Abstract
The QDR2 gene of Saccharomyces cerevisiae encodes a putative plasma membrane drug:H+ antiporter that confers resistance against quinidine, barban, bleomycin, and cisplatin. This work provides experimental evidence of defective K+ (Rb+) uptake in the absence of QDR2. The direct involvement of Qdr2p in K+ uptake is reinforced by the fact that increased K+ (Rb+) uptake due to QDR2 expression is independent of the Trk1p/Trk2p system. QDR2 expression confers a physiological advantage for the yeast cell during the onset of K+ limited growth, due either to a limiting level of K+ in the growth medium or to the presence of quinidine. This drug decreases the K+ uptake rate and K+ accumulation in the yeast cell, especially in the Δqdr2 mutant. Qdr2p also helps to sustain the decrease of intracellular pH in quinidine-stressed cells in growth medium at pH 5.5 by indirectly promoting H+ extrusion affected by the drug. The results are consistent with the hypothesis that Qdr2p may also couple K+ movement with substrate export, presumably with quinidine. Other clues to the biological role of QDR2 in the yeast cell come from two additional lines of experimental evidence. First, QDR2 transcription is activated under nitrogen (NH4+) limitation or when the auxotrophic strain examined enters stationary phase due to leucine limitation, this regulation being dependent on general amino acid control by Gcn4p. Second, the amino acid pool is higher in Δqdr2 cells than in wild-type cells, indicating that QDR2 expression is, directly or indirectly, involved in amino acid homeostasis.
The therapeutic potential of drugs is seriously limited by the manifestation of cellular drug resistance (14). The multidrug efflux transporters found in the plasma membrane of all living cells apparently recognize a wide variety of structurally and pharmacologically unrelated drugs and actively extrude them from the cytoplasm to the outer medium, providing protection from these compounds (1, 4, 14, 17, 29, 32, 37, 40). However, the molecular mechanisms behind the apparent promiscuity of these multidrug efflux pumps remain elusive and a topic of debate (17, 29, 32, 36, 37, 40). Moreover, the inspection of the genome sequences becoming available is revealing the intriguing presence of a large number of predicted multidrug transporters. In particular, this is the case for Saccharomyces cerevisiae (26, 30).
In the present work, we have further examined the biological function of the multidrug resistance (MDR) transporter encoded by the QDR2 gene, present in the plasma membrane of S. cerevisiae (47). According to the classification of Nelissen et al. (26), Qdr2p belongs to cluster I of the so-called DHA12 drug efflux family (EC 2.1.3) (30), including putative drug:H+ antiporters with 12 predicted membrane-spanning segments. Qdr2p confers yeast resistance against the antiarrhythmic and antimalarial drug quinidine, the herbicide barban, and the antitumour agents bleomycin and cisplatin (45, 47). Remarkably, at least six other members of the DHA12 family of S. cerevisiae drug:H+ antiporters, belonging to the same cluster I or to cluster II, are individually required for quinidine resistance (8, 9, 10, 27, 44, 45). Together with the stereoisomer quinine, the cinchona alkaloid quinidine is the antimalarial agent most widely used intravenously (46, 49). As an antiarrhythmic agent, quinidine blocks cardiac Na+ channels to alter action potential duration and slow conduction. It also blocks repolarizing cardiac K+ channels to prolong action potential duration (6). The role of all of the referred yeast MDR transporters of the major facilitator superfamily in alleviating the deleterious effects of quinidine and other compounds that are not usually present in the yeast cell natural environment appears to suggest that their physiological function may have nothing to do with broad chemoprotection. They may transport a still-unidentified specific physiological substrate, and drugs may be transported only fortuitously or opportunistically. However, experimental evidences indicated that Qdr2p is involved, directly or indirectly, in the active expulsion of quinidine from preloaded yeast cells (47).
In the present work, we show results supporting the involvement of Qdr2p in K+ uptake. QDR2 expression was found to confer a physiological advantage for the yeast cells under conditions leading to K+-limited growth, due either to a limiting level of K+ in the growth medium or to the presence of quinidine. Potassium uptake in S. cerevisiae occurs essentially through the plasma membrane proteins Trk1p and Trk2p, responsible for high- and moderate-affinity K+ transport, respectively (11, 21, 34). TRK1 appears to be the most important component of potassium uptake, as mutations of this gene result in a significant defect in potassium transport and the inability to grow under limiting concentrations of this cation (11, 21, 33, 34). Mutation of TRK2 aggravates the requirement for potassium of a Δtrk1 mutant. However, the viability of Δtrk1 Δtrk2 cells has revealed the existence of additional forms to transport potassium. Although the nature of this transport remains unclear, the activities of a nonselective cation channel and different nonspecific transporters have been postulated (3, 23, 38, 50). Here, we provide evidence suggesting that Qdr2p may be one of these predicted low-affinity uptake systems for K+. In a previous study, Qdr2p expression was found to reduce the decrease of intracellular pH registered in quinidine-stressed yeast cells suspended in slightly acidified medium (pH 5.5) optimal for yeast growth (47). Having this in mind, we examined the hypothesized role of quinidine in the dissipation of the H+ gradient across the plasma membrane and the involvement of QDR2 in this process.
In this work, we also searched for environmental conditions leading to the activation of QDR2 transcription in the hope that this information might contribute to the elucidation of the physiological role of this gene. QDR2 transcription, which is not stimulated in quinidine-stressed cells (47), is here demonstrated to be activated under nitrogen (NH4+) limitation or when the auxotrophic strain examined enters stationary phase due to leucine limitation in a Gcn4p-dependent way. Having in mind these expression results and the fact that S. cerevisiae Aqr1p, a drug:H+ antiporter highly homologous to Qdr2p (26, 30), was proposed to be involved in the excretion of amino acids (48), we also tested the effect of QDR2 expression on amino acid partition between the cell interior and the surrounding medium. The results suggest the involvement of Qdr2p in amino acid homeostasis, directly or indirectly promoting amino acid excretion.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
Saccharomyces cerevisiae strain BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) and the derived single deletion mutants from the Euroscarf collection, the BY4741 Δqdr2 and BY4741 Δgcn4 strains, were used in this study, as well as the amino acid prototrophic strain 23344c (MATα ura3) (24), provided by B. André (Université Livre de Bruxelles, Belgium). The DBY746 Δtrk1 Δtrk2 double mutant (trk1::LEU2 trk2::HIS3) was a gift from Joaquín Ariño and Amparo Ruiz (Departamento de Bioquímica i Biologia Molecular, Universitat Autónoma de Barcelona, Barcelona, Spain). This double mutant was constructed in the DBY746 background (MATα ura3 leu2 his3 trp1) (39). The DBY746 Δtrk1 Δtrk2 Δqdr2 strain was constructed in the present work, as described below.
The plasmid pYCG_QDR2, constructed during a previous study (47), using pFL38 as the cloning vector, was used in this work for complementation assays. Transformation of yeast cells was performed by the method of Gietz et al. (12).
