Abstract
A sequence database was created for the Leishmania N-acetylglucosamine-1-phosphate transferase (nagt) gene from 193 independent isolates. PCR products of this single-copy gene were analyzed for restriction fragment length polymorphism based on seven nagt sequences initially available. We subsequently sequenced 77 samples and found 19 new variants (genotypes). Alignment of all 26 nagt sequences is gap free, except for a single codon addition or deletion. Phylogenetic analyses of the sequences allow grouping the isolates into three subgenera, each consisting of recognized species complexes, i.e., subgenus Leishmania (L. amazonensis-L. mexicana, L. donovani-L. infantum, L. tropica, L. major, and L. turanica-L. gerbilli), subgenus Viannia (L. braziliensis, L. panamensis), and one unclassified (L. enriettii) species. This hierarchy of grouping is also supported by sequence analyses of selected samples for additional single-copy genes present on different chromosomes. Intraspecies divergence of nagt varies considerably with different species complexes. Interestingly, species complexes with less subspecies divergence are more widely distributed than those that are more divergent. The relevance of this to Leishmania evolutionary adaptation is discussed. Heterozygosity of subspecies variants contributes to intraspecies diversity, which is prominent in L. tropica but not in L. donovani-L. infantum. This disparity is thought to result from the genetic recombination of the respective species at different times as a rare event during their predominantly clonal evolution. Phylogenetically useful sites of nagt are restricted largely to several extended hydrophilic loops predicted from hypothetical models of Leishmania NAGT as an endoplasmic reticulum transmembrane protein. In silico analyses of nagt from fungi and other protozoa further illustrate the potential value of this and, perhaps, other similar transmembrane molecules for phylogenetic analyses of single-cell eukaryotes.
Many microorganisms speciate via clonal evolution. They replicate asexually, with genetic recombination as a rare event. A typical example among single-cell eukaryotes is the trypanosomatid protozoa (58, 59), which are mostly parasites, e.g., Leishmania spp. and Trypanosoma spp. Leishmania spp. live extracellularly in the digestive tracts of blood-sucking female sand flies of various species as their vectors and intracellularly in the macrophages of different mammalian hosts, i.e., human, canine, rodent, and other reservoir animals. The complexities of such unusual ecological niches undoubtedly contribute to Leishmania speciation.
A large body of biological, biochemical, immunological, and molecular data (7, 10, 23, 55) exists in the literature suggesting that the genus Leishmania consists of three groups (55) as follows: (i) subgenus Leishmania, which includes species complexes distributed in both the New World and the Old World, e.g., L. major [Leishmania (Leishmania) major], L. tropica, L. donovani-L. infantum, L. amazonensis-L. mexicana, and L. turanica-L. gerbilli; (ii) subgenus Viannia, whose members are restricted to the Neotropics, e.g., L. braziliensis [Leishmania (Viannia) braziliensis] and L. panamensis; and (iii) several unclassified species (3), e.g., L. enriettii. The pathogenic species listed above have long been subjected to diagnostic typing and phylogenetic analyses (15, 23, 49). They are referred to as species complexes due to subspecies heterogeneity, as shown in some population genetic analyses (26, 29, 51). The molecular “markers” and methodologies of choice for phylogenetic analyses have been reviewed and discussed in the context of genomic typing for integrating microbial taxonomy, phylogeny, population genetics, and clinical epidemiology (62).
Single-copy protein-coding genes have been used for phylogenetic analyses of Trypanosoma spp., e.g., GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (25, 56) but not extensively for Leishmania spp., e.g., several markers previously used with multilocus enzyme electrophoresis for L. donovani-L. infantum (41). We have begun to examine nagt, which encodes N-acetylglucosamine-1-phosphate transferase (NAGT), a microsomal transmembrane enzyme in the first step of N-linked glycan biosynthesis. N-glycosylation of Leishmania gp63 is associated with the stability of this zinc protease as a virulence factor (42). Wild-type L. amazonensis (LV78) (32, 36) and L. major (Friedlin) contain nagt genes in a single copy per haploid genome without paralogous genes or pseudogenes. Knockout mutants of nagt are nonviable unless rescued episomally, indicative of the gene's functional indispensability (12). Previously, nagt sequence heterogeneity compared favorably against restriction fragment length polymorphism (RFLP) assays of mitochondrial or kinetoplast and nuclear repetitive DNAs for groupings of >50 Leishmania isolates from one endemic area (1). Sequencing 12 PCR-amplified nagt genes revealed five genotypes within the species complexes expected in that region, i.e., L. infantum, L. tropica, L. major, and L. major variants.
