ||
| NtS1GFP-CDC20 | TCGACCTGCTTGCACTGAGCCGCGGCTCCTTCGTGTCCAC | agagtcg | ||
| ggatcctctagtgtttaaacc | DOC1-NS2 | GCAATATTTATACATAACTATACACGAAATCTC | ||
| NtS2GFP-CDC20 | ACCAGTTTGGCTAGTGCAGGGAGAGATTCTCGTTGCAacc | AGTTCATCGCTAGCTAGCctgcagccaaacagtgttcc | ||
| atgattacgccaagcttgc | DOC1-G1 | TATGCCAAGACATCGACATATATAG | ||
| PDS1-S1 | CCTCTGAACAGTTCCAACGGTATTCAAAGTAGGCTTTCA | DOC1-G4 | CGACGCTACGACGTGTAGCA | |
| GGCTGATAgctagggataacagggtaat | DOC1-I3 | ACGTCGATATCACCTCGCTC | ||
| PDS1-S2 | GTACATAGTATGCATGCGTCGTATTAGGCTTTATATTAC | F1-13myc DOC1 | GAAGGGTTCCACAGCATTGAGTTCCTCTCCCA | |
| AAGCACCCaggcatgcaagcttagatct | GTCCCGGATACGAaaaacgacggccagtgaattcg | |||
| PDS1-G1 | GCGGCAAATGAAACGCAATACC | F2-13myc DOC1 | ATCCGGATCCAAAAAAGCAATATTTATACATA | |
| PDS1-G4 | AGACAGAGGACCCCAGCAACAC | ACTATACACGAAAcatgattacgccaagcttgc | ||
| PDS1-I | TTCGGGACATGCGGTAGCTC | DOC1-rev1 (I3) | CTTCTCGCTCTTCGTCGATG | |
| PDS1-Bdb | CGAAGACACCCTGCGACTGCGTGCTATTCCTGCCGGCC | GS1-9myc DOC1 | TCCACAGCATTGAGTTCCTCTCCCAGTCCCGG | |
| AAG | TACGAaaaacgacggccagtgaattcg | |||
| PDS1-Cdb | CAGTCGCAGGGTGTCTTCGGGCTGAAGACAAGC | GS2-9myc DOC1 | AGCAATATTTATACATAACTATACACGAAATCT | |
| GS1-PDS1-HA | GTGAGGGATTGGATTCTAAGGACCTACATTCTCTATTAG | accatgattacgccaagcttgc | ||
| AC aaaacgacggccagtgaattcg | SEQ-GCYFO | CAAGAGTGCCATGCCCGAA | ||
| GS2-PDS1-HA | GTCGTGGCTAAATACATAACTTGTACATAGTATGCAT | SEQ-REV | CTGCAGGTCGACTCTAGAG | |
| GCGT accatgattacgccaagcttgc | G3.2 (GEN3) | CTCCAACTCGGCACTATTTTA | ||
| Fw-CDC16-cl | ATCCGTGTGCGTCTGCTTCC | G3 (GEN3) | TCGCAGACCGATACCAGGATC | |
| Rev-CDC16-cl | AAGTAACTGCAGAAGGGCGCCAGCAAGAAG | G2.1 (GEN3) | TGCCTCCAGCATAGTCGAAG | |
| F1-13myc CDC16 | GCAGGGCAGTACGTCATCTGATGAGGGTGATTCCATGG | V2PDC1P | GAACAAACCCAAATCTGATTGCAAGGAGAGTG | |
| ATATAGAGaaaacgacggccagtgaattcg | AAAGAGCCTT | |||
| F2-13myc CDC16 | CATATGTTTCAATTATGTATGTGGCTTCTATTAACGTAA | V3PDC1T | GACCAGACAAGAAGTTGCCGACAGTCTGTT | |
| CCTAAATcatgattacgccaagcttgc | GAATTGGCCTG | |||
| CDC16-fw2 (I3) | AACCACGGAAATCGAGCTAC |
Lowercase letters are regions of homology to the cassette containing a selectable marker.
Construction of gene deletion mutants.
A. gossypii open reading frames (ORFs) were replaced by cassettes carrying either the dominant marker GEN3 or CloNAT. The cassettes were obtained by PCR amplification of the selection markers from pGEN3 (52) or from pUC19NATPS (kindly provided by Dominic Hoepfner), using pairs of 65- to 76-nucleotide primers named S1 and S2 plus the gene name for amplification of GEN3 or NS1 and NS2 plus the gene name for amplification of the CloNAT marker (Table 3). The 20 to 22 lowercase letters of the primer sequences have homology to either end of the selection marker, and the 45 to 56 uppercase letters of the primers have homology to the sequences immediately upstream of the start codon or downstream of the stop codon, respectively. The PCR products were directly transformed into A. gossypii by electroporation. Heterokaryon strains that have a mixture of wild-type and mutant nuclei were selected as primary transformants on AFM-plus-G418 (or AFM-plus-CloNAT) plates. Correct ORF targeting was verified by PCR using the primers G1 and G4 (plus gene name) binding upstream and downstream of the deleted ORF, respectively, and the primers G2.1 (GEN) and G3 (GEN) binding inside the GEN marker or V2PDC1P and V3PDC1T binding inside the CloNAT marker. Spores from three independent heterokaryon mycelia each were incubated overnight under selective conditions and analyzed by microscopy.
Genomic DNA clones in replicating vectors.
Plasmids carrying the genes AgCDC23 (pAG6955), AgDOC1 (pAG1393), AgCDH1 (pAG7019), and AgPDS1 (pAG14820) were obtained from the genomic DNA pRS416 library used for sequencing of the A. gossypii genome (12). These plasmids can autonomously replicate in S. cerevisiae and A. gossypii. A genomic clone of AgCDC20 in pRS416 (pNS01) was constructed by PCR amplification of genomic DNA, including 800 bp upstream and 400 bp downstream of its ORF by using the primers Fw-CDC20-cl and Rev-CDC20-cl, respectively. The PCR product was phosphorylated with T4 polynucleotide kinase and cloned into the SmaI site of pRS416. The pNS01 plasmid was verified by sequencing. For cloning of AgCDC16 (pGVS1) DNA from bacterial artificial chromosome, clone bAG1591, used in the genome project, was amplified with primers Fw-CDC16-cl and Rev-CDC16-cl; the first one binds upstream of a genomic PstI site, and the second primer contains a PstI site. The 3.2-kb PstI fragment was first cloned into PstI-digested pBSII SK+ (Stratagene) with an in-frame inactivation of the ScaI site and, after amplification in Escherichia coli, into PstI-digested pRS415. The cloned AgCDC16 gene (pGVS1) was verified by sequencing.
Construction of tagged ORFs for fluorescence microscopy.
To generate A. gossypii strains expressing myc-tagged APC/C components, the primer pair F1-13myc and F2-13myc plus the gene name (Table 3) was used to generate PCR products. The 3′ ends of these primers have sequence homology to either end of the 13myc-ScURA3term-GEN3 cassette in plasmid pAGT146, kindly provided by Andreas Kaufmann. The 5′ ends have sequence homology to the end of the targeted ORF immediately upstream and several nucleotides downstream of the stop codon, respectively. The PCR products containing the 13-myc-GEN3 cassette were cotransformed into the yeast DHD5 strain with the plasmids pGVS1, pAG1393, and pAG6955 to obtain plasmids pGVS2 (AgCDC16-13myc), pGVS3 (AgDOC1-13myc), and pGVS4 (AgCDC23-13myc), respectively. The plasmids were amplified in E. coli, verified by sequencing, and transformed into A. gossypii to generate the strains GVS2, GVS3, and GVS4, respectively. The A. gossypii myc-tagged transformants were selected on Geneticin-G418 and then verified by PCR using the primers listed in Table 3 with the F1-13myc and F2-13myc primers for each gene construct. For construction of strain NSG11 carrying a genomic AgDOC1-9myc-GEN3 fusion allele, the primers GS1-9myc DOC1 and GS2-9myc DOC1 were used for amplification of the 9myc-KlIPP1term-GEN3 cassette in pG3-9Myc, kindly provided by Andreas Kaufmann. After cotransformation of the PCR product with pAG1393 (AgDOC1) into yeast DHD5 and verification of correct targeting, the plasmid with the AgDOC1-9myc-GEN3 allele was amplified in E. coli and cleaved to isolate a fragment with long flanking sequence homology to the genomic AgDOC1 locus for one-step gene replacement. Geneticin-G418-resistant primary transformants were sporulated to isolate homokaryotic transformants. Strain NSG11 did not lose the resistance marker after 6 days of nonselective growth and carried a correct gene replacement, as shown by PCR verification using the primer pair DOC1-I3/SEQ-REV and DOC1-G4/G3.2 (GEN) as well as by hybridization of an AgDOC1 and a Myc probe to cleaved genomic DNA.