Cells were batch cultured at 30°C, with orbital agitation (250 rpm) in growth media resulting from different nutrient or drug supplementation of the basal medium (BM) with the following composition (per liter): 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco), 20 g glucose, and 2.65 g (NH4)2SO4 (Merck). Strain BY4741 was grown in MM4 medium that resulted from BM supplementation with 20 mg methionine, 20 mg histidine, 60 mg leucine, and 20 mg uracil (all from Sigma). For the amino acid prototrophic strain 23344c, the growth medium used (MM5) resulted from BM supplementation with 20 mg uracil.
Ammonium phosphate-derived media were also used to test BY4741, DBY746 Δtrk1 Δtrk2, and derived strains' growth under K+ limitation (5). Ammonium phosphate basal medium contains, per liter, a mixture of 0.492 g MgSO47H2O (Merck), 0.02 g anhydrous CaCl2 (Panreac), 1.056 g (NH4)2HPO4 (Merck), 3.96 g (NH4)2SO4, 20 g glucose, 2 mg niacin, 2 mg pyridoxine, 2 mg thiamine, 2 mg pantothenate, 0.02 mg biotin, and the desired concentration of KCl (all from Sigma). For BY4741 growth, this medium was supplemented with 20 mg methionine, 20 mg histidine, 60 mg leucine, and 20 mg uracil (all from Sigma), while for the DBY746 Δtrk1 Δtrk2 strain, this medium was supplemented with 20 mg histidine, 60 mg leucine, 20 mg tryptophan, and 20 mg uracil (all from Sigma). To maintain selective pressure over the recombinant strains, the addition of uracil to this medium was carried out only to grow the host yeast cells.
Growth medium supplementation with quinidine (Sigma) was carried out as described below. Quinidine was dissolved in 1.4% ethanol, and the control assays were prepared with identical concentrations of the solvent.
Agarized solid media contained, besides the above-indicated ingredients, 20 g/liter agar (Iberagar) or 20 g/liter bacteriological agar no. 1 (Oxoid), used to meet the purity requirements of the ammonium phosphate medium.
QDR2 disruption in the S. cerevisiae DBY746 Δtrk1 Δtrk2 strain.
Disruption of QDR2 (YIL121w) in the S. cerevisiae DBY746 Δtrk1 Δtrk2 strain was performed as described previously, using a replacement cassette prepared before (47) to be used for the systematic inactivation of this gene in any S. cerevisiae strain, by creating longer homologous sequences on both sides of the loxP-kanMX4-loxP module.
Spot growth assays under K+ limitation and/or quinidine-induced stress.
Spot growth assays using BY4741 and BY4741Δqdr2 cells under K+ limitation were compared on agarized medium derived from ammonium phosphate supplemented with different KCl concentrations (0.4 to 20 mM) (5), in the presence or absence of quinidine (2.4 mM; Sigma). The cells used to inoculate the agar plates were mid-exponential cells grown in the absence of chemical stress. These cells were resuspended in sterile H2O to obtain suspensions with an optical density at 600 nm (OD600) of 0.05 ± 0.005 (mean ± standard deviation), and these and diluted cell suspensions were applied as spots (4 μl) onto the surface of the agarized media and incubated at 30°C for 3 to 5 days, depending on the cell growth rate.
K+ content of the yeast cells.
In order to determine the effect of quinidine on the K+ content of yeast cells, yeast cells were grown overnight in liquid MM4 medium. When cultures reached an OD600 of 0.5 to 0.6, cells were collected and resuspended in fresh medium supplemented or not supplemented with quinidine (4.1 mM). After overnight incubation, the K+ content of cells was determined by collecting them on Millipore filters, which were rapidly washed with 20 mM MgCl2. The cells were then extracted with acid and the extracts analyzed by atomic emission spectrophotometry (35). The experiments were repeated three to six times and the standard deviations calculated.
To test the K+ content of yeast cells incubated with different KCl concentrations, yeast cells were grown overnight in liquid ammonium phosphate medium supplemented with 10 mM KCl until the culture reached an OD600 of 0.5 to 0.6. Cells were collected and resuspended in fresh ammonium phosphate medium containing different KCl concentrations, either supplemented or not supplemented with quinidine (2.4 mM).
Rb+ (K+) uptake and K+ efflux experiments.
Rb+ was used as a K+ transport analogue (35). The time course of Rb+ uptake in cells grown in ammonium phosphate medium supplemented with 10 mM KCl and in the absence or presence of quinidine (2.4 mM) was studied. When the OD600 of the culture reached 0.3, the cells were centrifuged, washed, and suspended in the buffer, consisting of 10 mM MES [0.1 mM Mg2Cl, 2 mM Ca2Cl, and 2% (wt/vol) glucose, brought to pH 5.8 with Ca(OH)2]. When required, the uptake buffer was supplemented with quinidine (2.4 mM). Immediately, RbCl (35, 50, or 100 mM) was added to the medium (time zero) and cell samples were withdrawn at various times thereafter (35). The cells were then extracted with acid and the extracts analyzed for Rb+ by atomic emission spectrometry and treated as described above.
K+ efflux was determined in cells grown in MM4 medium up to an OD600 of 0.3. Cells were recovered by centrifugation, washed, and resuspended in MES buffer containing different quinidine concentrations. At different times, samples of cells were taken and treated as described before to determine the K+ content of the cells.
The experiments were repeated three times and the standard deviations calculated.
Extracellular acidification curves promoted by yeast cells in the presence of increasing concentrations of quinidine.
In order to compare the in vivo active exports of protons from energized cells of the BY4741 parental strain and the BY4741 Δqdr2 mutant strain, in the presence of increasing concentrations of quinidine, the external medium pH was followed for 10 min of incubation. Cells of both strains were grown in liquid MM4 medium, collected by centrifugation (8,000 rpm, 5 min, 4°C) when the culture reached an OD600 of 0.5 to 0.6, washed twice with distilled water, and incubated in sorbitol (20 g/liter) for 30 min to deenergize the cells and deactivate plasma membrane H+-ATPase. These cells were washed with water and resuspended in distilled water to obtain a dense cell suspension (OD600 of 20.0 ± 2.0). Acidification experiments were carried out in a water-jacketed cell of 10-ml capacity, at 30°C, containing 3.5 ml of water or quinidine solutions (from 0.3 to 4.1 mM) and 1 ml of the cellular suspension described above. The pH of the resulting suspension was adjusted to 5.50 ± 0.05, and 1 ml of 100 g/liter glucose (at pH 5.5) was added (to obtain a final concentration of 20 g/liter). The variation of extracellular medium pH was followed by potentiometry using a pH microelectrode (Metrohm 6.0204.000) attached to a pH meter (Metrohm 605) for 10 min.
The increase of the acidification ability of the wild-type or Δqdr2 strain by the expression of QDR2 from plasmid pYCG_QDR2 was also tested. Recombinant cells were grown in MM4 minimal medium without uracil.
Northern blot analysis.
