Abstract
We used differential display to select genes differentially expressed during differentiation of epimastigotes into metacyclic trypomastigotes in the protozoan parasite Trypanosoma cruzi. One of the selected clones had a sequence similar to that of the small-subunit (SSU) processome protein Sof1p, which is involved in rRNA processing. The corresponding T. cruzi protein, TcSof1, displayed a nuclear localization and is downregulated during metacyclogenesis. Heterologous RNA interference assays showed that depletion of this protein impaired growth but did not affect progression through the cell cycle, suggesting that ribosome synthesis regulation and the cell cycle are uncoupled in this parasite. Quantitative PCR (qPCR) assays of several SSU processome-specific genes in T. cruzi also showed that most of them were regulated posttranscriptionally. This process involves the accumulation of mRNA in the polysome fraction of metacyclic trypomastigotes, where TcSof1 cannot be detected. Metacyclic trypomastigote polysomes were purified and separated by sucrose gradient sedimentation. Northern blot analysis of the sucrose gradient fractions showed the association of TcSof1 mRNA with polysomes, confirming the qPCR data. The results suggest that the mechanism of regulation involves the blocking of translation elongation and/or termination.
Trypanosoma cruzi (7) is a flagellate protozoan parasite of the order Kinetoplastida. This parasite is the causal agent of Chagas' disease, which affects thousands of people every year in South and Central America, posing a major public health problem. During its complex life cycle, the parasite alternates between an insect and a mammalian host. Four developmental stages have been well characterized, and a number of intermediate forms are known (13). Epimastigotes and amastigotes are the replicative forms, whereas metacyclic trypomastigotes and bloodstream trypomastigotes are the infective, nonreplicative forms. The process by which epimastigotes differentiate into metacyclic trypomastigotes is called metacyclogenesis and takes place in the digestive tract of the insect vector. This event can be mimicked in vitro under chemically defined conditions (11).
Trypanosomatid genes are organized into long polycistronic units. Polycistronic RNAs are resolved into monocistronic mRNAs through coupled trans splicing and polyadenylation. Although this organization resembles bacterial operons, genes present in the same transcriptional unit may differ in their stage-specific expression. As yet, no typical RNA polymerase II promoters have been identified. Moreover, transfection experiments have shown that RNA polymerase II can initiate transcription in the absence of canonical promoter sequences. This lack of developmental control over transcription initiation strongly supports the hypothesis that trypanosome gene expression is controlled posttranscriptionally. Stage-specific expression is achieved by modulating mRNA half-life and controlling translation (9, 33). Regulatory mechanisms involve interactions between trans-acting factors and sequence elements in cis, mostly in the 3′ untranslated regions of mRNAs (15).
The rRNA genes of trypanosomatids are organized into tandem repeats, as in other eukaryotes (25). Synthesis begins in the nucleolus, where RNA polymerase I synthesizes a polycistronic 35S pre-rRNA. The rRNA genes in this primary transcript are separated by internal transcribed spacers (ITS) and flanked by external transcribed spacers (the 5′ETS and the 3′ETS). The 5S rRNA is transcribed independently by RNA polymerase III, outside the nucleolus (26). The newly synthesized 35S pre-rRNA associates with processing proteins to form the 90S preribosome particle, which is processed to generate the mature 18S, 5.8S, 24Sα, and 24Sβ rRNAs (22). This processing involves a sequence of specific cleavages by several enzymes and ribonucleoprotein nucleolar complexes (snoRNPs). These snoRNPs consist of large numbers of proteins associated with small nucleolar RNAs, which coordinate these events. The complete processing and assembly of ribosomes involve more than 200 proteins (helicases, nucleases, and transport and assembly factors) (18).
The U3 snoRNP, also known as the small-subunit (SSU) processome, is involved in the early cleavage of the 35S pre-rRNA. This complex consists of the U3 small nucleolar RNA of the C/D box snoRNA, with two conserved sequence elements (the C domain and the D domain) and 40S subunit processing factors (16). The SSU processome assembles during the transcription of the 35S pre-rRNA, within seconds of the completion of rRNA transcription (31). Cleavage of the 35S pre-rRNA releases the SSU processome and the 40S and 60S preribosomal particles (14). Tandem affinity purification of tagged processing proteins has led to the identification of more than 40 SSU processome proteins in Saccharomyces cerevisiae, only 7 of which are considered specific to this complex (Sof1p, Imp4p, Imp3p, Dhr1p, Rrp9p, Lcp5p, and Mpp10p) (16). Most of these proteins are also found in all eukaryotes, suggesting that synthesis mechanisms are strongly conserved (14).
The depletion of individual SSU processome proteins in yeast leads to a block in the G1 phase of the cell cycle (4). This observation is consistent with previous suggestions that ribosome processing and assembly are directly related to cell cycle progression, cell growth, and cell size control (27, 32). In nonreplicating metacyclic trypomastigote forms of T. cruzi, a decline in RNA polymerase I activity is accompanied by a decrease in ribosomal protein levels, nucleolus disassembly, and the arrest of parasites in a G1-like phase of the cell cycle (17).
We previously identified TcImp4, a protein very similar to the Imp4p protein of S. cerevisiae. This protein is nuclear and is downregulated in metacyclic trypomastigotes. Northern blot data suggest that TcImp4 gene expression is regulated by polysomal mobilization mechanisms (20). The differential mobilization of mRNAs to the polysomal fraction has been described as a mechanism of translational regulation in these parasites (2). Analysis of the proteins involved in rRNA processing in T. cruzi should provide new insight into the mechanisms controlling the ribosome assembly pathway and their role in gene expression and cell cycle regulation in this parasite.
We investigated the mechanisms regulating gene expression in T. cruzi further by identifying stage-specific genes by differential display. Nucleotide sequencing showed that one of the selected genes encoded a protein similar to Sof1p from Arabidopsis thaliana. We describe here the molecular characterization of this protein, TcSof1. We also show that SSU processome proteins are downregulated in metacyclic trypomastigotes. In addition, Northern blot analysis of polysome-associated mRNAs suggests that the mechanisms of regulation involved included the blocking of translational elongation.