Tagging of AgCdh1p and AgCdc20p by direct PCR targeting at the C terminus was not successful, most likely because epitope fusions to the highly conserved carboxy end of both proteins interfered with protein function. For tagging of the N terminus, a GEN3-AgBni1promoter-GFP cassette (kindly provided by Michael Köhli) was targeted to the start codon of the cloned genes. For AgCDH1, the PCR primers NtS1GFP-CDH1 and NtS2GFP-CDH1 were used and, for AgCDC20, the primers NtS1GFP-CDC20 and NtS2GFP-CDC20. PCR products were cotransformed into the DHD5 strain together with the plasmid pNS01 (AgCDC20) or pAG7019 (AgCDH1). Plasmids with 5′ green fluorescent protein (GFP) gene fusions were rescued and verified by enzymatic digestions, KpnI/XmnI for GFP-CDH1 (pNS015) and MluI/XbaI for GFP-CDC20 (pNS016). A. gossypii was transformed with pNS015 and pNS016, generating the strains NSG12 and NSG13, respectively. The heterokaryotic transformants were verified by PCR using the primer pairs SEQ-GCYFO/CDH1-I (for NSG12) and SEQ-GCYFO/CDC20-I (for NSG13).
Construction of the AgPDS1 destruction box deletion and the AgPDS1-HA fusion.
The pds1Δdb mutant allele was made using an overlap PCR approach which deleted 30 base pairs (124 to 153) in the ORF of AgPDS1. To construct the plasmid pAgpds1Δdb, a first PCR with the primer pair PDS1-G1/PDS1-Bdb and the cloned AgPDS1 gene (pAG14820) as the template produced a 478-bp fragment named product A. The second PCR with the primer pair PDS1-I/PDS1-Cdb and the same template produced a 434-bp product named product B. The primers PDS1-G1 and PDS1-I have homology a few hundred base pairs upstream and downstream, respectively, of the D box. The primer PDS1-Bdb has homology immediately upstream and downstream of the D-box coding sequence but lacks 10 codons of the D box. The PDS1-Cdb primer binds immediately downstream of the D-box coding sequence. The overlapping region of the products A and B was used in a third PCR as the template to produce an 889-bp-long fragment that has an in-frame deletion of the D box. This gel-purified PCR product was cotransformed into the DHD5 yeast strain together with MluI-digested pAG14820 DNA. MluI cuts only in the D-box nucleotide region of AgPDS1 in pAG14820. The gap-repaired plasmid containing the deleted D box was selected based on the plasmid-carrying URA3 marker, confirmed by sequencing the complete insert on both strands, and was named pNS09 (Agpds1Δdb).
In the next step, the 3′ end of the deletion allele in this plasmid was fused with a sequence coding for the hemagglutinin (HA) epitope. The 6HA-GEN3 cassette of pAGT145, kindly provided by Andreas Kaufmann, was amplified using the primers GS1-PDS1-HA and GS2-PDS1-HA. These primers have homology at their 5′ end to the AgPDS1 gene (upstream and downstream of the stop codon, respectively) and at their 3′ end to the 6HA-GEN3 marker. The PCR fragment was cotransformed with pNS09 into the DHD5 yeast strain, generating the plasmid pNS07 (Agpds1Δdb-6HA-GEN3). Verification of this construct was done by enzymatic digestion with SspI, by PCR verification using the primer pair G3.3 (GEN3)/PDS1-G4, and by DNA sequencing.
A. gossypii was transformed with either nondigested plasmid pNS07 or NheI/BssSI-cleaved plasmid, which generates a DNA fragment with long regions of homology to the genomic AgPDS1 locus to replace the wild-type allele by the HA-tagged destruction box deletion allele. In both cases, viable Geneticin-G418-resistant transformants were obtained. The nondigested plasmid can replicate in A. gossypii and can be maintained under selective conditions, indicating that expression of the destruction box deletion allele is not lethal in the presence of the wild-type allele. However, these transformants did not sporulate. One verified strain, NSG10, was selected for immunofluorescence. Two types of Geneticin-G418-resistant transformants were obtained with the linearized fragment. Transformants of the first type grew and sporulated like the wild type allowing the isolation of homokaryotic transformants, which maintained the resistance marker after 6 days of nonselective growth. These sporulating homokaryon strains were examined by PCR using oligonucleotides PDS1-I and PDS1-G1, followed by enzymatic digestion with MluI, a site that is lost if the D box is deleted. In all cases, the wild-type and not the deletion allele was present, indicating a dominant-negative effect of the deletion allele. One strain, NSG09, was rigorously tested with PCR verification and DNA hybridizations and showed at its AgPDS1 locus a wild-type allele fused in frame to 6HA-GEN3. The end of the transforming linear DNA fragment carrying the destruction box deletion was most likely degraded prior to or during gene replacement. The second type of heterokaryotic transformants obtained with the linear fragment did not sporulate, thus preventing the isolation of homokaryotic strains. Verifications by PCR of primary transformants with the primer pairs PDS1-G4/G 3.2 (GEN3) and Seq-Rev/PDS1-Cdb did not give conclusive results, and these transformants were not further investigated.
Immunofluorescence, Hoechst staining, and microscopy.
A. gossypii cells were processed for immunofluorescence as described for yeast cells (40), with slight modifications. Young mycelia containing approximately 75 to 100 nuclei were fixed for 1.0 h in 3.7% formaldehyde (Fluka) and digested in 1.0 mg/ml Zymolyase (Seikagaku Corporation) plus 1% beta-mercaptoethanol (Sigma) for 30 to 45 min before antibody incubation. Anti-myc, anti-HA, or anti-GFP and tubulin stainings were done sequentially so as to limit cross-reactivity, beginning with mouse anti-HA (Covance), mouse anti-myc (Santa Cruz Biotech), or rabbit anti-GFP (Molecular Probes) at 1/50, then Alexa 488 anti-mouse (Molecular Probes) or Alexa 488 anti-rabbit at 1/200, then rat antitubulin (YOL34; Serotec) at 1/50, and finally Alexa 568 anti-rat (Molecular Probes) at 1/200 with Hoechst (Molecular Probes) dye to visualize nuclei at 5 μg/ml. Antibody dilutions and washes were performed with phosphate-buffered saline plus 1.0 mg/ml immunoglobulin G-free bovine serum albumin (Sigma).
The microscope used for all fixed cell images (immunofluorescence and Hoechst stainings with cells mounted in standard fluorescent mounting medium containing 1 mg/ml p-phenylenediamine in 90% glycerol) was essentially as described by Hoepfner et al. (19) and consisted of an Axioplan 2 imaging microscope (Carl Zeiss) with a Plan Neofluar 100× Ph3 numerical-aperture 1.3 objective. It was equipped with a 75 W XBO and a 100 W HBO illumination source controlled by a MAC2000 shutter and filter wheel system (Ludl Electronics). The camera was a TE/CCD-1000PB back-illuminated cooled charge-coupled-device camera (Princeton Instruments). The following filter sets for different fluorophores were used: no. 10 for Alexa 488 and no. 20 for Rhodamine-Alexa 568 (Carl Zeiss) and no. 41018 for GFP (Chroma Technology, Brattleboro, VT). The excitation intensity was controlled with different neutral density filters (Chroma Technology). MetaMorph 4.6r9 software (Universal Imaging) controlled the microscope, camera, and Ludl controller and was used for processing images. All images presented here are maximum projections of >20× 0.5-μm step images acquired along the z axis to ensure that all hyphae were observed.