QDR2 mRNA levels under different growth conditions were assessed by Northern blot analysis. RNA extraction from yeast cells and Northern experiments were carried out as described before (47). The total RNA in each sample used for Northern blotting was approximately constant (20 μg, assessed by the measurement of A260). The specific DNA probe used to detect QDR2 transcripts was prepared by PCR amplification using the same primers as those used before (47), and the ACT1 mRNA level was used as an internal control. The probe was radiolabeled with a random-primed DNA labeling kit (Roche), using [α32 P]dCTP. Kodak BioMax MS (Amersham) films were exposed to nitrocellulose membranes and incubated with an intensifying screen at −70°C for approximately 1 to 4 days to obtain the hybridization signals. Their relative intensities in the autoradiograms were quantified by densitometry (Molecular Dynamics computing densitometer, ImageQuant software version).
Intracellular and extracellular concentrations of amino acids.
The intracellular concentration of amino acids was determined, as described by Klasson et al. (19), in BY4741 and BY4141 Δqdr2 cells harvested in mid-exponential growth phase and in early stationary phase of growth. The concentration of amino acids in the supernatants of the same culture samples was also determined. To assess intracellular amino acid pools, cells were harvested by centrifugation and the cell pellets were washed twice with 1.5 ml of water and resuspended in 1.5 ml of AA buffer (2.5 mM K2HPO4-KH2PO4 buffer at pH 6.0, 0.6 M sorbitol) with 10 mM glucose and 0.2 mM CuCl2. These cell suspensions were incubated for 10 min at 30°C with gentle shaking to allow Cu2+-induced plasma membrane permeabilization, followed by filtration (Whatman GF/F filters) of 1-ml aliquots. The filters were washed four times with the basal AA buffer without glucose or CuCl2, and the filtrates were combined and considered the total extracts. The concentrations of amino acids in total extracts and the corresponding supernatants were determined by high-performance liquid chromatography at the Amino Acid and Sequencing Service of ITQB, Oeiras, Portugal.
The values of the cell content for each amino acid are expressed as μmol of amino acid per unit of dry biomass.
RESULTS
QDR2 deletion increases the requirement for potassium, and this defective growth phenotype is aggravated under quinidine stress.
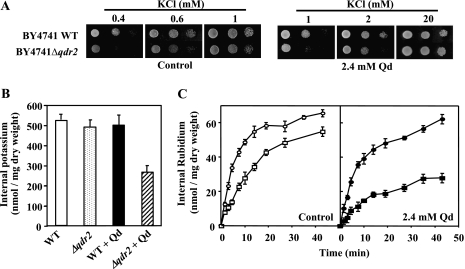
When cells of BY4741 or the respective mutant with the QDR2 gene deleted (Δqdr2) were spotted onto minimal agar medium with a saturating K+ concentration (20 mM), similar growth patterns were observed for both strains, either in the absence or in the presence of 2.4 mM of quinidine (Fig. 1A and data not shown). However, the Δqdr2 mutant showed a defective growth phenotype, either in the presence or in the absence of quinidine, at limiting K+ concentrations (Fig. 1A). This effect was observed in the presence of quinidine at 2 mM of K+ but became more evident with the decrease of K+ concentration in the growth medium, being maximal for the lowest concentration tested (0.4 mM). These results indicate that the presence of quinidine increases potassium requirements in both the wild type and the Δqdr2 mutant, this effect being severely aggravated in the Δqdr2 mutant.
FIG. 1.
QDR2 deletion increases the requirement for K+ by reducing K+ (Rb+) uptake, this effect being aggravated under quinidine stress. (A) Growth of S. cerevisiae BY4741 (parental strain) and of the Δqdr2 deletion mutant in agar plates with ammonium phosphate medium supplemented with increasing K+ concentrations, in the presence or absence of quinidine. Serial dilutions (1:1, 1:5, and 1:10, from left to right) of a cell suspension were spotted in agar plates, and growth was observed after 3 to 5 days of incubation, depending on growth kinetics. (B) Effect of quinidine on the K+ content of the parental strain BY4741 (white and black bars) and the Δqdr2 mutant (dotted and striped bars). Cells were grown as described in Materials and Methods and incubated in fresh medium, either in the absence (white and dotted bars) or in the presence (black and striped bars) of quinidine (4.1 mM). Data are means ± standard deviations (SD) for three independent experiments. (C) Rb+ uptake in the BY4741 parental strain (○ and •) and in the Δqdr2 mutant (□ and ▪), in the absence (control) or in the presence of 2.4 mM quinidine. Cells of the wild type and the mutant were grown and treated as described in Materials and Methods. At time zero, RbCl (35 mM) was added to the uptake buffer. Then, cell samples were taken and internal Rb+ was determined. Data are means ± SD for three independent experiments. WT, wild type; Qd, quinidine.
Quinidine decreases internal K+ levels and inhibits K+ (Rb+) uptake, these effects being aggravated in the Δqdr2 mutant.
The growth differences registered between the parental strain and Δqdr2 cells under K+-limiting conditions (Fig. 1A) suggested that the capacity to transport K+ may be altered in the mutant strain, this defect being aggravated by the presence of quinidine. To check this possibility, we determined the internal K+ content, the time course of Rb+ (a K+ analogue) uptake, and the K+ loss process in wild-type and Δqdr2 cells either in the absence or in the presence of the drug. The lack of QDR2 slightly reduced the K+ content of the yeast cell compared with the parental strain. However, in quinidine-stressed cells, the values of internal K+ measured in the Δqdr2 mutant were about one-half of those determined for the wild-type strain (Fig. 1B).
A reduction of KCl concentration in the incubation medium resulted in a decrease of the internal concentration in both strains but led essentially to the same conclusions (data not shown). Results from Rb+ transport assays indicate that the K+ (Rb+) uptake rate in the Δqdr2 mutant is below the value determined for the wild-type strain (Fig. 1C) and that this defect is significantly increased by quinidine (Fig. 1C). These observations suggest that quinidine affects potassium uptake into the yeast cells and that Qdr2p plays an important role in the maintenance of physiological levels of K+, especially in quinidine-stressed cells, by increasing K+ uptake. This idea was reinforced by results from K+ efflux experiments, since they showed that neither the lack of QDR2 nor the addition of quinidine to the buffer has a significant effect on K+ loss (data not shown).
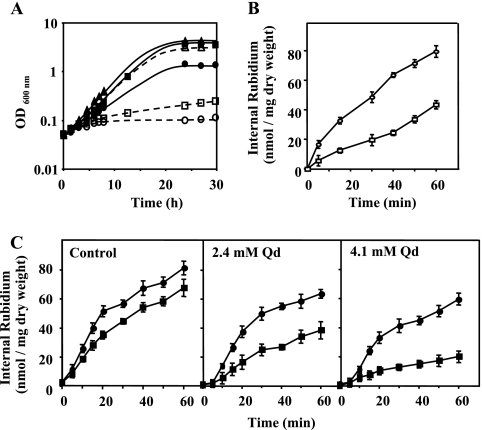
Increased K+ uptake due to QDR2 expression and quinidine-induced inhibition of K+ uptake are independent of the Trk1p/Trk2p system.