MATERIALS AND METHODS
Cells.
T. cruzi clone Dm28c (10) epimastigote forms were grown at 28°C in liver infusion tryptose medium supplemented with 10% fetal bovine serum. Cells were differentiated in vitro into metacyclic trypomastigotes under chemically defined conditions (TAU3AAG medium), as previously described. Metacyclic trypomastigotes were purified by DEAE-51 cellulose chromatography (11). Procyclic forms of Trypanosoma brucei (Lister 427) strain 29-13 (35) were grown in SDM-79 medium (10% fetal bovine serum) in the presence of 50 μg/ml hygromycin and 15 μg/ml G418 (19).
RNA extraction and Northern blot analysis.
Polysome-associated RNAs were prepared as described by Goldenberg et al. (21). Total RNA was obtained with an RNeasy kit (QIAGEN). RNA present in the sucrose gradient fractions was isolated by phenol-chloroform extraction, followed by ethanol precipitation. Nucleic acids were blotted onto Hybond N+ membranes (Amersham). Probes were labeled with [α-32P]dCTP by nick translation (Amersham) and purified on Sephadex G-50 columns. Membranes were hybridized and washed at high stringency according to the manufacturer's instructions.
Differential display and cloning.
Polysomal RNAs (2 μg) were converted to cDNA by incubation with oligo(dT)15 and ImProm-II reverse transcriptase and reagents (Promega) for 2 h at 42°C. Samples were purified by centrifugation through a Microcon YM-30 filter (Millipore). The cDNA was amplified by PCR, using a combination of poly(A)-complementary poly(T)11 primers with C, G, and A terminators and random primers in the presence of [α-33P]dCTP. Random primers were AAGCTTGATTGCC (AP1), AGCTTCGACTGT (AP2), GCTTTGGTCAG (AP3), GGTACTCCAC (AP4), GTTGCGATG (AP5), GCAATCGATG (AP6), and ATTCCTTCGG (AP7). PCRs contained cDNA (100 ng), 0.4 U Taq DNA polymerase, reaction buffer (Promega), 2 μM deoxynucleoside triphosphate, 0.2 μM (T)11N, random primers, and 5 μCi [α-33P]dCTP. PCR conditions were as follows: 40 cycles of 94°C for 30 seconds, 40°C for 2 min, 72°C for 30 seconds, and 72°C for 5 minutes. Amplified fragments were separated by electrophoresis in 6% polyacrylamide gels, which were then dried and placed against an X-ray film. The selected fragments were reamplified with the same primers and inserted into pDK101 (28).
The complete coding sequence of TcSof1 in the Dm28c clone was obtained using synthetic primers based on the CL Brener sequence (Tc00.1047053508461.390) (forward, 5′ATGGTGAAGGTGGAAAACCATC3′; reverse, 5′TCTAGATAAATTAAGAGACTGATAAAC3′) to amplify the gene. PCR conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 30 seconds, and 72°C for 2 min and a final extension step of 72°C for 7 min. The PCR products were inserted into pGEM-T Easy vector (Promega). Positive clones were purified with a QIAprep Spin Miniprep kit (QIAGEN) and sequenced in an ABI 3100 automatic sequencer (Applied Biosystems). Internal primers (forward, 5′GGTCAGTGGAGCGTTTGATGGGGG3′; reverse, 5′TTATACCATTAATATGTCCCTGAAAAACG3′) were synthesized for complete sequencing. The sequence was analyzed and the database searched with Lasergene software (DNASTAR Inc.) and the BLAST algorithm.
Production of polyclonal antiserum and Western blot analysis.
The TcSof1 coding region was amplified by PCR; primers used were 5′GGAGCATGCCATGGTGAAGGTGAAAAC3′ and 5′CCGAAACTGCAGTTAATTAAGAGAC3′. Amplified fragments were inserted into the pQE31 vector (QIAGEN) and used to transform Escherichia coli strain M15. Production of the recombinant protein was induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Inclusion bodies containing the recombinant protein were isolated, and the recombinant protein was purified by electroelution. Polyclonal antiserum was obtained from New Zealand White rabbits, as previously described (12). Total protein extracts were prepared by resuspending phosphate-buffered-saline (PBS)-washed parasites in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (106 parasites/μl). Proteins were fractionated by SDS-PAGE in 10% polyacrylamide gels, transferred onto nitrocellulose membranes (Amersham), and probed with the polyclonal antiserum, as described elsewhere (12).
Immunofluorescence assays and transmission electron microscopy.
Epimastigote forms were washed and resuspended at a density of 107 cells/ml in PBS. Cells were added to the poly-l-lysine-coated slides, which were then incubated at room temperature for 20 min. Parasites were fixed by incubation in 4% paraformaldehyde for 5 min and were then washed twice in 0.1 M glycine (pH 8.6) in PBS, for 2 min each. Cells were made permeable by incubation for 5 min with 0.075% Triton X-100 in PBS. Parasites were blocked by incubation overnight with 1% bovine serum albumin in PBS and then incubated for 1 h with polyclonal antiserum against TcSof1. Parasites were washed and incubated for 45 min with 1% bovine serum albumin in PBS, 10 μg propidium iodide, and anti-rabbit fluorescein isothiocyanate-conjugated antibody (Sigma). Cells were examined with a Nikon E600 microscope. For immunocytochemical procedures, cells were prepared as previously described (20) and observed under a Zeiss 900 transmission electron microscope.
Interference RNA and fluorescence-activated cell-sorting (FACS) analyses.