We have expanded the nagt database to a cumulative total of 238 independent isolates largely from the Old World. Phylogenetic analyses of the 26 divergent sequences obtained produced results consistent with additional sequence data from other single-copy genes, i.e., segregation of the genotypes into subgenera and species complexes. Different species complexes vary significantly in subspecies divergence. The incongruity of this divergence with the extent of species distribution may bear on host-dependent selection for Leishmania evolutionary adaptation. The disparity among species complexes in the homozygosity/heterozygosity ratio is notable. This is hypothesized to result from their rare genetic recombination at different times during clonal evolution. The extended fourth hydrophilic loop of Leishmania NAGT, as an endoplasmic reticulum (ER) transmembrane molecule, is rich in phylogenetically useful sites. Trypanosoma and fungal nagt genes are also informative for resolving their taxonomic relationships.
MATERIALS AND METHODS
Leishmania isolates and culture.
Independent Leishmania isolates were generously provided by colleagues, either as promastigotes (total, 115) or as DNA samples (total, 78). The origins of these isolates are worldwide, but they are mostly from the Old World (Fig. 1). Leishmania species names were provided by the donors for ∼100 samples, including many well-known strains typed by different methodologies. See Table S1 in the supplemental material for a complete list of the isolates, from which 77 were selected to build the nagt sequence database (65 isolates are listed in Table 1) (see also http://66.99.255.20/cms/micro/sample%20list.pdf). The Leishmania promastigotes needed for DNA isolation were grown at ∼25°C in HEPES-buffered (pH 7.4) medium 199 (Sigma) with 10 to 20% heat-inactivated fetal bovine serum plus penicillin and streptomycin (100 units and 100 μg/ml, respectively). Two L. tropica (allele combination I/II) isolates (HA06 and U41) from Turkey (1) were cloned on agar plates. Four colonies were grown as cloned populations for the present study.
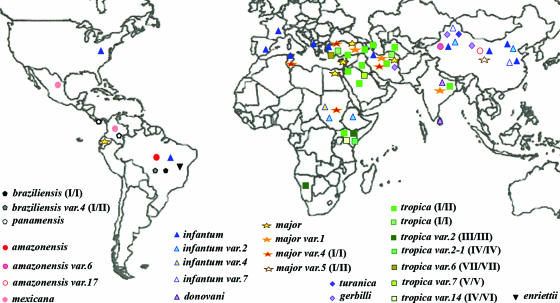
FIG. 1.
Geographic distribution of 26 Leishmania nagt gene variants (var.). The map contains information from a grand total of 238 Leishmania isolates by including those studied previously (1, 36, Leishmania DB). The 26 variants are grouped by RFLP and sequence analyses as subspecies genotypes (color coded) within each of the 7 species complexes indicated by different symbols: circle, L. amazonensis-L. mexicana; triangle, L. donovani-L. infantum; square, L. tropica; star, L. major; diamond, L. turanica-L. gerbilli; inverted triangle, L. enriettii; pentagon, L. braziliensis-L. panamensis. Note that the triangle for L. infantum in the United States denotes three isolates from the foxhounds of a hunting club where visceral leishmaniasis appears to have been transmitted among these locally raised animals by unknown mechanisms but not to the human population (50).
TABLE 1.