Since the most important K+ uptake system in yeast, involving Trk1p and Trk2p, is active in the BY4741 Δqdr2 mutant, we have deleted the QDR2 gene in a DBY746 double mutant with the TRK1 and TRK2 genes deleted. This triple mutant exhibited defective growth at limiting K+ concentrations compared with the Δtrk1 Δtrk2 strain, the growth phenotype becoming clearer as the K+ concentration in the growth medium decreased to 0.5 mM (Fig. 2A). Based on K+ (Rb+) transport assays, these results can be explained on the basis of the impaired K+ (Rb+) transport displayed by Δtrk1 Δtrk2 Δqdr2 cells compared with that displayed by the cells of the double mutant parental strain, this difference being more important at the lowest concentration of RbCl tested (50 mM) (Fig. 2B and C). The results indicate that increased growth at limiting K+ concentrations and increased K+ (Rb+) influx observed in yeast strains expressing QDR2 are independent of the main K+ transport system encoded by TRK1 and TRK2 genes. Having in mind the important effect of quinidine in reducing K+ uptake and K+ accumulation in yeast, we also tested the effect of this drug on the Δtrk1 Δtrk2 Δqdr2 mutant strain (Fig. 2C). The results indicate that quinidine is not essentially acting via an inhibition of the Trk1p/Trk2p system (Fig. 1C). Indeed, the dose-dependent effect of quinidine in reducing K+ uptake was also observed in the mutant lacking the main transport systems for K+ uptake, Trk1p and Trk2p, the quinidine-induced inhibition of K+ (Rb+) uptake being more drastic for the mutant with the QDR2 gene deleted (Fig. 2C).
FIG. 2.
Increased K+ (Rb+) uptake due to QDR2 expression and inhibition of K+ (Rb+) uptake by quinidine are independent of the Trk1p/Trk2p system. (A) Effect of QDR2 expression on growth curves of the S. cerevisiae DBY746 Δtrk1 Δtrk2 mutant (full line and black symbols) and the corresponding DBY746 Δtrk1 Δtrk2 Δqdr2 mutant (dotted line and white symbols) under potassium limitation. The growth media were prepared using ammonium phosphate medium supplemented with 0.5 mM (• and ○), 5 mM (□ and ▪), and 100 mM (▴ and ▵) of KCl. The cells used as inoculum were exponential-phase cells cultivated in the presence of 100 mM of KCl. (B and C) Effect of QDR2 expression on the time course of Rb+ uptake in the DBY746 Δtrk1 Δtrk2 mutant (○ and •) and in the respective Δtrk1 Δtrk2 Δqdr2 mutant (□ and ▪), in the presence or absence of quinidine. At time zero, (B) 50 mM RbCl or (C) 100 mM RbCl was added to the uptake buffer, in the absence (B and C, control) or presence of quinidine (C, 2.1 mM and 4.2 mM). Cell samples were taken during incubation and internal Rb+ was determined, as described in Materials and Methods. Data are means ± standard deviations for three independent experiments. Qd, quinidine.
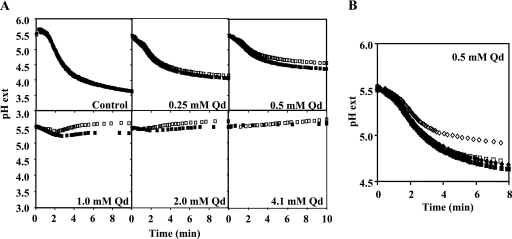
Quinidine reduces the rate of extracellular acidification by yeast cells, and this effect is diminished by QDR2 expression.
Results from a previous study indicated that intracellular pH (pHi) is fairly constant and close to neutrality in exponential yeast cells either expressing or not expressing the QDR2 gene (47). However, in that same study, we found that the presence of quinidine (4.1 mM) leads to an internal acidification (down to pH 6.3) of yeast cells suspended in the acidic growth medium (initial pH of 5.5) suitable for fungal growth. Moreover, the lack of QDR2 implicated a more drastic internal acidification (down to pH 5.2) of cells challenged with identical quinidine concentrations.
The intracellular pH regulation is essentially explained by the action of the plasma membrane H+-pumping ATPase Pma1p (41). Considering that the result of the in vivo activity of this H+ pump and of the passive H+ influx through the plasma membrane can be assessed by the acidification of the external medium (31), the acidification curves promoted by the wild-type strain BY4741 and the Δqdr2 mutant, in the presence of increasing concentrations of quinidine, were compared (Fig. 3A). This drug was found to reduce the rate of extracellular acidification by yeast cells suspended in an aqueous glucose solution supplemented with quinidine. The inhibition of the rate of H+ active efflux was dose dependent, and for concentrations equal to or above 1.0 mM of quinidine, no net efflux was detected for the Δqdr2 cells while the wild-type cells were still able to acidify the extracellular medium, although with severely inhibited kinetics. Although inhibited, a net proton extrusion was registered at 0.25 and 0.5 mM of quinidine for wild-type and Δqdr2 strains, but the H+ efflux in the presence of the drug by yeast cells expressing QDR2 was higher. In the absence of quinidine, the extracellular H+ acidification curves registered for the wild type and the Δqdr2 mutant cells were, however, apparently coincident. The expression of the recombinant plasmid pYCG_QDR2 in BY4741 Δqdr2 cells restored the rate of H+ efflux in the presence of quinidine to values similar to those observed for BY4741 transformed with the cloning vector pFL38 (Fig. 3B). Moreover, Qdr2p-increased expression was also able to increase the parental strain H+ efflux rate under identical quinidine stress (Fig. 3B). These observations reinforce the idea that the QDR2 gene is implicated in the maintenance of pH homeostasis across the plasma membrane in the presence of quinidine.
FIG. 3.
QDR2 expression increases external acidification rate affected by quinidine. (A) Dose-dependent inhibition by quinidine of external medium acidification promoted by energized cells of the BY4741 parental strain (▪) and the BY4741 Δqdr2 mutant strain (□). (B) Complementation of the reduced ability of energized cells of the BY4741 Δqdr2 deletion mutant (⋄ and ♦) to promote extracellular acidification in the presence of quinidine stress compared with that of the BY4741 parental strain (□ and ▪) by expressing the QDR2 gene from plasmid pYCG_QDR2. Mutant cells with QDR2 deleted or the respective wild-type strain was transformed either with the recombinant plasmid pYCG_QDR2 (▪ and ♦) or with the cloning vector pFL38 (□ and ⋄), and experimental assays were carried out under conditions identical to those used for panel A. Values are representative of two independent experiments that gave rise to similar results. Qd, quinidine; pH ext, extracellular pH.
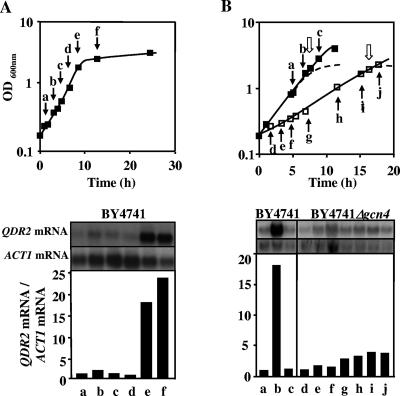
QDR2 transcription is activated when the auxotrophic strain BY4741 enters stationary phase due to leucine limitation.