Bioinformatic analysis.
Multiple protein alignments were performed with sequences from S. cerevisiae (15), A. gossypii (12), Saccharomyces bayanus, Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii, Saccharomyces castellii (10, 26), Kluyveromyces waltii (25), and Kluyveromyces lactis (13). These sequences can be retrieved from the Saccharomyces Genome Database maintained at Stanford University (http://genome-www.stanford.edu/Saccharomyces/). Clustal W software (http://www.ebi.ac.uk/clustalw/) was used to align the protein sequences. To search for APC domains and motifs (WD repeats, C box, D box, and KEN box) Swiss-Prot (http://www.expasy.ch/sprot) was used as a primary source of information for the S. cerevisiae orthologues. This facilitated visual screening to find known and new motifs in the multiple alignments. We gained confirmative and also additional information by separately aligning orthologues from some species with nonduplicated genomes (A. gossypii, K. waltii, and K. lactis) and those from other species with duplicated genomes.
RESULTS
The APC/C and the targeting proteins Cdh1p and Cdc20p are present and key domains are conserved in A. gossypii.
To begin to evaluate the function of the APC/C in A. gossypii, we searched for homologues of the targeting subunits Cdh1p and Cdc20p as well as the core components of the APC/C in the A. gossypii genome (Fig. 1 and Table 4). We identified 14 possible APC/C subunits in A. gossypii with amino acid homology ranging from 22% to 53% identity compared to S. cerevisiae, with 8 of the subunits with less than 38% identity. The A. gossypii APC/C and coactivator genes are present in syntenic positions in the genome compared to their positions in the yeast genome, supporting that these are functional homologues of the APC/C despite the low identity to yeast proteins. The targeting factors AgCdh1p and AgCdc20p were somewhat more similar to the yeast homologues, with identity levels of 66% and 59%, respectively, and the genes were also in syntenic positions. In most cases, the predicted A. gossypii APC/C protein was 5 to 15% shorter in length than the S. cerevisiae protein.
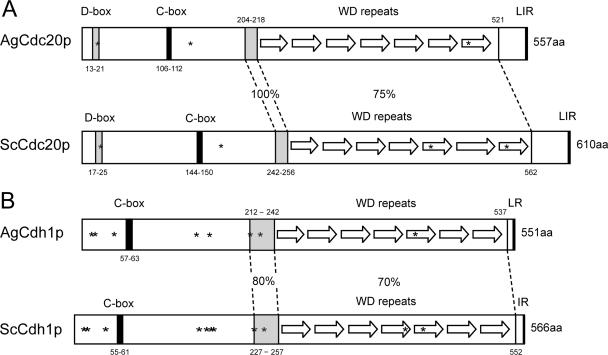
FIG. 1.
Comparison of homologous domains and sequence features in APC/C cofactors from A. gossypii and S. cerevisiae. (A) AgCdc20p (AFL014C) and ScCdc20p (YGL116W) are 59% identical on the amino acid level. The positions of a D box, a C box, and seven WD repeats are shown for each protein, and notably, a stretch of 15 identical amino acids (aa) precedes the WD repeats. Asterisks mark potential CDK phosphorylation sites (S20, S136, and S493 in AgCdc20p and S24, S173, S439, and S534 in ScCdc20p). These sites are conserved in eight yeast species and A. gossypii except S439 (in the fifth WD repeat). In all nine Cdc20p orthologues, the last amino acid is arginine preceded by two hydrophobic amino acids. (B) AgCdh1p (AFL007C) and ScCdh1p (YGL003C) are 66% identical, with the highest homology in the WD repeat region and the preceding 21 amino acids. All nine analyzed Cdh1 orthologues terminate with an arginine preceded by a hydrophobic amino acid. The eight asterisks in AgCdh1p mark CDK phosphoryation consensus sites (T12, S16, S44, T144, T161, S212, S224, and S421), all of which are conserved in eight yeast orthologues. In ScCdh1p, the homologous sites are phosphorylated with the possible exceptions of S42 and S227 (16a, 57). The three additional consensus CDK sites in ScCdh1p (S169, T173, and S418) are most likely not phosphorylated. From the 11 non-CDK phosphorylation sites identified in ScCdh1p by those authors, only 5 (S38, S172, S193, S225, and S556) are conserved in A. gossypii and other yeast Cdh1 sequences. LIR, LR, and IR refer to amino acids.
TABLE 4.
Comparison of APC/C core factors and coactivator subunits from S. cerevisiae and A. gossypiia
| Core factor or subunit | % Identity | Protein length (aa)
|
|
|---|---|---|---|
| A. gossypii | S. cerevisiae | ||
| Apc1 | 35 | 1,670 | 1,748 |
| Apc2 | 33 | 710 | 853 |
| Apc4 | 22 | 716 | 652 |
| Apc5 | 24 | 667 | 685 |
| Apc9 | 32 | 257 | 265 |
| Apc11 | 53 | 148 | 165 |
| Cdc16 | 53 | 708 | 840 |
| Cdc23 | 49 | 614 | 626 |
| Cdc26 | 25 | 119 | 124 |
| Cdc27 | 48 | 657 | 758 |
| Doc1 | 45 | 250 | 283 |
| Mnd2 | 25 | 263 | 368 |
| Swm1 | 22 | 146 | 170 |
| Cdh1 | 66 | 522 | 566 |
| Cdc20 | 59 | 558 | 610 |
| Ama1 | 47 | 573 | 562 |
Data from http://www.yeastgenome.org and reference 35. aa, amino acids.
Both AgCdc23p and AgDoc1p, the two core subunits we have analyzed below, have conserved sequence features supporting their function in the APC/C. In AgCdc23p, nine out of nine tetratricopeptide repeats, which are important for associations between APC/C core subunits, are conserved compared to those in the budding yeast homologue (29). AgDoc1p clearly has a “DOC domain” conserved which is found in various proteins associated with ubiquitin protein ligase activity (16). The conservation of domains in the targeting subunits AgCdc20p and AgCdh1p is summarized in Fig. 1. These proteins contain seven WD repeat motifs which mediate protein-protein interactions and thus interact with substrates and specifically D boxes (6, 28). Homology within these repeats is high between the A. gossypii and S. cerevisiae homologues (73 to 83% identity for Cdh1p and 66 to 90% identity for Cdc20p) and the orthologues of seven other yeast species. The 15 amino acids preceding the WD repeat region and the last three amino acids of all orthologues are even more highly conserved. Additionally, a D box is present in AgCdc20p and three CDK phosphorylation consensus sites (Fig. 1A) which are conserved in all known yeast orthologues. All ScCdh1p regulatory CDK phosphorylation sites are conserved in AgCdh1p and in other yeast orthologues (see legend to Fig. 1B). Furthermore, both AgCdh1p and AgCdc20p have C boxes (conserved in Cdc20 family members), which are regions thought to link these targeting factors to the APC/C (39, 43). When combined, it appears as if A. gossypii cells have the basic subunits of the APC/C to direct regulated protein degradation. However, given the low identity between most of the factors and the differences between uninucleated cell division and multinucleated, asynchronous nuclear division, it is likely that some aspects of APC/C function or regulation have diverged since budding yeast and A. gossypii shared a common ancestor.
Cells lacking APC/C core subunits and targeting subunits arrest in late stages of mitosis.
Mutants were generated with deletions in components of the APC/C and evaluated for defects in cell cycle progression. All mutants created by gene targeting in A. gossypii are initially made as heterokaryons because only a subset of nuclei in the multinucleated hyphae is transformed. The heterokaryons produce uninucleated spores carrying either the wild-type or the mutated allele. Spores can be germinated under selection to produce homokaryons in which all nuclei contain the mutation; however, these spores may also contain small amounts of wild-type protein packaged from the heterokaryon. This pool of “maternal” protein may sustain some growth in the presence of an ultimately deleterious deletion. If the APC/C plays a role in degrading factors required for promoting anaphase and mitotic exit in A. gossypii, an arrest or delay in mitosis would be expected in mutant cells lacking the APC/C components.