Three fragments of TbSof1 and two fragments of TbImp4 covering the entire lengths of these genes were PCR amplified and inserted into the p2T7-177 vector (21). T. brucei DNA was extracted in phenol-chloroform and 100 ng used for PCR amplification. The forward and reverse primers used contained restriction sites for BamHI and HindIII. The TbSof1 fragments (nucleotides 41 to 459, 389 to 727, and 803 to 1221) were amplified with the following primers: TbSof1iRNAF1 (5′CGCGGATCCGGACCAAGGATCGGAACGG3′), TbSof1iRNAR1 (5′CCCAAGCTTCCGCCAGGGGTTCTACTTTG3′), TbSof1iRNAF2 (5′CGCGGATCCGGGACAAGGTGGTTAAGATGTGG3′), TbSof1iRNAR2 (5′CCCAAGCTTTGCAACACATCTCCAAAACAACC3′), TbSof1iRNAF3 (5′CGCGGATCCATATGAGGATACCGGGACGCC3′), and TbSof1iRNAR3 (5′CCCAAGCTTGGCACTGCGGATGGCTTTAG3′). The TbImp4 fragments (nucleotides 10 to 234 and 407 to 851) were amplified with the following primers: TbImp4iRNAF1 (5′CGCGGATCCAGCGTTATTCGGCAGCGTAAG3′), TbImp4iRNAR1 (5′CCCAAGCTTAGCTTTGGCGTACTCGTCGTC3′), TbImp4iRNAF2 (5′CGCGGATCCTAGTGGTTTTGCAGGAATCTCAGG3′), and TbImp4iRNAR2 (5′CCCAAGCTTCTCCGCTTCTTCGCTGTGTTC3′).
Procyclic forms of the T. brucei 29-13 cell line were then transformed with 10 μg of NotI-linearized plasmid. Cells were transfected by electroporation in 4-mm cuvettes, using a Bio-Rad GenePulser II electroporator, with two pulses of 1,600 V and 25 μF. After 24 h, 5 μg/ml phleomycin was added for the selection of transfected parasites. Transfectants were cloned by serial dilution in 24-well plates. The synthesis of double-stranded RNA (dsRNA) was induced by adding 2 μg/ml of tetracycline to exponentially growing cultures at time zero and adding an additional 1 μg/ml of tetracycline on each subsequent day. Growth curves were obtained over a period of 8 to 9 days after the induction of RNA interface (RNAi). For FACS analysis, cells were fixed in methanol, stained with propidium iodide, and analyzed by flow cytometry, as previously described (24). Parasites were resuspended at a concentration of 106 parasites/ml and incubated with 5 μg/ml propidium iodide at room temperature for 30 min. FACS was performed with a BD FACSCalibur system equipped with an FL-2 detector. Data were analyzed with WinMDI version 2.8 software.
Real-time PCR.
Two-step real-time reverse transcription-PCR assays were performed in an ABI PRISM 7000 sequence detection system (Applied Biosystems). For a 25-μl reaction mixture, we added 10 ng of cDNA and the recommended concentration of SYBR green master mix (Applied Biosystems). The specific primers were added at concentrations of 200 nM in all cases. Primers were as follows: TcL9F (5′ CCTTCACTGCCGTTCGTTGGTTTG 3′), TcL9R (5′ATGCGAGAGTGCCGTGTTGATGGT 3′), TcH2BF (5′ CGGTGGTGCGCGTCAACAAGAAGC 3′), TcH2BR (5′ CCAGGTCCGCCGGCAGCACGAG 3′), TcSof1F (5′ GGTCAGTGGAGCGTTTGATGGGGG 3′), TcSof1R (5′ AAGCGTAGAATCCAACGATC 3′), TcDhr1F (5′ GGAATTGTTATCGGTACTGTCTGGTGC 3′), TcDhr1R (5′ TGAGCGACCCAAGAGGATGTAAGG 3′), TcImp3F (5′ AGGACCGTGAAGACTACCGCAGG 3′), TcImp3R (5′ GAAACGCGGGGTCCCTAACTTG 3′), TcMpp10F (5′ CAAACTAGAGGACCGCATTCGCC 3′), TcMpp10R (5′ GTAGTAGAAGTTGCTGAGGGCGTCG 3′), TcRrp9F (5′ ATGTTGTGCCTCTTGATGATTCCAG 3′), TcRrp9R (5′ CTTCACCACCGCCGTCTCGTC 3′), TcImp4F (5′ GACGTTGGCACGATGTCTGAGC 3′), TcImp4R (5′ CTCGGCGTCGTCCATCTCAAG 3′), TcLcp5F (5′ GATGGTGTCGGCACTTGGCTAC 3′), and TcLcp5R (5′ TTGCACGCATCAGTCGTACTTGTTC 3′).
PCR conditions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Thermal dissociation confirmed that reverse transcription-PCR generated a single amplicon. We used a standard curve method, based on cycle threshold values, to assess the expression of the genes studied. We used triplicate 1:2 dilutions of known concentrations of cDNA to generate curves extending from 10 ng to 78.1 pg of cDNA. Dilution series of eight known concentrations (10 ng to 78.1 pg) of cDNA were used in triplicate. A standard curve was generated for each of the genes studied and for both control genes. We calculated cDNA concentration by dividing the value obtained for the gene under investigation by the value obtained for each of the control genes. Differences in expression are reported, using epimastigotes as the reference population.
Sucrose density gradient separation and Northern blot analyses.