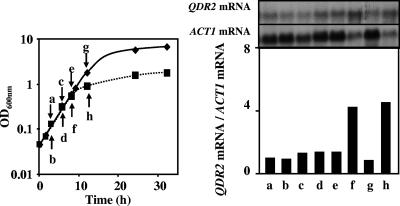
QDR2 transcription was highly activated (18-fold) when cells of S. cerevisiae BY4741 entered the stationary phase of growth in MM4 medium, due to leucine limitation (Fig. 4A). Leucine is one of the amino acids that have to be added to the growth medium due to BY4741 auxotrophies, and the concentration added (60 mg/liter) is growth limiting (data not shown). Moreover, the high levels of QDR2 mRNA present in leucine-limited cells returned to basal levels when fresh leucine was added to the culture with leucine-starved cells (Fig. 4B).
FIG. 4.
Gcn4-dependent activation of QDR2 transcription when the auxotrophic strain BY4741 enters stationary phase due to leucine limitation. (A) Relative levels of QDR2 mRNA in S. cerevisiae BY4741 cells during growth in MM4 liquid medium. Cell samples were harvested at the times indicated by the letters in the figure. Relative ratios of QDR2 mRNA to ACT1 mRNA were obtained by densitometry of the autoradiograms. The basal QDR2 mRNA relative value of exponential cells at an OD600 of 0.3 ± 0.05 was set as 1. (B) Relative levels of QDR2 mRNA in S. cerevisiae BY4741 (▪) and BY4741 Δgcn4 (□) cells during growth in MM4 medium. Cell samples were harvested at different times of cultivation, as indicated by the black arrows and the letters, during exponential growth in early stationary phase and 1 hour following the addition of fresh leucine to starved cells, as indicated by the white arrows. The growth curves in medium without extra leucine supplementation are represented by dotted lines. Relative ratios of QDR2 mRNA to ACT1 mRNA were obtained by densitometry of the autoradiograms. The basal QDR2 mRNA relative value of exponential cells at an OD600 of 0.3 ± 0.05 was set as 1. Values are representative of two independent experiments.
The strong activation of QDR2 transcription registered in BY4741 cells approaching the stationary phase of growth, due to leucine limitation, was not detected in the respective mutant with the GCN4 gene deleted (Fig. 4B). This gene encodes the transcription factor Gcn4p, which has been implicated in the activation of several genes responsive to amino acid starvation (16, 25). These results indicate that QDR2 transcription is activated in response to leucine limitation, this activation being dependent on the presence of Gcn4p.
QDR2 transcription is activated under NH4+ limitation but not under carbon limitation.
The activation of QDR2 expression in yeast cells under nitrogen source (NH4+) limitation was also registered in the auxotrophic yeast strain examined in the present work (data not shown) and in strain 23344c (Fig. 5). This prototrophic strain was used to avoid the presence of amino acids in the growth medium which are necessary for the auxotrophic strain BY4741. Results from Northern blotting experiments indicate that the levels of QDR2 transcripts increased fivefold when the 23344c cell population approached the stationary phase of growth due to ammonium limitation (initial concentration of ammonium sulfate, 0.0265 g/liter), compared with the values for cells grown with a saturating ammonium concentration (Fig. 5).
FIG. 5.
QDR2 transcription is activated under NH4+ limitation. Relative levels of QDR2 mRNA in cells of the amino acid prototrophic strain S. cerevisiae 23344c during growth in MM5 medium where the concentration of (NH4)2SO4 was saturating (2.65 g/liter) (♦) or limiting (0.0265 g/liter) (▪). Cell samples were harvested at different incubation times as indicated. Relative QDR2 mRNA/ACT1 mRNA ratios were obtained by densitometry of the autoradiograms. Values are representative of two independent experiments.
The eventual transcription activation of QDR2 under carbon source limitation was also investigated. Under glucose-limiting conditions (initial concentration of 0.05%, wt/vol), S. cerevisiae BY4741 growth proceeded with a specific growth rate below the value possible at a glucose-saturating concentration (2%, wt/vol) until the exhaustion of the carbon source. However, no modification of the mRNA level from QDR2 was observed when cells entered the stationary phase due to carbon limitation (data not shown). Altogether, the results indicate that the activation of QDR2 transcription in cells entering the stationary phase of growth cannot be attributed to nutrient limitation, in general, but is specific for nitrogen source limitation.
Intracellular amino acid pool size depends on QDR2 expression.
In light of the expression patterns registered for QDR2 under nitrogen source/leucine limitation and of the role in the excretion of amino acids postulated for the AQR1 gene (48), we compared the amino acid concentrations in cells of BY4741 and the Δqdr2 mutant, grown in the absence of quinidine. The amino acid pool sizes were determined in cells harvested in the middle of the exponential growth phase and when the culture reached an OD600 of 2, corresponding to entrance in the stationary phase of growth due to leucine limitation and to the consequent activation of QDR2 transcription (Table 1 and Fig. 4). The intracellular concentrations of 18 amino acids were higher in the mutant with QDR2 deleted than in the parental strain (Table 1). The registered differences were, in general, more evident in cells entering the stationary phase of growth due to leucine limitation, when QDR2 is overexpressed. Exceptions were registered only for exponential cells and essentially for the three amino acids that were added to the culture medium due to the auxotrophies of the tested yeast strain. The amino acids levels determined in the supernatants of the two cultures tested, at concentrations within the sensitivity levels of the method used, are consistent with the notion that QDR2 expression alters amino acid partition between the yeast cell interior and the extracellular medium, leading to the increase of their concentrations in Δqdr2 cells (data not shown).
TABLE 1.
Concentrations of amino acids in Saccharomyces cerevisiae BY4741 and BY4741 Δqdr2 cellsa
| Amino acid | Amino acid concn (μmol/g of cells [dry wt]) in:
|
|||
|---|---|---|---|---|
| Exponential phase
|
Early stationary phase
|
|||
| WT cells | Δqdr2 cells | WT cells | Δqdr2 cells | |
| Alanine | 37.0 | 49.0 | 123.9 | 200.1 |
| Arginine | 32.5 | 50.4 | 52.4 | 95.8 |
| Asparagine | 11.8 | 18.4 | 21.2 | 29.6 |
| Aspartate | 9.3 | 18.0 | 22.5 | 25.9 |
| Cysteine | 15.1 | 20.2 | 9.6 | 15.4 |
| Glutamine | 5.7 | 9.7 | 21.2 | 41.8 |
| Glycine | 4.3 | 6.5 | 8.1 | 9.2 |
| Isoleucine | 1.3 | 2.2 | 2.6 | 4.9 |
| Lysine | 11.3 | 17.1 | 29.8 | 31.0 |
| Phenylalanine | 1.7 | 1.6 | 2.7 | 4.9 |
| Serine | 4.4 | 7.9 | 11.5 | 12.3 |
| Threonine | 4.3 | 7.2 | 9.4 | 19.7 |
| Tryptophan | 11.3 | 11.6 | 5.7 | 8.4 |
| Tyrosine | 0.8 | 1.8 | 5.4 | 15.0 |
| Valine | 6.1 | 9.1 | 9.8 | 18.8 |
| Histidine* | 11.4 | 8.9 | 29.2 | 41.9 |
| Leucine* | 2.5 | 2.2 | 1.3 | 2.5 |
| Methionine* | 1.0 | 1.2 | 6.5 | 10.7 |
Cells were harvested in mid-exponential growth phase or when the culture entered the stationary phase of growth (OD600 of 2) due to leucine limitation. The amino acids corresponding to BY4741 auxotrophies are indicated by an asterisk. WT, wild type.