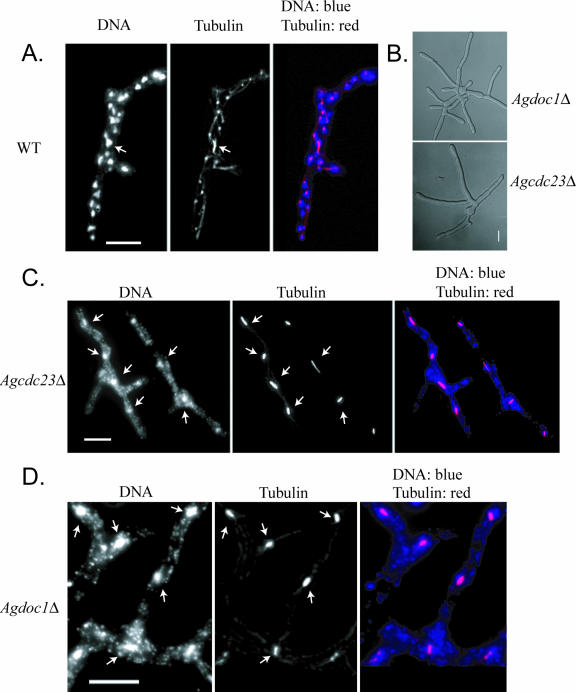
Deletion of either AgCDC23 or AgDOC1 led to a “late lethal” terminal phenotype in which cells reached the size of small mycelia with 4 to 7 nuclei (which in wild-type cells would have 10 to 30 nuclei) but could not develop further (Fig. 2 and Table 5). Nuclear density was low compared to that of the wild type, and nuclei in these mutant strains were often fragmented and had elongated mitotic spindles (>80% of nuclei compared to 13% in the wild type; n = 150 nuclei). We determined the nuclear division stage of nuclei in these cells by measuring the length of mitotic spindles and comparing them to wild-type spindle lengths in normally proliferating cells. A spindle length of more than 2.8 μm was considered anaphase, based on previous work looking at the nuclear dynamics and kinetics of anaphase by time-lapse microscopy (2). In wild-type cells, 42% of nuclei with a mitotic spindle were in anaphase and the anaphase spindle was 4.8 ± 0.41 μm long (mean ± standard deviation), with lengths ranging from 2.8 to 9.5 μm (46 anaphase spindles, 110 mitotic spindles). In contrast, the anaphase spindle length for Agcdc23 mutants at the terminal growth stage was 3.9 ± 0.26 μm (n = 112) and for Agdoc1 mutants 3.8 ± 0.36 μm (n = 58). However, the overall proportions of nuclei in anaphase were similar in both mutants compared to that of the wild type (243 total spindles for Agcdc23 and 134 for Agdoc1). The shortened average anaphase spindle length suggests that the nuclei in these mutants can enter anaphase, but spindles fail to completely elongate and nuclei are challenged to exit mitosis (Table 5). Based on these phenotypes, we predict that the A. gossypii APC/C does in fact regulate nuclear division and that there must be some factor(s) that has to be degraded by the APC/C for completion of mitosis in these multinucleated cells.
FIG. 2.
Cells lacking AgCDC23 and AgDOC1 are inviable and arrest in mitosis. (A) Spores from wild-type (WT) A. gossypii cells were grown for 15 h at 30°C and processed for antitubulin immunofluorescence and DNA staining. (B) Brightfield images depicting “late lethal” arrest point as small mycelia from Agdoc1Δ and Agcdc23Δ mutants. (C and D) Spores from Agcdc23Δ heterokaryons (RNSN01) and Agdoc1Δ heterokaryons (RNSN02), respectively, were grown under selection for 15 h at 30°C and processed for antitubulin immunofluorescence and DNA staining. Arrows point to examples of mitotic nuclei. Bar, 10 μm.
TABLE 5.
Summary of terminal phenotypes of A. gossypii wild-type and homokaryotic deletion mutants
| Straina | No. of nuclei in 80% of the myceliab | Maximum no. of nuclei in a myceliumb | % of nuclei with a mitotic spindle | % of nuclei with one or two spindle poles | Total no. of spindles measured | No. of spindles in metaphasec | No. of spindles in anaphasec | Anaphase spindle length (μm)c
|
|
|---|---|---|---|---|---|---|---|---|---|
| Range | Mean ± SD | ||||||||
| WT | 15-30 | 70 | 13 | 87 | 110 | 64 | 46 | 2.8-9.5 | 4.83 ± 0.41 |
| cdc23Δ mutantd | 4-7 | 10 | 81 | 19 | 243 | 131 | 112 | 2.8-9.0 | 3.9 ± 0.26 |
| doc1Δ mutantd | 4-7 | 9 | 83 | 17 | 134 | 86 | 58 | 2.8-7.7 | 3.8 ± 0.36 |
| cdh1Δ mutant | 1-4 | 7 | 58 | 42 | 142 | 95 | 47 | 2.8-5.0 | 3.6 ± 0.40 |
| cdc20Δ mutant | 1-2 | 3 | 90 | 10 | 117 | 66 | 41 | 2.8-5.9 | 3.8 ± 0.43 |
| pds1Δ mutant | 1-2 | 3 | 79 | 21 | 92 | 36 | 56 | 2.8-9.5 | 4.2 ± 0.37 |
Spores were inoculated in AFM (plus CloNAT for the cdc23Δ and doc1Δ deletion mutants and plus G418 for the cdh1Δ, cdc20Δ, and pds1Δ deletion mutants). After growth for 14 h at 30°C, mycelia were washed and processed for immunofluorescence with antitubulin antibody. Fifty independent mycelia were analyzed for the wild type (WT), and more than 100 mycelia were analyzed for each mutant strain.
Nuclei were visualized with Hoechst dye. Many nuclei, however, appeared to be fragmented in deletion mutant cells, and in these cases, the number of nuclei per mycelium was determined based on the number of spindles and spindle poles observed by tubulin immunofluorescence.
Spindles with lengths between 1.5 and 2.8 μm are considered metaphase spindles. Spindles longer then 2.8 μm are considered anaphase spindles. The length measurements include the spindle pole bodies, so metaphase spindles are slightly longer than the diameter of nuclei, which is 2.23±0.21 μm (2).
The mycelia produced from mutants generated with the CloNAT resistance cassette had two classes of phenotypes. Seventy percent of the mycelia reached a terminal phenotype, with a high frequency of anaphase spindles, and these measurements are presented in this table. Thirty percent stopped growing, with a high percentage of G1 nuclei as observed in the wild type. These mycelia very likely originated from wild-type spores in the primary heterokaryotic transformants, which carry relatively high concentrations of the CloNAT resistance protein. This protein packaged in the spores will support limited resistance to and growth in the presence of the drug, even in the absence of the gene encoding this product. Such background was not observed in mutants selected with G418.