T. cruzi polysomes were purified and separated on sucrose gradients as described by Brecht and Parsons (6), with modifications. Exponentially growing cultures of 5 × 108 epimastigote cells and 109 metacyclic trypomastigotes were lysed in lysis buffer (300 mM KCl, 10 mM MgCl2, 10 mM Tris-HCl [pH 7.4], 10% NP-40, and 2 M sucrose) and centrifuged at 16,000 × g for 5 min at 4°C. The supernatant (400 μl) was layered onto 10% to 50% sucrose density gradients prepared in TKM300 buffer plus inhibitors (300 mM KCl, 10 mM MgCl2, 10 mM Tris-HCl [pH 7.4], 100 μg/ml cycloheximide, 10 μM E-64, 1 mM phenylmethylsulfonyl fluoride) and centrifuged at 4°C for 1 h or 2 h at 38,000 rpm in a Beckman SW41 rotor. For the EDTA assay, the supernatant was treated with 100 mM of EDTA for 30 min on ice before layering onto sucrose density gradient. For the puromycin assay, exponentially growing cultures of epimastigotes (5 × 108 cells) and metacyclic trypomastigotes (109 cells) were preincubated with 2 mM of puromycin for 1 h at 28°C. The cells were incubated in high-salt lysis buffer (500 mM KCl, 2 mM MgCl2, 10 mM Tris-HCl [pH 7.4], 10% NP-40, 2 M sucrose, and 10 mM puromycin) and centrifuged at 16,000 × g for 5 min at 4°C. The supernatant (400 μl) was layered onto 10% to 50% sucrose density gradients prepared in TKM500 buffer plus inhibitors (500 mM KCl, 2 mM MgCl2, 10 mM Tris-HCl [pH 7.4], 10 μM E-64, 1 mM phenylmethylsulfonyl fluoride) and centrifuged at 4°C for 2 h at 38,000 rpm in a Beckman SW41 rotor. After centrifugation, 500-μl fractions were collected, using the ISCO gradient fractionation system.
In vivo labeling of T. cruzi proteins and immunoprecipitation.
Epimastigote and metacyclic trypomastigote cells (109) were incubated in 5 ml of TAU3AAG (supplemented with 10 mM glucose and 200 μCi of [35S]methionine) for 2 h at 28°C. After this time, 5 × 108 cells were colleted (zero time) and the remaining were harvested and incubated in 5 ml of liver infusion tryptose medium (for epimastigotes) or TAU3AAG medium (for metacyclic trypomastigote) for 30 min at 28°C before lysis in 1 ml NP-40 lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.0], and 1% NP-40) for 1 h at 4°C. Protein A-Sepharose beads (Sigma) were incubated with each respective antiserum overnight at 4°C and blocked with 5% skim milk in PBS. Protein extracts corresponding to 5 × 108 cells were incubated with the Sepharose beads for 2 hours at room temperature, washed twice in PBS-1% Tween, and boiled for 5 min in SDS-PAGE loading buffer.
Nucleotide sequence accession number.
The sequence determined in this study was deposited in GenBank under accession number AY738725.
RESULTS
Cloning of the TcSof1 gene.
We used differential display to identify genes differentially expressed in metacyclic trypomastigotes. The polysomal RNA fraction from epimastigotes and metacyclic forms was used in amplification assays. The DNA fragments preferentially amplified from the metacyclic polysomal fraction were selected for further analysis. We identified 46 bands corresponding to differentially expressed genes, which were then reamplified, cloned, and sequenced. The sequences formed seven clusters, with 17 sequences remaining unclustered. Most of these sequences displayed various degrees of similarity to hypothetical proteins. However, only three were found to be similar to known proteins or specific motifs (not shown). One of these sequences was very similar to the sequence of proteins bearing the conserved WD40 motif. The complete coding sequence was obtained from the T. cruzi GeneDB database. Primers for amplification of the full-length gene from T. cruzi Dm28c were designed based on the CL Brener clone sequence. The full-length gene was amplified by PCR, inserted into the pGEM-T Easy (Promega) vector, and sequenced. The predicted protein displayed 58% identity at the amino-acid level to the Sof1p protein of Arabidopsis thaliana and 55% identity to the homologous protein in S. cerevisiae. The Sof1p protein is a component of the SSU processome, or U3snoRNP complex, involved in the early steps of rRNA processing. The gene identified, TcSof1, is a single-copy gene with an open reading frame of 1,335 nucleotides encoding a predicted protein of 50.4 kDa. Sequence analysis showed that most of this protein displays the conserved WD structure. The specific Sof1 domain has no known function and is found in the carboxy-terminal region (see Fig. S7 in the supplemental material).
Stage-specific expression of TcSof1.
The coding sequence of TcSof1 was inserted into the pQE30 expression vector, which was then used to transform E. coli M15. The 50-kDa recombinant protein was recovered as a fusion polypeptide with a carboxy-terminal histidine tag, after induction with IPTG. This tag made it possible to purify the recombinant protein by affinity chromatography on nickel columns (QIAGEN). Two bands of approximately 50 kDa in size were purified; the identity of the purified recombinant protein was confirmed by mass spectrometry (not shown). The recombinant protein was used to prepare a polyclonal antiserum in rabbits. The polyclonal antiserum was used to analyze the expression of TcSof1 during metacyclogenesis and amastigogenesis.
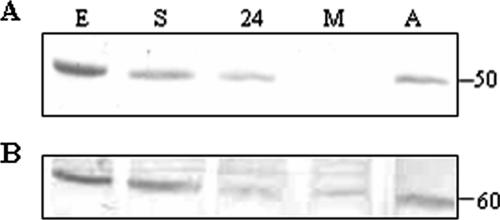
On Western blots of cellular extracts from differentiating parasites, a band of 50 kDa was detected, corresponding to the expected molecular mass of TcSof1 (Fig. 1A). This protein was observed in the epimastigote forms and in differentiating parasites but was not detected in metacyclic trypomastigotes. Expression of TcSof1 was restored soon after the parasites began to differentiate into amastigotes. After 24 h of in vitro amastigogenesis, when intermediate, round cells were observed, a signal corresponding to TcSof1 was clearly detected. A signal corresponding to an 80-kDa protein was detected in the metacyclic trypomastigote extracts. However, this signal may result from nonspecific recognition or cross-reaction with another protein bearing the conserved WD motif.
FIG. 1.
(A) Western blot analysis of protein extracts from epimastigotes (E), epimastigotes under nutritional stress (S) and after 24 h of differentiation (24), metacyclic trypomastigotes (M), and trypomastigotes after 24 h of amastigogenesis (A), probed with antiserum against TcSof1 protein (1/500 dilution). (B) The same membrane was probed with antiserum against PEPCK, to control for protein loading.
Immunolocalization of TcSof1.