DISCUSSION
The mechanisms behind the apparent promiscuity of the multidrug efflux pumps in different organisms and the presence of a multitude of different efflux pumps that may provide protection against compounds that are not usually present in their natural environment remain elusive and a topic of debate. In the present work, we show evidence supporting the notion that the expression of the yeast multidrug resistance transporter Qdr2p plays an important role in the maintenance of physiological levels of K+ in the cell. This biological function becomes crucial under environmental conditions leading to K+-limited growth, due either to the presence of limiting levels of K+ in the growth medium or to the presence of quinidine.
The presence of quinidine was found to strongly affect the K+ uptake rate and K+ requirements in the yeast cells. This quinidine-induced inhibition of K+ uptake was also observed in a strain with TRK1 and TRK2 genes deleted. Therefore, the major transport system for K+ uptake is not the sole or the main direct target of the drug. The results also indicate that increased K+ availability improves yeast growth in the presence of quinidine, in agreement with a physiological advantage of cells expressing the QDR2 gene under quinidine stress. Indeed, QDR2 expression leads to an increase of the K+ (Rb+) uptake rate, this biological activity being crucial to counteract the deleterious effect of quinidine.
The uptake of K+ has been linked to a number of very important cellular processes, in particular the regulation of intracellular acidification. The activities of the major K+ influx transporters, Trk1p and Trk2p, are important in setting the pHi (51) since H+ pumping by Pma1p is electrogenic and K+ is the major return current in yeast (41). It is thus conceivable that the difficulty of the Δqdr2 mutant to accumulate K+ in the presence of quinidine could impair this mutant's ability to counteract cytosolic acidification as the result of decreased H+ extrusion. Consistent with this hypothesis, we demonstrated before that, for the same quinidine concentration, Δqdr2 exponential cells exhibit an average pHi (5.2) significantly below the average pHi of wild-type exponential cells (6.3) (47). The protective effect exerted by QDR2 expression against quinidine may in part be related to its role in controlling pHi and the internal K+ concentration in the presence of the drug. The maintenance of pHi within a permissive range for the functionality of sensitive cellular systems is crucial for cell physiology, due to its effects on proteins and biochemical reactions.
The data obtained in this study are consistent with quinidine acting as an uncoupler that dissipates the H+ gradient across the plasma membrane. Consistent with this hypothesis, we found that the well-established uncoupler carbonyl cyanide 3-chlorophenylhydrazone (CCCP) has an effect that mimics the action of quinidine on the inhibition of extracellular medium acidification by yeast. Moreover, QDR2 expression was also found to play a role in counteracting this effect of CCCP (unpublished results). However, although CCCP was used at low concentrations (5 to 20 μM), expected to lead to the dissipation of H+ gradient across the plasma membrane without significantly affecting mitochondrial respiration, the anticipated effect of this uncoupler on the Δqdr2 phenotype under K+ limitation could not be clearly demonstrated (unpublished results), presumably due to the much higher toxicity of CCCP which may have masked the observation of the clear phenotype registered for quinidine.
Other possible clues to the biological function of QDR2 in the yeast cell may come from two lines of experimental evidence obtained during this work. First, it appears that the expression of QDR2 not only increases the permeability of the yeast cell to K+ but also increases cell permeability to the nonspecific efflux of amino acids. This conclusion is suggested by the larger amino acid pool in Δqdr2 cells, with the QDR2 gene deleted, than in wild-type cells. This observation is also in agreement with the proposed role for the multidrug transporter Aqr1p, a Qdr2p homologue, in the excretion of amino acids (48). However, the effect of QDR2 expression on amino acid homeostasis is not specific for a narrow range of amino acids but was registered for at least 18 different amino acids, raising doubts about the specific and direct role of Qdr2p as an amino acid exporter.
The second remarkable observation is that QDR2 transcription was highly activated when yeast cells entered the stationary phase of growth, due either to the nitrogen source (NH4+) limitation or to leucine limitation, for the auxotrophic strain examined. Moreover, this regulation is dependent on general amino acid control by Gcn4p. Translation of GCN4 mRNA is derepressed in amino acid-deprived cells, leading to transcriptional induction of nearly all genes encoding amino acid biosynthetic enzymes (16). Although the uptake and accumulation of amino acids from the external medium seem to be irreversible, amino acids are excreted into the medium whenever they are overproduced above a given threshold by yeast cells (13). This can occur when growth is arrested under conditions where amino acid synthesis can continue and amino acid efflux is mediated by mechanisms that are distinct from the characterized uptake systems (13). Interestingly, the first proposed outward-pumping system for amino acid excretion was the multidrug resistance transporter of the major facilitator superfamily, Aqr1p (48), belonging to the same cluster I of the 12-spanner drug:H+ antiporter DHA12 family (26, 30, 40). However, this same role was hypothesized before for ATR1 (13, 18), a member of the 14-spanner drug:H+ antiporter DHA14 family (26, 30). ATR1 is required for yeast resistance to aminotriazole (18), a competitive inhibitor of the HIS3 gene product, an imidazoleglycerolphosphate dehydratase (20, 42), causing histidine starvation. Interestingly, Gcn4p also controls the expression of ATR1 through a DNA element related to the yAP-1 recognition element (7). Also, in the case of the QDR2 promoter region, there is no established DNA binding site for Gcn4p [the sequence TGA(G/G)TCA]. Using the YEASTRACT database (http://www.yeastract.com) (43), five cis-acting replication element-like sequences were found. Since Gcn4p was proven to compete with Sko1p for these binding sites in the HAL1 promoter region (28), it is possible that they may also be used to bind Gcn4p to the QDR2 promoter region. Moreover, there are three yAP-1 sequences in the QDR2 promoter region also recognized by Gcn4p and Yap1p (7).
The interpretation of results of amino acid partition between the cell interior and the surrounding medium is complicated by the possible modification of processes involving amino acid synthesis and the degradation and mobilization of stored nitrogen sources under nitrogen starvation. Nevertheless, the results shown in this work are consistent with the presence of a larger amino acid pool in Δqdr2 cells. The indirect (or direct) role of Qdr2p in promoting this apparently nonspecific efflux of amino acids may become crucial to avoid abnormal large amounts of amino acids resulting from deregulated metabolism that may be deleterious to cell physiology, as is the case during growth arrest due to leucine limitation. Remarkably, a recently published work has established a link between K+ limitation, ammonium toxicity, and amino acid excretion (15). According to the indication of this microarray analysis, complemented by metabolite profiling, under limiting potassium concentrations, toxic amounts of ammonium might enter yeast via potassium channels, as observed in other organisms. When experiencing ammonium toxicity, yeast responds by excreting amino acids which may constitute a rudimentary ammonia detoxification mechanism in the yeast cell (15).