Twenty six nagt variants within known species complexes of Leishmania recognized by analyses of 193 independent isolatesa
| nagt genotypesb | WHO Codec | Diseased | nagt genotypesb | WHO Codec | Diseased | |
|---|---|---|---|---|---|---|
| L. amazonensis/mexicana | HOM/TN/90/FMH | CL | ||||
| complex (5)e | 13 Variant 5 (I/II) (1) | HOM/CN/99/Gansu-Wangg | VL | |||
| 1 L. amazonensis (1) | RAT/BR/72/LV78g | CL | ||||
| 2 Variant 6 (1) | HOM/CN/75/Kashi-BTO13 | VL | L. turanica/gerbilli | |||
| 3 Variant 17 (1) | HOM/CN/97/Gansu-LUO | VL | complex (21) | |||
| 4 L. mexicana (2) | HOM/CO/94/1182g | CL | 14 L. turanica (15) | RHO/CN/99/KMA2g | CL | |
| HOM/MX/84/ISTE GS | CL | RHO/CN/92/Qi Daig | CL | |||
| L. donovani/infantum | RHO/IR/04?/IR14 | CL | ||||
| complex (91) | 15 L. gerbilli (6) | RHO/CN/62/20g | CL | |||
| 5 L. infantum (63) | HOM/TR/00/OG-VLg | VL | IMON/CN/97/KMP3 | N/A | ||
| HOM/TR/03/Adana #7 | CL | |||||
| HOM/GR/70?/GH5 | CL | L. tropica complex (48) | ||||
| CANL/IR/04/IR2B | VL? | 16 L. tropica (I/II) (22) | HOM/TR/95/MACg | CL | ||
| HOM/FR/80/189 | VL? | HOM/SY/00/Am | CL | |||
| HOM/ES/81/260 | VL? | HOM/AZ/74/K27 | CL | |||
| HOM/CN/80/801 | VL | HOM/IQ/73/LRC L32 | CL | |||
| HOM/TN/90/DREP13 | CL | HOM/IR/94/X54 | CL | |||
| HOM/BR/82/BA-2, C1 | VL | HOM/SA/91/WR2044 | VL (asymptomatic) | |||
| 6 Variant 2 (15) | HOM/CN/50?/Bman | VL | HOM/JO/90-91?/Jh33 | CL | ||
| HOM/SD/03?/VL2 | VL | HOM/SU/60/BTO11 | CL | |||
| 7 Variant 4 (1) | HOM/KE/84/NLB323g | VL | HOM/KE/81/NLB 030Bg | VL | ||
| 8 Variant 7 (7) | HOM/CN/93/KXG-LIUg | CL | HOM/IN/76/UR6g | VL | ||
| IWUI/CN/87/KXG65 | N/A | 17 L. tropica (I/I) (9) | HOM/TR/04/EP94 | CL | ||
| HOM/CN/89/Shandong | VL | HOM/IR/03/IR1 | HIV-CL/VL | |||
| 9 L. donovani (5) | HOM/IN/96/JD | VL | CAN/IR/02/IR3C | VL ? | ||
| HOM/LK/03/H9 | CL | 18 Variant 2 (III/III) (7) | IROS/NA/87/8688g | N/A | ||
| L. major complex (23) | HOM/NA/84/K1 | CL | ||||
| 10 L. major (9) | HOM/IL/80/Friedling | CL | 19 Variant 2-1 (IV/IV) (1) | HOM/KE/84/NLB297Ag | ? | |
| HOM/TR/96/DK | CL | 20 Variant 6 (VII/VII) (6) | HOM/TR/03/EP82g | CL | ||
| HOM/EG/95/Dr. B | CL | 21 Variant 7 (V/V) (1) | HOM/IQ/91/WR1095g | VL (asymptomatic) | ||
| HOM/IR/??/IR173 | CL | 22 Variant 14 (IV/VI) (2) | HOM/KE/85/NLB545g | ? | ||
| HOM/EQ/87/G-09 | CL | L. braziliensis/panamensis (4) | ||||
| 11 Variant 1 (6) | HOM/TR/94/HKg | CL | 23 L. braziliensis (I/I) (1)f | HOM/BR/75/M2904g | CL | |
| HOM/IR/02/Iran 9 | CL | 24 Variant 4 (I/II) (1) | HOM/BR/75/M2903 | CL | ||
| HOM/IN/05?/RMP-240 | PKDL | 25 L. panamensis (2) | HOM/PA/71/LS94 | CL | ||
| 12 Variant 4 (I/I) (7) | HOM/TR/93/HA | VL | HOM/CO/00?/140 | CL | ||
| HOM/TR/93/SYg | CL | L. enriettii (1) | ||||
| HOM/IR/64/Iran8 | CL | 26 enriettii (1) | CAV/BR/45/enriettiig | CL | ||
| RHO/SD/02?/CL1 | CL |
Listed are 65 representatives of 77 sequenced.
Variant 1 to 17, the number of nucleotide substitutions against an arbitrarily selected reference sequence in each species complex. I to VII, different alleles in L. major, L. tropica, and L. braziliensis complex. Numbers in parentheses represent the actual number of isolates.