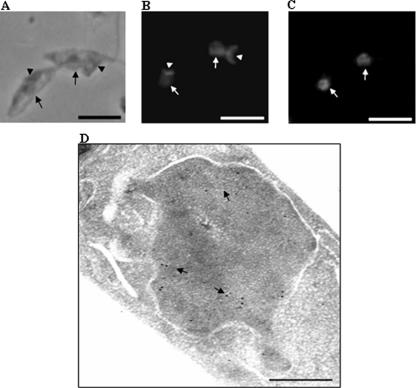
In yeast, the SSU processome complex assembles on the nascent pre-rRNA and the initial cleavage steps take place in the nucleolus. Immunofluorescence studies were carried out to determine the cellular distribution of TcSof1 in T. cruzi epimastigote forms. Labeling was observed throughout the nucleus (Fig. 2C). This result was confirmed by ultrastructural immunocytochemistry, using the same anti-TcSof1 serum (Fig. 2D).
FIG. 2.
Immunolocalization of T. cruzi TcSofI in epimastigote forms. (A) Phase-contrast microscopy. (B) Propidium iodide staining. (C) Immunofluorescence analysis showing dispersed labeling of the nucleus. (D) Ultrastructural immunocytochemistry confirmed the labeling in the nucleus, with gold particles distributed throughout the nucleus. (A, B, and C) Arrowheads indicate the kinetoplast structure, and arrows indicate the nucleus. (A, B, and C) Bar, 10 μm. (D) Bar, 0.25 μm.
RNA interference and growth inhibition.
Recent studies have indicated that ribosome synthesis is tightly coupled to growth and cell cycle regulation. The depletion of SSU processome proteins leads to cell cycle arrest in the G1 phase. Metacyclic trypomastigotes have low levels of RNA polymerase II activity and are arrested in a G1-like phase of the parasite cell cycle. Accordingly, our results show that rRNA processing proteins are also downregulated in these forms, raising the question of whether ribosome synthesis and the cell cycle are also linked in the parasite. RNA interference was used to deplete the cells of TbSof1 and TbImp4 orthologs. T. cruzi lacks the machinery required for RNA interference, so these experiments were carried out in T. brucei, using the orthologous genes as targets. Various fragments spanning the entirety of these genes were inserted into p2T7-177 and tested as double-strand probes. The Sof1-IB (Fig. 3A) and -IC (not shown) and Imp4-IB (Fig. 3D) constructs clearly inhibited growth, in at least three different experiments, following tetracycline induction. Parasites transfected with constructs expressing green fluorescent protein (GFP) dsRNA alone were used as controls (Fig. 3F). Growth inhibition was consistent with the absence of an mRNA signal on Northern blots (Fig. 3B and E), thereby confirming the silencing of both genes and the need for the products of both these genes for parasite growth. We checked that the growth defect observed was due to abnormal ribosome synthesis by sucrose density gradient centrifugation to isolate the polysomal fraction of RNAi parasites. The tetracycline-induced parasites had lower polysome levels and accumulated 60S subunits due to defective 40S subunit synthesis (see Fig. S8 in the supplemental material). The total DNA content of the parasites was analyzed by FACS. Parasites on the fifth day of RNAi silencing were stained with propidium iodide and compared with noninduced cells. Similar results were obtained for both induced and noninduced cells, showing that there was no specific accumulation in a particular phase of the cell cycle in response to the depletion of both 18S rRNA processing proteins (Fig. 3C).
FIG. 3.
TbSof1 RNA interference inhibits growth in procyclic forms of T. brucei. RNAi cell lines expressing dsRNA fragments of TbSof1 (A) and TbImp4 (D) were grown in medium with (+tet) or without (−tet) tetracycline. Cell number was monitored daily. Results for one of at least three independent assays are shown. (B and E) Northern blot analysis of parasites after various periods of tetracycline induction. (C) Parasites transfected with a construct expressing a GFP dsRNA were used as a control. (F) FACS analysis of the DNA content of procyclic forms on the fourth day with and without tetracycline. The graphs for one of three experiments for tetracycline-induced and noninduced cells have been superimposed.
Posttranscriptional coregulation of SSU processome proteins.
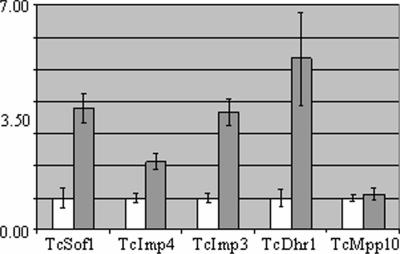
There is strong evidence for coregulation of the transcription of genes involved in ribosome synthesis in most eukaryotes. Synthesis of the TcImp4 protein is known to be reduced in metacyclic trypomastigotes. We show here that TcSof1 protein levels decrease during T. cruzi metacyclogenesis. This is consistent with the coregulation of expression of specific SSU processome genes. Using the yeast SSU processome genes as probes, we searched the GeneDB database for T. cruzi orthologs. We identified 40 putative orthologs, including the seven proteins previously described as specific to U3snoRNP (see Table S1 in the supplemental material). We analyzed the production of these SSU processome-specific proteins during metacyclogenesis by quantitative PCR (qPCR), using the polysomal RNA fraction from epimastigote and metacyclic trypomastigote forms. Unexpectedly, mRNA levels for most of these specific genes were found to be higher in the metacyclic trypomastigote forms. Accordingly, real-time PCR showed higher levels of mRNA in the polysomal fraction for the TcSof1, TcImp4, TcImp3, and TcDhr1 genes (Fig. 4). Conversely, no variation was found in levels of expression of the TcMpp10 gene. The results for TcLcp5 and TcRrp9 were not considered reliable, due to the presence of secondary bands in the PCR products. Results were normalized with respect to expression of the ribosomal L9 gene (Fig. 4) and the histone H2B gene (see Fig. S9 in the supplemental material).
FIG. 4.
Quantitative PCR analysis of SSU processome complex genes. Polysomal RNA fractions of epimastigotes (white bars) and metacyclic trypomastigotes (gray bars) were analyzed. Standard deviations for triplicate experiments are shown. Results were normalized against those for the L9 ribosomal protein.