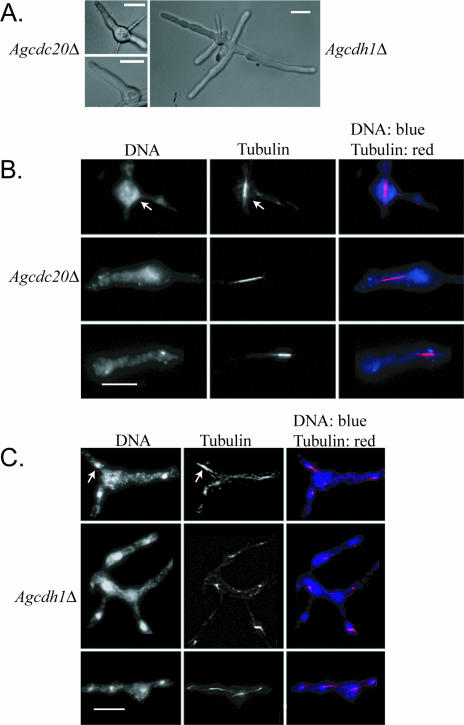
AgCDH1 and AgCDC20 were then deleted, and null mutants were characterized to determine whether they may function to target substrates to the APC/C. Both of these APC targeting factors were also essential, and mutants stopped growing as young germlings, with clear defects in nuclear division (Fig. 3 and Table 5). Agcdc20 cells arrest with only one or two germ tubes containing fragmented nuclei and a single elongated spindle (90% of nuclei with a spindle compared to 13% of those in the wild type; n = 150) and with an anaphase spindle length of 3.8 ± 0.43 μm (41 spindles) (Table 5). Only 35% of mitotic nuclei were in anaphase compared to 42% of those in the wild type, suggesting a possible, slight delay in entering anaphase (117 total mitotic nuclei). Furthermore, the maximum spindle length measured was only 5.9 μm compared to 9.5 μm in the wild type. The length and appearance of the spindles suggest that the nuclei are challenged in progressing through anaphase and exiting mitosis in the absence of AgCdc20p (Fig. 3A and B and Table 5).
FIG. 3.
Cells lacking AgCDC20 or AgCDH1 are inviable and arrest in mitosis. (A) Brightfield images depicting arrest as uni- or bipolar germlings for Agcdc20Δ and small mycelia for Agcdh1Δ. (B) Spores from Agcdc20Δ heterokaryons (NSG01) were incubated under selection for 15 h at 30°C and processed for tubulin and DNA staining. After this incubation time, single mitotic spindles are still present in the arrested germlings but nuclei appear fragmented. (C) Spores from Agcdh1Δ heterokaryons (NSG02) were incubated under selection for 15 h prior to tubulin and DNA staining. The arrow points to an example of a mitotic spindle. Bar, 10 μm.
Agcdh1 mutants grow somewhat larger than Agcdc20 cells and form small germlings with several branches (Fig. 3A). Like those in Agcdc20 mutants, however, nuclei in Agcdh1 cells are fragmented and blocked predominantly in mitosis (Fig. 3C and Table 5). Compared to wild-type cells, where only 13% of nuclei have mitotic spindles, 58% of Agcdh1 mutant spindles appeared mitotic (>150 nuclei) and of these only 33% were in anaphase compared to 42% in the wild type. Agcdh1Δ cells had an anaphase spindle length of 3.6 ± 0.40 μm and a maximum length of 5.0 μm (based on 47 spindles) (Table 5), suggesting that, as with the Agcdc20 mutants, there may be a problem entering and progressing through anaphase in cells lacking AgCdh1p. Most germlings of Agcdh1 had one to four spindles, but cells could be found with up to seven spindles, suggesting that at least two to three rounds of nuclear division can occur prior to the lethal arrest; however, it is possible that these rounds of division are promoted by “maternal” stores of AgCdh1p from the germinating spore. It is also conceivable that Agcdh1 mutants have elevated levels of AgCdc20p that can partially compensate, since in other systems Cdh1p homologues actually target Cdc20p for degradation (21). Thus, AgCdh1p, AgCdc20p, and the predicted APC/C components AgCdc23p and AgDoc1p contribute to normal nuclear division in A. gossypii cells and, based on the length of spindles at arrest, promote progression through anaphase. Given that the levels of the mitotic cyclin AgClb1/2p appear quite stable across the nuclear division cycle, these data suggest that factors other than this cyclin must be targeted for degradation by the APC/C to progress through mitosis.
AgPds1p, a securin homologue, is involved in nuclear progression in A. gossypii.
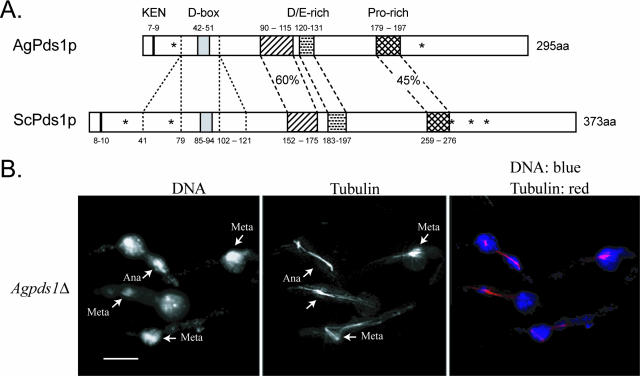
In addition to the mitotic cyclin, the APC/C is essential in S. cerevisiae for the degradation of one other factor, the securin protein Pds1p. ScPds1p degradation, which is mediated primarily by ScCdc20p, triggers timely anaphase entry once all of the chromosomes are properly attached and aligned on the spindle, and this function appears to be conserved throughout eukaryotes. Securins limit the activity of separases, which dissolve sister chromatid cohesion for anaphase entry, and degradation of securins at the metaphase/anaphase transition in turn liberates separase (33). The genes for AgPds1p and ScPds1p are in a syntenic position, but these homologues are only 29% identical and AgPds1p is almost 80 amino acids shorter in length, due mainly to two missing stretches of amino acids flanking the potential D box (Fig. 4A). It does, however, have a predicted D box and KEN box, suggesting that it could be degraded by the APC/C in A. gossypii (Fig. 4A). Despite the low sequence identity, does AgPds1p function as a mediator of anaphase entry in these multinucleated cells?
FIG. 4.
(A) Comparison between AgPds1p (AGR083W) and ScPds1p (YDR113C). The percent identity between the two proteins is 29%. N-terminal protein destruction motifs (KEN box and D box) are noted in addition to three conserved regions in the middle of Pds1p, one highly enriched in negatively charged amino acids (aa) and one enriched for proline, all of which are conserved in yeast species. A major difference between Pds1p orthologues of Saccharomyces species and A. gossypii is the absence of 39 amino acids upstream and 20 amino acids downstream of the D box. Pds1 orthologues from K. waltii and K. lactis also lack blocks of amino acids of similar sizes in this region. The asterisks in ScPds1p mark the positions of five CDK consensus phosphorylation sites (T27, S71, S277, S292, and T304); only two (T27 and S292) are conserved in AgPds1p (T23 and S213) and in seven other yeast orthologues. (B) Spores from Agpds1Δ heterokaryons (NSG03) were incubated under selection for 18 h at 30°C and processed for DNA and tubulin staining. Cells arrested as germlings with one germ tube, most of which carried one nucleus, arrested in either metaphase (Meta) or anaphase (Ana) (arrows denote different stage nuclei). Bar, 10 μm.
Deletion of AgPDS1 demonstrated that it is an essential gene and that it is required for normal nuclear cycle progression in A. gossypii. Agpds1 deletion mutants arrested growth as germlings with generally a single elongated spindle (79% of mitotic nuclei; n > 150) (Fig. 4B and Table 5). This is a more severe phenotype than that of Scpds1 null mutant cells, which are viable but temperature sensitive and at elevated temperatures attempt to enter additional rounds of the cell cycle without chromosome segregation (53, 54). Nuclei were frequently fragmented in Agpds1 mutants (often only one spindle end showed DNA staining), and over 60% of spindles appeared to be in anaphase compared to only 42% in the wild type. The anaphase spindle length was 4.2 ± 0.37 μm, and the maximal length observed was 9.5 μm (based on 56 spindles) (Fig. 4B and Table 5). The proportion of anaphase nuclei combined with the spindle length suggests that Agpds1 deletion mutants may prematurely enter anaphase, potentially before proper chromosome alignment, and then progress through to late anaphase/telophase but are challenged in exiting mitosis and in initiating the next round of division. Thus, the AgPds1p homologue is clearly involved in nuclear division and could be an essential substrate for the A. gossypii APC/C.
AgPds1p levels vary depending upon the stage of the nuclear division cycle.