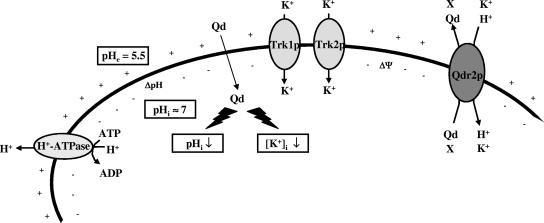
Although the dominant model in MDR literature is still the drug pump model, the results of the present work support the altered partitioning model (36, 37). This model considers that the altered expression of an MDR transporter may lead to altered ion transport, indirectly affecting intracellular accumulation of the drug by perturbing plasma membrane potential and/or intracellular pH. According to the model postulated here (Fig. 6), the putative drug:H+ antiporter Qdr2p may also be capable of coupling K+ ions to drive the export of its physiological substrate(s), as hypothesized before for the amino acid:H+ symport, by amino acid permeases (50). It is also possible that Qdr2p catalyzes directly the fortuitous export of specific drugs, in particular quinidine.
FIG. 6.
A model for the possible relationship between Qdr2p activity, K+ transport, and the effect of quinidine on the yeast cell. It is proposed that the drug:H+ antiporter Qdr2p may also be capable of coupling K+ ions to drive the export of its physiological substrate(s) (X). The presence of quinidine leads to impaired K+ uptake and the acidification of the cell interior by reducing H+ efflux from energized cells. In such a situation, the K+ uptake coupled with QDR2 expression would help the cell to counteract these deleterious effects. Qdr2p also reduces the internal concentration of quinidine by promoting, directly or indirectly, the active expulsion of the drug (47). Although the eventual antiport of quinidine with K+/H+ mediated by Qdr2p is considered fortuitous or opportunistic, the natural substrates(s) (X) for Qdr2p is (are) still unclear. Our model also involves Qdr2p, either directly or indirectly, in the extrusion of amino acids from the cell. See text for details. Qd, quinidine; ΔΨ, membrane potential.
The viability of Δtrk1 Δtrk2 cells has indicated the existence of additional forms for transporting potassium into the yeast cell that do not require the Trk1p/Trk2p transport system. However, the nature of this transport remains to be elucidated, although the nonspecific uptake of potassium occurring through sugar and amino acid permeases has been postulated (22, 23, 50). Based on a number of evidences, Wright et al. (50) have considered that amino acid:H+ symporters may not have an absolute requirement for H+ as a cosubstrate and may be capable of coupling K+ ions to drive amino acid uptake. The results obtained during this work are consistent with the hypothesis that the putative drug:H+ antiporter Qdr2p may also be capable of coupling K+ ions to drive the export of its physiological substrate and of drugs, in particular quinidine. It is, however, difficult to believe that the yeast cells have evolved a gene like QDR2, together with several others, whose normal physiological role is to export a compound not found in its natural environment. The hypothesis that Qdr2p may be directly involved in K+ uptake is reinforced by the fact that, when QDR2 was deleted in the Δtrk1 Δtrk2 background, growth deficiency and decreased K+ uptake in this Δqdr2 triple mutant under K+ limitation were still observed, indicating that K+ uptake due to QDR2 expression is independent of the Trk1p/Trk2p system. The results also show that even after deletion of TRK1, TRK2, and QDR2, there is still K+ transport, indicating that other transporters may be involved in K+ uptake. It is usually accepted that several nonspecific transporters may be involved in the low-affinity transport of K+ observed in the Δtrk1 Δtrk2 mutant (22). The existence of a yeast nonselective cation channel responsible for the low-affinity K+ uptake was postulated before, although the identity of the protein(s) responsible for this inwardly rectifying pathway is not known (2, 3). Amino acid or sugar transporters could be involved in this process (22, 50) together with Qdr2p and, eventually, other plasma membrane multidrug resistance transporters. Since quinidine strongly decreases the uptake of K+, presumably by acting as an uncoupler at the plasma membrane, the involvement of Qdr2p in the uptake of this essential ion confers a physiological advantage to quinidine-stressed cells. This biological activity may be the mechanism behind the role of Qdr2p as a determinant of resistance to the antiarrhythmic and antimalarial drug tested.
Acknowledgments
We thank J. Ariño and A. Ruiz for the yeast strains.
This research was supported by FEDER, “Fundação para a Ciência e Tecnologia” (FCT), the POCTI Programmes (contracts POCTI/BME/46526/2002 and PDCT/BIO/56838/2004 and grants to R. C. Vargas [SFRH/BD/924/2000], S. Tenreiro [SFRH/BPD/5649/2001], and M. C. Teixeira [SFRH/BPD/14484/03]), and grant BFU2005-06388-C04-03 from Ministerio de Educación y Ciencia, España (J.R.).
Footnotes
Published ahead of print on 22 December 2006.