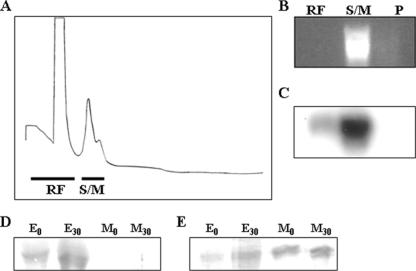
We also investigated the profile and the distribution of some processome messages in the polysomes of epimastigotes and metacyclic trypomastigotes after sucrose density gradient (10% to 50%) centrifugation. Epimastigote extracts displayed a typical polysome profile (Fig. 5A), whereas metacyclic parasites had shorter polysomes, present at lower concentrations (Fig. 5E). We confirmed that fast sedimenting forms on sucrose gradients corresponded to polysomes, by showing that a characteristic polysome dissociation profile was obtained when extracts were treated with EDTA before centrifugation (see Fig. S10 in the supplemental material). RNA was extracted from pooled fractions of the gradient and analyzed by Northern blotting with the TcSof1 and TcImp4 probes. In epimastigotes, only a small fraction of the TcSof1 mRNA was found associated with polysomes. Most of the mRNA was found in the ribosome-free and monosome fractions (Fig. 5C). We checked the integrity of the epimastigote polysomal RNA, using the H2B gene as a probe (Fig. 5D).
FIG. 5.
Polysome profiles of epimastigote (A) and metacyclic trypomastigote (E) forms. The positions of the 40S and 60S subunits, the 80S ribosome monomer, and polysomes are indicated in the sucrose density gradient. Fractions were collected and pooled (RF, ribosome free; S/M, subunits and monomers; P, polysomes), and RNA was purified. Northern blots of epimastigote fractions (B and C) and metacyclic trypomastigote fractions (F and G) analyzed with the radioactively labeled TcSof1 gene probe are shown. (D) The membrane shown in panel C was stripped and reprobed with an H2B histone gene probe.
A completely different pattern was observed for metacyclic trypomastigotes, in which as much as 70% of the mRNA was found to be associated with polysomes (Fig. 5G). The results obtained were consistent with qPCR results showing that TcSof1 mRNA was preferentially associated with polysomes in metacyclic trypomastigote forms. The same specific association with metacyclic trypomastigote polysomes was obtained when TcImp4 was used as a probe (not shown). To get a better resolution of heavier polysomes, sucrose gradients were run for a shorter time in order to avoid pelleting of heavy polysomes. Northern blots with the TcSof1 probe showed the same pattern as previously described (see Fig. S11C in the supplemental material). However, it has been described that EDTA treatment could dissociate other mRNA complexes that would cosediment with polysomes. As another control, metacyclic trypomastigote extracts were preincubated with puromycin, a translation inhibitor. Puromycin treatment disrupted polysomes (Fig. 6A), and the collected fractions were pooled and total RNA extracted (Fig. 6B). Northern blots with the TcSof1 probe showed that after puromycin treatment, most of the mRNA was present in the monosome fraction (Fig. 6C).
FIG. 6.
Polysome profile of metacyclic trypomastigote forms after puromycin treatment (A). Fractions were collected and pooled (RF, ribosome free; S/M, subunits and monomers; P, polysomes), and RNA was purified (B). Northern blot analysis of metacyclic trypomastigote fractions (C) hybridized with the radioactively labeled TcSof1 gene probe is shown. SDS-PAGE analysis of immunoprecipitated labeled proteins from epimastigote (E) and metacyclic trypomastigote (M) soluble fractions with the anti-TcSof1 (D) and anti-PEPCK (E) sera is shown. Proteins were separated in a 10% SDS-PAGE gel, and the gel was dried and exposed to X-ray films for 5 days.
The association of the TcSof1 transcripts with polysomes from metacyclic trypomastigotes, together with the absence of the corresponding protein, could be the result of rapid protein degradation in this parasite form. To test this hypothesis, we performed pulse-chase experiments followed by immunoprecipitation with the TcSof1 polyclonal antiserum. Epimastigote and metacyclic trypomastigote forms were incubated in TAU3AAG medium in the presence of [35S]methionine for 2 hours. Soluble protein extracts were prepared at 0 and 30 minutes after labeling and immunoprecipitated with the TcSof1 antiserum. An antiserum against phosphoenolpyruvate carboxykinase (PEPCK) was used as a control in parallel experiments. Immunoprecipitated proteins were resolved by SDS-PAGE. A labeled band corresponding to the 50-kDa TcSof1 protein was observed in the epimastigote extracts while a very faint signal could be observed in the metacyclic trypomastigote extracts (Fig. 6D). This faint signal could be due to the presence of approximately 5% epimastigote forms in the metacyclic cultures. The anti-PEPCK serum detected the expected 35-kDa band in both extracts (Fig. 6E).
DISCUSSION
We have cloned and characterized a T. cruzi ortholog of Sof1p, a component of the SSU processome (U3snoRNP) in yeast. The function of this protein in the processing complex is still not known. The presence of a WD domain involved in protein-protein interactions suggests a role in the processome complex assembly. The SSU processome is a multiprotein complex highly conserved through evolution. We identified putative orthologs for most of the components previously described for yeast, suggesting that the mechanisms involved in pre-rRNA processing are also conserved in T. cruzi. The SSU processome assembles on the nascent pre-rRNA in the nucleolus soon after the initiation of transcription (31). Given the dynamics of this process, we expected TcSof1 to accumulate in the nucleolus. Instead, the TcSof1 signal was clearly distributed throughout the nucleus, accumulating preferentially at the periphery.