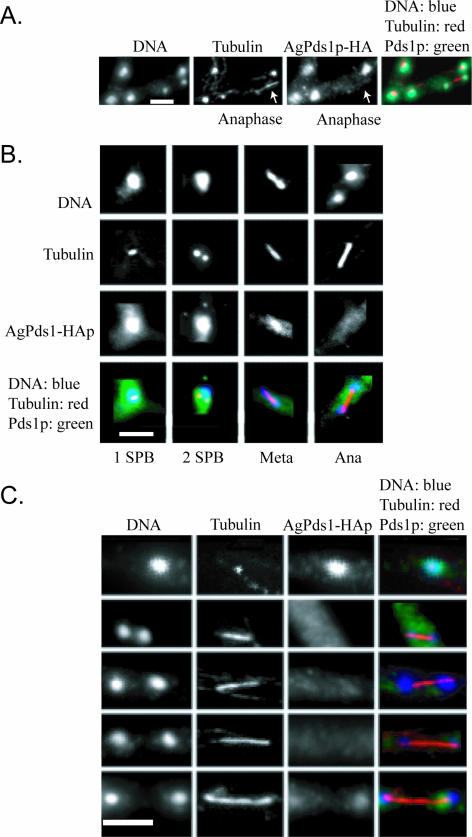
If AgPds1p is in fact a target of the APC/C in A. gossypii, we predicted that AgPds1p would be present in early stages (S/G2/early M) of the cell cycle, when sister chromatid cohesion must be maintained, but absent in late stages, as nuclei progress through anaphase and exit of mitosis. To assay when AgPds1p was present, a strain expressing an HA-tagged protein from the endogenous promoter was analyzed by immunofluorescence. AgPds1p-6HA was present as a diffuse nuclear signal in nuclei with a single spindle pole body (SPB), duplicated SPBs, and metaphase spindles (Fig. 5A and B) (SPBs can be detected when microtubules are visualized by immunofluorescence). It colocalized with the spindle in a fraction of metaphase nuclei but was also still observed in the nucleoplasm, and then levels dropped substantially in anaphase nuclei. Some faint HA signal was present in 10% of anaphase nuclei, and the amount of AgPds1p-6HA detected increased as spindles elongated (Fig. 5C). When levels of HA-tagged protein were assayed in lysates from mycelia where nuclei were synchronized in G2/M by nocodazole and then released, the level of AgPds1p-6HA similarly diminished but was not eliminated completely when 50% of nuclei were in anaphase (data not shown). Thus, the majority of AgPds1p must be degraded at the metaphase-to-anaphase transition (potentially, the pool bound to the separase homologue AgEsp1p), but some limited protein can still be observed in 10% of anaphase nuclei.
FIG. 5.
AgPds1p is present in nuclei but diminishes in anaphase. Spores from a strain expressing AgPds1p-HA (NSG09) were grown under selective conditions and then processed for immunofluorescence. (A) Segment of a branched hypha showing four nonmitotic nuclei and one anaphase nucleus (arrow). Only the nonmitotic nuclei contain AgPds1p-HA. (B) AgPds1p-HA localization in different nuclear cycle stages showing its presence in nuclei with one or two spindle pole bodies and in metaphase (Meta) nuclei but not in anaphase (Ana) nuclei. (C) Hyphae were released from a nocodazole block to follow AgPds1p-HA through anaphase in a large number of nuclei. Weak AgPds1p-HA signals are observed only in maximally extended spindles (bottom panels). Bar, 5 μm.
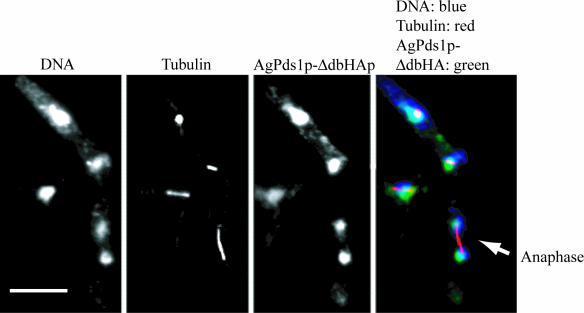
To test whether the disappearance of AgPds1p in anaphase is essential and whether it is likely mediated by the APC/C, we constructed Agpds1 mutants lacking the destruction box motif. The plasmid-coded HA-tagged destruction box deletion allele Agpds1Δdb-6HA (strain NSG10) expresses a securin which lacks the predicted destruction box motif TRMPLASKDRN (Fig. 4). To evaluate whether this deletion of the destruction box stabilized AgPds1p during the nuclear cycle, plasmid-derived AgPds1Δdb-6HAp was localized along with tubulin. The mutant protein could be readily seen in nuclei of all division stages, including anaphase (Fig. 6), in contrast to the wild-type AgPds1p, which was essentially absent in anaphase (Fig. 5B). Additionally, in regions of mycelia where AgPds1Δdbp was clearly observed in anaphase nuclei, there were many aberrantly large nuclei with mitotic spindles, suggesting that stabilized AgPds1p leads to a mitotic delay and/or defects. It is likely that AgPds1p is an important target of the APC/C in A. gossypii and that its stability may contribute to the lethality of Agcdc20, Agcdh1, and/or APC/C deletion mutants. As outlined in Materials and Methods, it was not possible to isolate a homokaryotic strain in which the wild-type AgPDS1 allele was replaced by the destruction box deletion allele. Only when both alleles are simultaneously expressed can the multinucleated mycelia grow, but such coexpression inhibits sporulation. This strongly suggests that the AgPds1Δdbp allele is a dominant-negative allele in terms of sporulation and that AgPds1p levels must be carefully regulated in this process.
FIG. 6.
AgPds1Δdbp persists in anaphase nuclei. Hyphae containing a plasmid expressing AgPds1Δdb-6HA (NSG10) were scraped from a selective plate, vortexed in liquid medium to break apart the mycelia, grown under selection for 15 h, and processed for tubulin and HA epitope staining. The arrow highlights one anaphase nucleus with nondegraded AgPds1Δdb-6HA. Bar, 10 μm.
APC/C subunits and targeting factors are nuclear in all stages of the cell cycle.
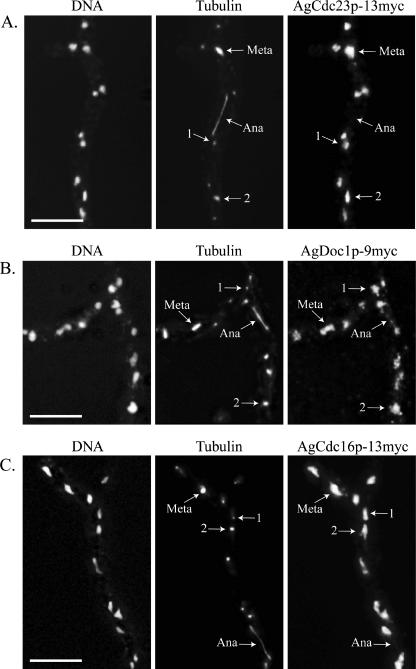
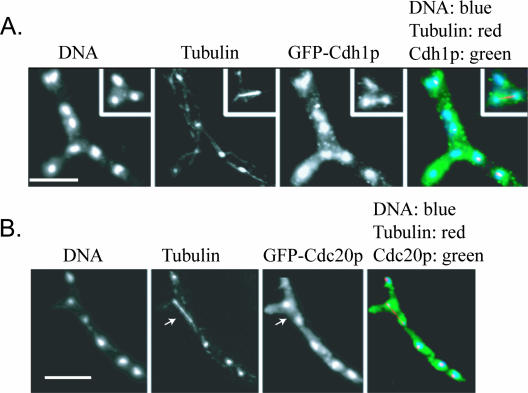
AgPds1p is at least one factor that is likely to be degraded for normal nuclear division in A. gossypii, and this degradation seems to be limited to the metaphase-to-anaphase transition. Given that nuclei are asynchronously dividing in A. gossypii, this raises the question as to how the APC/C may be activated only in the subset of nuclei at the meta-anaphase boundary and presumably inactive in neighboring nuclei that are in other stages. It is conceivable that the APC/C and/or its targeting factors AgCdh1 and AgCdc20 may vary in their localization through the different periods of the nuclear cycle. The predicted core APC/C subunits AgCdc23p, AgDoc1p, and AgCdc16p were found in all nuclei of all nuclear division cycle stages when localized by immunofluorescence (Fig. 7). This suggests that the APC/C is present in all nuclei and specifically activated in anaphase, leading to the prediction that the targeting factors AgCdc20p and AgCdh1p would fluctuate in their localization across the cell cycle. Remarkably, however, GFP-AgCdh1p and GFP-AgCdc20p were also found to be nuclear and present in all cell cycle stages when expressed from the AgBNI1 promoter and localized by immunofluorescence using an anti-GFP antibody (Fig. 8). This antibody did not produce any nonspecific nuclear staining in untagged strains (see Fig. S1 in the supplemental material). We speculate then that any cell cycle-specific substrate degradation must be due to the regulation of APC/C activity, which either impacts its ability to interact with potential substrates or alters how factors within the APC/C itself associate, but not the presence or absence of factors in certain nuclei.