Host/reservoirs: CAN, dog (Canis familiaris); CANL, wolf (C. lupus pallipos); CAV, guinea pig (Cavia sp.); HOM, Homo sapiens; IMON, sand fly (Phlebotomus mongolensis); IROS, sand fly (P. rossi); IWUI, sand fly (P. major wui); RAT, Rattus rattus; RHO, great gerbil (Rhobomys opimus). Countries of origin: AZ, Azerbaijan; BR, Brazil; CN, China; CO, Colombia; EG, Egypt; EQ, Ecuador; ES, Spain; FR, France; GR, Greece; IL, Israel; IN, India; IQ, Iraq; IR, Iran; JO, Jordan; KE, Kenya; LK, Sri Lanka; MX, Mexico; NA, Namibia; PA, Panama; SA, Saudi Arabia; SD, Sudan; SU, Soviet Union; SY, Syria; TN, Tunisia; TR, Turkey. 45 to 05, year of isolation. ?, information not provided in writing by the donors.
PKDL, post-kala-azar dermal leishmaniasis; N/A, not applicable.
Numbers of isolates examined by RFLP analyses and/or sequencing.
Sequence obtained from GeneDB.
Strains used for sequencing the fc gene and other single-copy genes. fc gene sequences were also obtained from L. donovani (HOM/IN/00/IN0041J) and L. infantum variant 2 (HON/CN/90/901), which are not listed.
PCR, RFLP, and Southern blotting analyses.
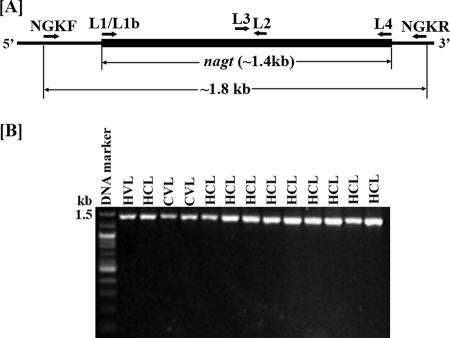
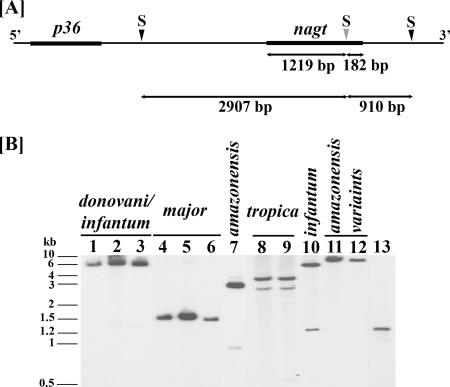
These assays were carried out as previously described (1). The ∼1.4-kb nagt and other single-copy genes (see below) were PCR-amplified from genomic DNAs (see Table S2 in the supplemental material for all genes examined, their chromosomal location, PCR conditions used, and products expected; and see Table S3 in the supplemental material for the PCR primers used). nagt was readily PCR-amplified with the L1 (or L1b for the Viannia group)/L4 primer pair as described previously (1) from all 193 samples, except for two, i.e., L. amazonensis variant 6 and variant 17. An alternative NGKF/NGKR primer set was designed for PCR assays of these two samples to obtain the nagt gene-containing ∼1.8 kb, which was either cloned directly into pGEM-T Easy (Promega) or subjected to nested PCR for nagt with the L1/L4 primer set (Fig. 2A). All PCR-amplified nagt genes were first evaluated for new variants by restriction mapping using species- and subspecies-unique sites determined from available sequences by BioEdit (24) (see Fig. 4A). Samples for PCR amplification of other single-copy genes were selected from those already genotyped as nagt variants. For Southern blotting analyses, isolated DNAs (5 μg each) were digested with intragenic single cutters to evaluate nagt as a single-copy gene (Fig. 3A). The digests were alkali-transferred to a nylon membrane (Hybond-N+; Amersham) and probed with [alpha-32P]dCTP-labeled nagt (∼1.4-kb complete open reading frame [ORF]), PCR-amplified from L. amazonensis (Table 1, genotype 1). Hybridization was carried out under stringent conditions (1).
FIG. 2.
PCR amplification and sequencing strategies for the Leishmania nagt gene (A) and the PCR-amplified ∼1.4 kb nagt gene (B) from genomic DNAs of representative isolates. The L1/L4 or L1b/L4 primer set designed from the very 5′ and 3′ ends of the nagt gene (A) PCR-amplifies the complete ∼1.4-kb ORF for all 193 genomic DNA samples (B), except for two, for which the nagt gene was obtained by nested PCR of products from the NGKF/NGKR primer set designed from the 5′ and 3′ flanks (A) (data not shown). L2 and L3 were designed as additional primers in combination with L1 or L1b and L4 for sequencing both strands of the PCR products to completion. H, human; C, canine.
FIG. 4.