A similar distribution has been observed in HeLa cells for the human orthologs, including the Imp3, Imp4, and Mpp10 SSU processome proteins (29). This discrepancy may result from the assembly of the U3snoRNP complex in the nucleoplasm before its transfer to the nucleolus. Recent studies have also questioned whether Sof1 is really specific to the U3snoRNP complex, because it seems to assemble on the 35S pre-RNA independently of U3snoRNA (3). SSU processome proteins shuttle between the nucleus and the cytoplasm, and Sof1p is the protein with the highest rates of shuttling. Heterokaryon fusion assays, using GFP-tagged proteins, showed that this shuttling was not coupled to pre-rRNA synthesis or preribosome particle export, leaving the functional relevance of this shuttling unresolved (29).
Ribogenesis in yeast requires the coordination of RNA polymerase I and II transcription, preribosome assembly, and pre-rRNA processing (23). Ribosome biosynthesis has recently been shown to be linked to cell size and cell cycle control, although the mechanisms underlying this linkage remain unclear (14, 32). In yeast, ribosome biogenesis is not regulated during mitosis, but SSU processome protein depletion results in cell cycle arrest in the G1 phase (4), suggesting a strong link between rRNA processing, ribosome assembly, and progression to S phase. In T. cruzi, metacyclic trypomastigote cells do not divide and the transcription of rRNA and ribosomal protein genes is downregulated (1, 17). This results in the arrest of the cell cycle in a G1-like phase in metacyclic cells. Our data, and those from previous studies, show that at least two rRNA-processing protein genes are also negatively regulated, suggesting that there may be a link between ribosome synthesis and processing and the cell cycle. In heterologous RNAi experiments, we depleted T. brucei procyclic replicating cells of the SSU processome proteins TbSof1 and TbImp4. FACS analysis of total DNA content showed no difference between depleted and control cells, suggesting that these proteins affect cell growth but are not required for cell cycle progression. At the onset of metacyclogenesis, parasites are subjected to nutritional stress, triggering differentiation. Cells stop replicating, but the proteins involved in ribosome synthesis are downregulated only in the final steps of differentiation. This suggests that cell cycle progression and ribosome synthesis are uncoupled in this parasite, at least during the differentiation process.
The TcSof1 gene was isolated as an mRNA abundant in the polysomal RNA fraction of metacyclic trypomastigotes. To our surprise, we found that TcSof1 protein was negatively regulated in these forms, showing opposite patterns of mRNA and protein dynamics. Proteins from a given multiprotein complex or metabolic pathway are often coregulated; we therefore used qPCR to investigate the levels of mRNA production for U3snoRNP-specific proteins. Most of the corresponding genes analyzed displayed expression patterns similar to that of TcSof1, with mRNA levels most abundant in the polysomal RNA fraction and significant accumulation in metacyclic forms. Accumulation of this kind was also observed for another gene related to the SSU processome (TcImp4). An analysis of the polysome profile after sucrose density gradient separation and the association of mRNAs with polysomal fractions clearly showed that, in infective forms, most of the TcSof1 mRNA is associated with polysomes. Similar results were obtained for the TcImp4 mRNA, and consistent results were obtained in at least three independent experiments for each gene. Based on the qPCR results, we can assume that several SSU processome-specific proteins share the same regulation mechanism. Pulse-chase experiments showed that the differential expression of the TcSof1 protein is not controlled at the level of protein degradation since de novo protein synthesis was not detected in metacyclic trypomastigotes. An association of mRNA with polysomes in the absence of protein accumulation has been described for several proteins in eukaryotic cells, and the mechanisms involved operated after the initial steps of translation blocking elongation or termination (8, 34).
These types of translational repression mechanisms following translation initiation have been described as typical of micro-RNA-mediated control (5, 30). T. cruzi has no functional RNAi machinery, mainly due to the absence of Dicer- and Drosha-like ribonucleases. However, an Argonaute-like protein has been annotated in the sequence of the parasite genome, so we cannot exclude the possibility of repression mechanisms based on noncoding RNAs. The results presented herewith strongly suggest that expression of TcSof1 and probably other SSU processome proteins is regulated at the level of translation elongation or termination steps. Translational control mechanisms, acting mostly after the initiation of translation, may facilitate the rapid activation or inactivation of mRNAs, enabling the parasite to adapt rapidly to different environments. The molecular mechanisms underlying the regulation of SSU processome proteins and the factors involved are currently being investigated, and these studies may provide insight into posttranscriptional regulation in kinetoplastid parasites.
Supplementary Material
Acknowledgments
We thank Nilson Fidêncio for skillful technical assistance, Paulo Arauco for DNA sequencing, and Noreen Williams for providing the RNAi vector.
This work received financial support from Fundação Oswaldo Cruz (FIOCRUZ), Programa de Núcleos de Excelência (PRONEX/Fundação Araucária), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq). S.C.N., A.R.A., L.M., M.A.K., S.G., and B.D. are research fellows from CNPq.