FIG. 7.
The predicted A. gossypii APC/C subunits AgCdc23p, AgDoc1p, and AgCdc16p localize to nuclei in all stages of the nuclear division cycle. (A) AgCdc23p-13myc (GVS4), (B) AgDoc1p-9myc (NSG11), and (C) AgCdc16p-13myc (GVS2) cells were grown for 16 h at 30°C under selective conditions and processed for anti-myc and antitubulin immunofluorescence. In AgDOC1p-13myc (GVS3), a signal was also observed for all nuclear cycle stages. Arrows highlight nuclei in different stages of the division cycle, where “1” indicates a nucleus with a single SPB and “2” indicates a nucleus with duplicated SPBs. Meta, metaphase; Ana, anaphase. Bar, 10 μm.
FIG. 8.
APC activators AgCdh1p and AgCdc20p localize to all nuclei independent of the nuclear cycle stage. (A) Immunostained hyphae expressing N-terminally GFP-labeled AgCdh1p (NSG12). (B) Immunostained hyphae expressing N-terminally GFP-labeled AgCdc20p (NSG13). All nuclei, including anaphase nuclei (arrows), show anti-GFP immunofluorescence. The nuclei of nontransformed hyphae were not stained (see Fig. S1 in the supplemental material). Bar, 10 μm.
DISCUSSION
With these experiments, we have begun to evaluate the role of protein degradation during the nuclear cycle in multinucleated A. gossypii cells. We found that components of the APC/C and the activator proteins AgCdh1p and AgCdc20p are required for growth and nuclear cycle progression. This study was initiated because of the previous observation that the mitotic cyclin AgClb1/2p does not appear to be degraded in A. gossypii (14). Here, we have demonstrated that there is an essential role for APC/C-mediated degradation and that there are other important substrates of the APC/C that must be eliminated from A. gossypii nuclei in a timely manner. AgPds1p, a securin homologue, is one factor whose levels must be precisely controlled such that both deletion and stabilization of AgPds1p lead to strong phenotypes. Given that mitosis is asynchronous in A. gossypii, the APC/C must be regulated in a nuclear autonomous manner so that its activation is limited in time and space. We discuss here ways in which the APC/C in A. gossypii may have evolved to adapt to a syncytial and asynchronous division cycle.
All of the APC/C mutants that we generated showed specific defects in mitosis and impacted the progression through anaphase and/or exit from mitosis. Our data suggest that one key substrate of the APC/C is AgPds1p. We first characterized the function of wild-type AgPds1p and then evaluated its possible regulation by the APC/C by mutating its destruction box. In budding yeast, ScPds1p has a paradoxical role of binding and inhibiting ScEsp1p, the separase that dissolves sister chromatid cohesion for anaphase, and also promoting the nuclear localization of ScEsp1p. Remarkably, yeast cells lacking Pds1p are viable but restricted for growth at high temperatures, presumably because ScEsp1p can enter the nucleus through an alternative route to trigger dissolution of cohesion (1, 24).
One possible reason Agpds1 deletion mutants fail to grow is that AgEsp1p lacks such an alternate route, so it may never properly be localized to nuclei. In addition to impairing chromatid separation, this could lead to a failure in the activation of AgCdh1p and AgSic1p, both of which may respond to a signal originating from active AgEsp1p to activate the AgCdc14 early-anaphase release network (FEAR). Conversely, AgEsp1p may be overactive due to the loss of AgPds1p, and this leads to a lack of tension across a spindle, blocking paths that promote anaphase independent of AgPds1p. Future study of the behavior of AgEsp1p should distinguish between these possibilities and lend insight into how anaphase entry may be regulated in A. gossypii.
Just as the loss of AgPds1p is deleterious, timely degradation of this protein is also likely required for normal nuclear division. Agpds1Δdb homokaryon mutants were not recovered, and this allele was dominant in heterokaryon cells, suggesting that the levels of AgPds1p must be tightly regulated. Unregulated and stable AgPds1p could act to prevent activation of AgCdh1p by dephosphorylation (through failure to activate AgEsp1p and the FEAR network). It is not possible with the tools currently available in A. gossypii to determine whether AgPds1p degradation is important only for sporulation or whether it is also essential in the vegetative cycle and the likely cause of lethality of some of the APC/C mutants. Regions of heterokaryon hyphae with high levels of AgPds1Δdbp show an increase in metaphase nuclei, suggesting that at least some degradation of AgPds1p is important for normal progression. If, in fact, future experiments show that AgPds1p degradation is not essential, then other essential targets of the APC/C remain to be identified and possible targets to test are the S-phase cyclins AgClb5/6p and AgClb3/4p.
We would predict based on the presence of WD motifs that AgPds1p is targeted for degradation by AgCdc20p and/or AgCdh1p. Notably, both AgCDH1 and AgCDC20 are essential genes in contrast to yeast, where ScCdh1p and ScCdc20 are partially redundant and thus nonessential. Why are these targeting factors unable to functionally complement each other in A. gossypii? Lack of redundancy between proteins is often explained by differences in either the time of expression or the subcellular localization; however, both AgCdh1p and AgCdc20p, as well as the core APC/C, coexist in space and time. This lack of complementation could likely be explained by significant substrate specificity differences between these factors. There may be less redundancy in a multinucleated nuclear division cycle than in a uninucleated cell division cycle because the presence of many nuclei in one cytoplasm may essentially act as a buffer, where some mistakes in division are tolerated in the system because many other nuclei are there to compensate.
Our characterization of the APC/C, its activators, and a substrate points to a role for regulated protein degradation in mitotic progression in A. gossypii, despite the lack of oscillation in the pool of mitotic cyclins in this system. We proposed previously that AgClb1/2p degradation was replaced with direct inhibition by AgSic1p (14). We thought that because new cyclin protein is continually supplied to nuclei from the cytoplasm, this influx may be challenging to continually eliminate by proteolysis. The data we present here, however, show that there is still some role in A. gossypii for regulated proteolysis of at least some factors, including AgPds1p. In Drosophila embryos, there is precedent for spatially and functionally distinct pools of the APC/C and Cdh1/Cdc20 that help to regulate when and what substrates are degraded (20). In contrast, the core factors of the APC/C examined here are uniformly present in all nuclei in A. gossypii, but presumably, the whole complex is not constitutively active. This raises the question of how APC/C proteolysis activity is spatially and temporally limited to only mitotic/G1 nuclei in these multinucleated and asynchronous cells.