REFERENCES
- 1.Balzi, E., and A. Goffeau. 1995. Yeast multidrug resistance: the PDR network. J. Bioenerg. Biomembr. 27:71-76. [DOI] [PubMed] [Google Scholar]
- 2.Bertl, A., J. Ramos, J. Ludwig, H. Lichtenberg-Frate, J. Reid, H. Bihler, F. Calero, P. Martinez, and P. O. Ljungdahl. 2003. Characterization of potassium transport in wild-type and isogenic yeast strains carrying all combinations of trk1, trk2 and tok1 null mutations. Mol. Microbiol. 47:767-780. [DOI] [PubMed] [Google Scholar]
- 3.Bihler, H., C. L. Slayman, and A. Bertl. 2002. Low-affinity potassium uptake by Saccharomyces cerevisiae is mediated by NSC1, a calcium-blocked non-specific cation channel. Biochim. Biophys. Acta 1558:109-118. [DOI] [PubMed] [Google Scholar]
- 4.Bolhuis, H., H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1997. Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21:55-84. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, M., J. Ramos, and A. Rodríguez-Navarro. 1981. Potassium requirements of Saccharomyces cerevisiae. Curr. Microbiol. 6:295-299. [Google Scholar]
- 6.Chen, F. S., and D. Fedida. 1998. On the mechanism by which 4-aminopyridine occludes quinidine block of the cardiac K+ channel, hKv1.5. J. Gen. Physiol. 111:539-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, S. T., E. Tseng, and W. S. Moye-Rowley. 1997. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 272:23224-23230. [DOI] [PubMed] [Google Scholar]
- 8.Delling, U., M. Raymond, and E. Schurr. 1998. Identification of Saccharomyces cerevisiae genes conferring resistance to quinoline ring-containing antimalarial drugs. Antimicrob. Agents Chemother. 42:1034-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.do Valle Matta, M. A., J. Jonniaux, E. Balzi, A. Goffeau, and B. van den Hazle. 2001. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene 272:111-119. [DOI] [PubMed] [Google Scholar]
- 10.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sá-Correia, and P. Briza. 2002. Dtr1p, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaber, R. F., C. A. Styles, and G. R. Fink. 1988. TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2848-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grenson, M. 1992. Amino acid transporters in yeast: structure, function and regulation, p. 219-245. In J. De Pont (ed.), Molecular aspects of transport proteins. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 14.Hayes, J. D., and C. R. Wolf. 1997. Molecular genetics of drug resistance. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 15.Hess, D. C., W. Lu, J. D. Rabinowitz, and D. Botstein. 2006. Ammonium toxicity and potassium limitation in yeast. PLoS Biol. 4:e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407-450. [DOI] [PubMed] [Google Scholar]
- 17.Jungwirth, H., and K. Kuchler. 2006. Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett. 580:1131-1138. [DOI] [PubMed] [Google Scholar]
- 18.Kanazawa, S., M. Driscoll, and K. Struhl. 1988. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol. Cell. Biol. 8:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klopotowski, T., and A. Wiater. 1965. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazoleglycerolphosphate dehydratase. Arch. Biochem. Biophys. 112:562-566. [DOI] [PubMed] [Google Scholar]
- 21.Ko, C. H., and R. F. Gaber. 1991. TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:4266-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, H., C. H. Ko, T. Herman, and R. F. Gaber. 1998. Trinucleotide insertions, deletions, and point mutations in glucose transporters confer K+ uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madrid, F., M. J. Gomez, J. Ramos, and A. Rodriguez-Navarro. 1998. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273:14838-14844. [DOI] [PubMed] [Google Scholar]
- 24.Marini, A., S. Soussi-Boudekou, S. Vissers, and B. André. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelissen, B., R. D. Wachter, and A. Goffeau. 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21:113-134. [DOI] [PubMed] [Google Scholar]
- 27.Nunes, P. A., S. Tenreiro, and I. Sá-Correia. 2001. Resistance and adaptation to quinidine in Saccharomyces cerevisiae: role of QDR1 (YIL120w), encoding a plasma membrane transporter of the major facilitator superfamily required for multidrug resistance. Antimicrob. Agents Chemother. 45:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascual-Ahuir, A., F. Posas, R. Serrano, and M. Proft. 2001. Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J. Biol. Chem. 276:37373-37378. [DOI] [PubMed] [Google Scholar]
- 29.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6:446-451. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., M. K. Sliwinski, B. Nelissen, A. Goffeau, and M. H. Saier, Jr. 1998. Unified inventory of established and putative transporters within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 430:116-125. [DOI] [PubMed] [Google Scholar]
- 31.Portillo, F., and R. Serrano. 1989. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur. J. Biochem. 186:501-507. [DOI] [PubMed] [Google Scholar]
- 32.Prasad, R., and S. Panwar. 2004. Physiological functions of multidrug transporters in yeast. Curr. Sci. 86:62-73. [Google Scholar]
- 33.Ramos, J., R. Alijo, R. Haro, and A. Rodriguez-Navarro. 1994. TRK2 is not a low-affinity potassium transporter in Saccharomyces cerevisiae. J. Bacteriol. 176:249-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos, J., P. Contreras, and A. Rodriguez-Navarro. 1985. A potassium transport mutant of Saccharomyces cerevisiae. Arch. Microbiol. 143:88-93. [Google Scholar]
- 35.Ramos, J., R. Haro, and A. Rodríguez-Navarro. 1990. Regulation of potassium fluxes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1029:211-217. [DOI] [PubMed] [Google Scholar]
- 36.Roepe, P. D. 2000. What is the precise role of human MDR1 protein in chemotherapeutic drug resistance? Curr. Pharm. Des. 6:241-260. [DOI] [PubMed] [Google Scholar]
- 37.Roepe, P. D., L. Y. Wei, M. M. Hoffman, and F. Fritz. 1996. Altered drug translocation mediated by the MDR protein: direct, indirect, or both? J. Bioenerg. Biomembr. 28:541-555. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz, A., M. del Carmen, M. A. Sanchez-Garrido, J. Arino, and J. Ramos. 2004. The Ppz protein phosphatases regulate Trk-independent potassium influx in yeast. FEBS Lett. 578:58-62. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz, A., L. Yenush, and J. Ariño. 2003. Regulation of ENA1 Na+-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell 2:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sá-Correia, I., and S. Tenreiro. 2002. The multidrug-resistance transporters of the major facilitator superfamily, six years after disclosure of Saccharomyces cerevisiae genome sequence. J. Biotech. 98:215-226. [DOI] [PubMed] [Google Scholar]
- 41.Serrano, R. 1991. Transport across yeast vacuolar and plasma membranes, p. 523-585. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Struhl, K., and R. W. Davis. 1977. Production of a functional eukaryotic enzyme in Escherichia coli: cloning and expression of the yeast structural gene for imidazoleglycerolphosphate dehydratase (his3). Proc. Natl. Acad. Sci. USA 74:5255-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teixeira, M. C., P. Monteiro, P. Jain, S. Tenreiro, A. R. Fernandes, N. P. Mira, M. Alenquer, A. T. Freitas, A. L. Oliveira, and I. Sá-Correia. 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 1:D446-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenreiro, S., P. A. Nunes, C. A. Viegas, M. S. Neves, M. C. Teixeira, M. G. Cabral, and I. Sá-Correia. 2002. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292:741-748. [DOI] [PubMed] [Google Scholar]
- 45.Tenreiro, S., R. C. Vargas, M. C. Teixeira, C. Magnani, and I. Sá-Correia. 2005. The yeast multidrug transporter Qdr3 (Ybr043c): localization and role as a determinant of resistance to quinidine, barban, cisplatin, and bleomycin. Biochem. Biophys. Res. Commun. 327:952-959. [DOI] [PubMed] [Google Scholar]
- 46.Trampuz, A., M. Jereb, I. Muzlovic, and R. M. Prabhu. 2003. Clinical review: severe malaria. Crit. Care 7:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vargas, R. C., S. Tenreiro, M. C. Teixeira, A. R. Fernandes, and I. Sá-Correia. 2004. Saccharomyces cerevisiae yeast multidrug transporter Qdr2p (Yil121wp): localization and function as a quinidine resistance determinant. Antimicrob. Agents Chemother. 48:2531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velasco, I., S. Tenreiro, I. L. Calderon, and B. André. 2004. Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryot. Cell 3:1492-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warhurst, D. C., J. C. Craig, I. S. Adagu, D. J. Meyer, and S. Y. Lee. 2003. The relationship of physico-chemical properties and structure to the differential antiplasmodial activity of the cinchona alkaloids. Malar. J. 2:26-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright, M. B., J. Ramos, M. J. Gomez, K. Moulder, M. Scherrer, G. Munson, and R. F. Gaber. 1997. Potassium transport by amino acid permeases in Saccharomyces cerevisiae. J. Biol. Chem. 272:13647-13652. [DOI] [PubMed] [Google Scholar]
- 51.Yenush, L., J. M. Mulet, J. Arino, and R. Serrano. 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 21:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]