Footnotes
Published ahead of print on 8 December 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Atwood, J. A., III, D. B. Weatherly, T. A. Minning, B. Bundy, C. Cavola, F. R. Opperdoes, R. Orlando, and R. L. Tarleton. 2005. The Trypanosoma cruzi proteome. Science 309:473-476. [DOI] [PubMed] [Google Scholar]
- 2.Avila, A. R., S. F. Yamada-Ogatta, V. da Silva Monteiro, M. A. Krieger, C. V. Nakamura, W. de Souza, and S. Goldenberg. 2001. Cloning and characterization of the metacyclogenin gene, which is specifically expressed during Trypanosoma cruzi metacyclogenesis. Mol. Biochem. Parasitol. 117:169-177. [DOI] [PubMed] [Google Scholar]
- 3.Bax, R., H. R. Vos, H. R. Raue, and J. C. Vos. 2006. Saccharomyces cerevisiae Sof1p associates with 35S pre-rRNA independent from U3 snoRNA and Rrp5p. Eukaryot. Cell 5:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, K. A., and S. J. Baserga. 2004. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell 15:5038-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braat, A. K., N. Yan, E. Arn, D. Harrison, and P. M. Macdonald. 2004. Localization-dependent oskar protein accumulation; control after the initiation of translation. Dev. Cell 7:125-131. [DOI] [PubMed] [Google Scholar]
- 6.Brecht, M., and M. Parsons. 1998. Changes in polysome profiles accompany trypanosome development. Mol. Biochem. Parasitol. 97:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Chagas, C. 1909. Nova tripanosomiase humana. Estudos sobre a morfolojia e o cyclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolójico de nova morbidade do homem. Mem. Inst. Oswaldo Cruz 1:159-218. [Google Scholar]
- 8.Clark, I. E., D. Wyckoff, and E. R. Gavis. 2000. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr. Biol. 10:1311-1314. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras, V. T., T. C. Araujo-Jorge, M. C. Bonaldo, N. Thomaz, H. S. Barbosa, M. N. Meirelles, and S. Goldenberg. 1988. Biological aspects of the Dm 28c clone of Trypanosoma cruzi after metacyclogenesis in chemically defined media. Mem. Inst. Oswaldo Cruz 83:123-133. [DOI] [PubMed] [Google Scholar]
- 11.Contreras, V. T., J. M. Salles, N. Thomas, C. M. Morel, and S. Goldenberg. 1985. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 16:315-327. [DOI] [PubMed] [Google Scholar]
- 12.Dallagiovanna, B., C. Plazanet-Menut, S. F. Y. Ogatta, A. R. Ávila, M. A. Krieger, and S. Goldenberg. 2001. Trypanosoma cruzi: a gene family encoding chitin-binding-like proteins is posttranscriptionally regulated during metacyclogenesis. Exp. Parasitol. 99:7-16. [DOI] [PubMed] [Google Scholar]
- 13.De Souza, W. 2002. Basic cell biology of Trypanosoma cruzi. Kinetoplastid Biol. Dis. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dlakic, M. 2005. The ribosomal subunit assembly line. Genome Biol. 6:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Orso, I., J. G. De Gaudenzi, and A. C. Frasch. 2003. RNA-binding proteins and mRNA turnover in trypanosomes. Trends Parasitol. 19:151-155. [DOI] [PubMed] [Google Scholar]
- 16.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elias, M. C. Q. B., R. Marques-Porto, E. Freymuller, and S. Schenkman. 2001. Transcription rate modulation through the Trypanosoma cruzi life cycle occurs in parallel with changes in nuclear organization. Mol. Biochem. Parasitol. 112:79-90. [DOI] [PubMed] [Google Scholar]
- 18.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 3:313-318. [DOI] [PubMed] [Google Scholar]
- 19.Foldynova-Trantirkova, S., Z. Paris, N. R. Sturm, D. A. Campbell, and J. Lukes. 2005. The Trypanosoma brucei La protein is a candidate poly(U) shield that impacts spliced leader RNA maturation and tRNA intron removal. Int. J. Parasitol. 35:359-366. [DOI] [PubMed] [Google Scholar]
- 20.Fragoso, S. P., C. Plazanet-Menut, M. A. Carreira, M. C. Motta, B. Dallagiovana, M. A. Krieger, and S. Goldenberg. 2003. Cloning and characterization of a gene encoding a putative protein associated with U3 small nucleolar ribonucleoprotein in Trypanosoma cruzi. Mol. Biochem. Parasitol. 126:113-117. [DOI] [PubMed] [Google Scholar]
- 21.Goldenberg, S., J. M. Salles, V. T. Contreras, M. P. Lima Franco, A. M. Katzin, W. Colli, and C. M. Morel. 1985. Characterization of messenger RNA from epimastigotes and metacyclic trypomastigotes of Trypanosoma cruzi. FEBS Lett. 180:265-270. [DOI] [PubMed] [Google Scholar]
- 22.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 23.Granneman, S., and S. J. Baserga. 2005. Ribosome biogenesis: of knobs and RNA processing. Curr. Opin. Cell Biol. 17:281-286. [DOI] [PubMed] [Google Scholar]
- 24.Hammarton, T. C., J. Clark, F. Douglas, M. Boshart, and J. C. Mottram. 2003. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 278:22877-22886. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez, R., F. Diaz de Leon, and M. Castañeda. 1988. Molecular cloning and partial characterization of ribosomal RNA genes from Trypanosoma cruzi. Mol. Biochem. Parasitol. 27:275-280. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez, R., S. Martinez-Calvillo, R. Hernandez-Rivas, and E. Gomez. 1993. Trypanosoma cruzi ribosomal RNA genes: a review. Biol. Res. 26:109-114. [PubMed] [Google Scholar]
- 27.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovalic, D., J. H. Kwak, and B. Weisblum. 1991. General method for direct cloning of DNA fragments generated by the polymerase chain reaction. Nucleic Acids Res. 19:4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leary, D. J., M. P. Terns, and S. Huang. 2004. Components of U3 snoRNA-containing complexes shuttle between nuclei and the cytoplasm and differentially localize in nucleoli: implications for assembly and function. Mol. Biol. Cell 15:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 31.Osheim, Y. N., S. L. French, K. M. Keck, E. A. Champion, K. Spasov, F. Dragon, S. J. Baserga, and A. L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16:943-954. [DOI] [PubMed] [Google Scholar]
- 32.Rudra, D., and J. R. Warner. 2004. What better measure than ribosome synthesis? Genes Dev. 18:2431-2436. [DOI] [PubMed] [Google Scholar]
- 33.Teixeira, S. M. R., L. V. Kirchhoff, and J. E. Donelson. 1995. Post-transcriptional elements regulating expression of mRNAs from the amastin/tuzin gene cluster of Trypanosoma cruzi. J. Biol. Chem. 270:22586-22594. [DOI] [PubMed] [Google Scholar]
- 34.Wilheilm, J. E., and C. A. Smibert. 2005. Mechanisms of translational regulation in Drosophila. Biol. Cell 97:235-252. [DOI] [PubMed] [Google Scholar]
- 35.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. Regulated processive transcription of chromatin by T7 RNA polymerase in Trypanosoma brucei. Nucleic Acids Res. 99:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.