The ubiquitous localization of the APC/C suggests that the control of degradation is regulated at the level of the substrate rather than by the APC/C itself. This could be, such as is observed in the SCF degradation system in budding yeast, through cell cycle-regulated modification, such as phosphorylation of the substrate. Thus, the APC/C will always be ready and potentially active but substrates themselves will dictate their timely destruction. Modifications to proteins which trigger their destruction would likely be nuclear limited given the asynchronous pattern of division. Modification events could be regulated by pathways such as the spindle assembly checkpoint and thus be generated as a result of chromosome attachment or tension on the spindle and may therefore remain limited to within nuclei. If this type of a nuclear intrinsic event leads to the marking of a protein for recognition by the APC/C, then this may ensure that a neighboring nucleus in S or G2 does not prematurely degrade a given substrate. Evaluation of the function and localization of proteins in the spindle assembly checkpoint, such as Mad2 and those proteins thought to communicate tension status in spindles such as Ipl/Aurora kinase and kinetochore proteins, would be a first approach to testing whether these components play a normal role in marking the timely degradation of proteins for anaphase progression. Future study of the APC/C in A. gossypii should yield insights into how protein degradation can be limited in time and space in large, multinucleated hyphal cells.
Supplementary Material
Acknowledgments
We thank A. Kaufmann, D. Hoepfner, and M. Koehli for plasmids, Rachel Shakked for her assistance in constructing two deletion mutants, and Philipp Knechtle for advice and helpful discussions.
This work was supported by a grant from the Swiss National Science Foundation (3100A0-100734) to P.P. and A.S.G., by the Roche Foundation, and by a National Science Foundation postdoctoral fellowship to A.S.G.
Footnotes
Published ahead of print on 8 December 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Agarwal, R., and O. Cohen-Fix. 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 16:1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti-Segui, C., F. Dietrich, R. Altmann-Johl, D. Hoepfner, and P. Philippsen. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975-986. [DOI] [PubMed] [Google Scholar]
- 3.Altmann-Jöhl, R., and P. Philippsen. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69-80. [DOI] [PubMed] [Google Scholar]
- 4.Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563-4575. [DOI] [PubMed] [Google Scholar]
- 5.Burton, J. L., and M. J. Solomon. 2001. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 15:2381-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, J. L., V. Tsakraklides, and M. J. Solomon. 2005. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol. Cell 18:533-542. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, C. W., M. Enquist-Newman, and D. O. Morgan. 2005. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr. Biol. 15:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, C. W., and D. O. Morgan. 2002. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell Biol. 4:880-887. [DOI] [PubMed] [Google Scholar]
- 9.Castillo-Lluva, S., T. Garcia-Muse, and J. Perez-Martin. 2004. A member of the Fizzy-related family of APC activators is regulated by cAMP and is required at different stages of plant infection by Ustilago maydis. J. Cell Sci. 117:4143-4156. [DOI] [PubMed] [Google Scholar]
- 10.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Cohen-Fix, O., and D. Koshland. 1999. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 13:1950-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 13.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 14.Gladfelter, A. S., A. K. Hungerbuehler, and P. Philippsen. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 16.Grossberger, R., C. Gieffers, W. Zachariae, A. V. Podtelejnikov, A. Schleiffer, K. Nasmyth, M. Mann, and J. M. Peters. 1999. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem. 274:14500-14507. [DOI] [PubMed] [Google Scholar]
- 16a.Hall, M. C., E. N. Warren, and C. H. Borchers. 2004. Multi-kinase phosphorylation of the APC/C activator Cdh1 revealed by mass spectrometry. Cell Cycle 3:e38-e44. [DOI] [PubMed] [Google Scholar]
- 17.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 18.Hilioti, Z., Y. S. Chung, Y. Mochizuki, C. F. Hardy, and O. Cohen-Fix. 2001. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr. Biol. 11:1347-1352. [DOI] [PubMed] [Google Scholar]
- 19.Hoepfner, D., A. Brachat, and P. Philippsen. 2000. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell 11:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, J., and J. W. Raff. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, J. N., I. Park, E. Ellingson, L. E. Littlepage, and D. Pellman. 2001. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J. Cell Biol. 154:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, S. W., P. M. Mirabito, P. C. Scacheri, and N. R. Morris. 1995. The Aspergillus nidulans bimE (blocked-in-mitosis) gene encodes multiple cell cycle functions involved in mitotic checkpoint control and mitosis. J. Cell Sci. 108:3485-3499. [DOI] [PubMed] [Google Scholar]
- 23.Jaspersen, S. L., J. F. Charles, and D. O. Morgan. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9:227-236. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, S., M. Segal, D. J. Clarke, and S. I. Reed. 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152:27-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 26.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 27.King, R. W., R. J. Deshaies, J. M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 274:1652-1659. [DOI] [PubMed] [Google Scholar]
- 28.Kraft, C., H. C. Vodermaier, S. Maurer-Stroh, F. Eisenhaber, and J. M. Peters. 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18:543-553. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, J. R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257-259. [DOI] [PubMed] [Google Scholar]
- 30.Lim, H. H., P. Y. Goh, and U. Surana. 1998. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8:231-234. [DOI] [PubMed] [Google Scholar]
- 30a.Mohr, C. 1997. Ph.D. thesis. University of Basel, Basel, Switzerland.
- 31.Morris, N. R. 1976. A temperature-sensitive mutant of Aspergillus nidulans reversibly blocked in nuclear division. Exp. Cell Res. 98:204-210. [DOI] [PubMed] [Google Scholar]
- 32.Murray, A. W. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221-234. [DOI] [PubMed] [Google Scholar]
- 33.Nasmyth, K. 2005. How do so few control so many? Cell 120:739-746. [DOI] [PubMed] [Google Scholar]
- 34.O'Donnell, K. L., A. H. Osmani, S. A. Osmani, and N. R. Morris. 1991. bimA encodes a member of the tetratricopeptide repeat family of proteins and is required for the completion of mitosis in Aspergillus nidulans. J. Cell Sci. 99:711-719. [DOI] [PubMed] [Google Scholar]
- 35.Passmore, L. A. 2004. The anaphase-promoting complex (APC): the sum of its parts? Biochem. Soc. Trans. 32:724-727. [DOI] [PubMed] [Google Scholar]
- 36.Passmore, L. A., E. A. McCormack, S. W. Au, A. Paul, K. R. Willison, J. W. Harper, and D. Barford. 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 38.Pfleger, C. M., and M. W. Kirschner. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655-665. [PMC free article] [PubMed] [Google Scholar]
- 39.Pfleger, C. M., E. Lee, and M. W. Kirschner. 2001. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 15:2396-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pringle, J. R., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 41.Schott, E. J., and M. A. Hoyt. 1998. Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics 148:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwab, M., A. S. Lutum, and W. Seufert. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90:683-693. [DOI] [PubMed] [Google Scholar]
- 43.Schwab, M., M. Neutzner, D. Mocker, and W. Seufert. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20:5165-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirayama, M., A. Toth, M. Galova, and K. Nasmyth. 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402:203-207. [DOI] [PubMed] [Google Scholar]
- 45.Shirayama, M., W. Zachariae, R. Ciosk, and K. Nasmyth. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thornton, B. R., K. C. Chen, F. R. Cross, J. J. Tyson, and D. P. Toczyski. 2004. Cycling without the cyclosome: modeling a yeast strain lacking the APC. Cell Cycle 3:629-633. [PubMed] [Google Scholar]
- 48.Thornton, B. R., and D. P. Toczyski. 2003. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5:1090-1094. [DOI] [PubMed] [Google Scholar]
- 49.Tinker-Kulberg, R. L., and D. O. Morgan. 1999. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 13:1936-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visintin, R., S. Prinz, and A. Amon. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278:460-463. [DOI] [PubMed] [Google Scholar]
- 51.Wäsch, R., and F. R. Cross. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418:556-562. [DOI] [PubMed] [Google Scholar]
- 52.Wendland, J., Y. Ayad-Durieux, P. Knechtle, C. Rebischung, and P. Philippsen. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381-391. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto, A., V. Guacci, and D. Koshland. 1996. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 133:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.Ye, X. S., R. R. Fincher, A. Tang, A. H. Osmani, and S. A. Osmani. 1998. Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA. Mol. Biol. Cell 9:3019-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeong, F. M., H. H. Lim, C. G. Padmashree, and U. Surana. 2000. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell 5:501-511. [DOI] [PubMed] [Google Scholar]
- 57.Zachariae, W., M. Schwab, K. Nasmyth, and W. Seufert. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282:1721-1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.