Restriction maps constructed to discriminate 26 nagt genotypes (A) and AccI digests of PCR-amplified nagt genes to separate four Leishmania subgenus groups (B). The RFLP maps were based on 26 nagt gene sequence variants and verified by evaluating 193 independent samples for subgenera, species, and subspecies genotyping or differentiation. AccI (A) and BbeI (Bb) RFLPs of the PCR-amplified nagt gene differentiate species complexes L. amazonensis-L. mexicana, L. donovani-L. infantum, L. major, L. tropica, L. turanica-L. gerbilli, L. braziliensis-L. panamensis, and L. enriettii. Restriction sites shown to discriminate species and subspecies genotypes are A, AccI; Al, AlwI; Av, AvaII; B, Bsp1286I; Bb, BbeI; Bm, BmgBI; Br, BsrBI, H, HinfI; Hp, HpyCH4III; M, MseI; Na, NaeI; Nc, NciI; S, StyI; Sg, SgrAI; T, Tsp509 I; Ts, Tse I. *, indicates the restriction site for one of the two alleles due to heterozygosity, as indicated by Roman letters. Not shown in the two L. braziliensis genotypes are many TseI sites, except for the one which discriminates the two.
FIG. 3.
Leishmania nagt gene as a single-copy gene in representative genotypes. (A) Genomic SacII (S) map of the L. amazonensis nagt gene and its flanking regions. Note a single intragenic SacII site (true for all nagt genotypes of the Leishmania subgenus) near the 3′ end of the nagt ORF (thick line) at nt position 1219 (gray arrow head) and two flanking sites (black arrow head) [placed on the basis of sequences known for this chromosomal region of L. amazonensis (36)] at ∼2.9 kb upstream and ∼0.9 kb downstream of the intragenic SacII site. The p36 gene is another ORF ∼2.3 kb upstream of nagt, shown here for orientation. (B) Southern blot analysis of SacII-restricted genomic DNAs from representative Leishmania genotypes showing nagt as a single-copy gene. SacII digests of genomic DNAs were run in 0.8% agarose gel for Southern blotting analyses with the PCR-amplified L. amazonensis nagt gene as the probe. All samples gave at least one strong signal after a short exposure and no more than two after prolonged exposure (not shown). Note that the small variation among different samples in signal intensity is due to slightly unequal loading of the genomic DNAs. The two positive SacII fragments of the L. amazonensis nagt gene are exactly as expected in size and in intensity as mapped (panel A) for genomic DNA (lane 7) and for PCR products (lane 13). Lanes 1 to 3, L. donovani, L. infantum variant 7, and L. chagasi (infantum); lanes 4 to 6, L. major variant 1, L. major variant 4, and L. major; lane 7, L. amazonensis; lanes 8 to 9, L. tropica (I/II); lane 10, L. infantum; lanes 11 and 12, L. amazonensis variant 17 and L. amazonensis variant 6; lane 13, PCR-amplified L. amazonensis nagt gene. See Table 1 for L. amazonensis, L. infantum, and L. donovani designated as genotypes 1, 5, and 9, respectively.
PCR cycle sequencing.
Seventy-seven PCR products of the 1.4-kb nagt gene were cycle sequenced in both strands to completion by using four separate primers (L1 to L4 or L1b to L4 for the Viannia group) (Fig. 2A) via commercial service facilities. Other single-copy genes were similarly sequenced. See Table S3 in the supplemental material for a list of the sequencing primers used for nagt and other single-copy genes encoding ferrochelatase, prostaglandin F2 alpha-synthase, zeta-crystalline/NADPH oxidoreductase, and dihydrofolate reductase-thymidylate synthase, i.e., genes fc, pgfs, p36, and dhfr-ts, respectively. The nagt gene-containing ∼1.8 kb (Lnagt+; see Table S2 in the supplemental material) cloned in pGEM-T was sequenced with T7 and SP6 vector-specific primers plus L2 and L3. Clones of dhfr-ts and dhfr-ts+ (see Table S2 and S3 in the supplemental material) were similarly obtained and sequenced. The DNA sequences obtained were verified against their electrochromatograms for both strands, assembled, and aligned by using Clustal X (57).
Phylogenetic analyses.
nagt sequences were used in equal lengths of 1,324 bp (nucleotide [nt] 37 to 1360) or 441 amino acids (aa 13 to 453), except for two, i.e., the L. turanica and L. enriettii nagt genes with a codon deletion and insertion, respectively. Phylogenetic analyses of the aligned sequences were done as described previously (17, 18) by using different algorithms of Clustal X (57), BioEdit (24), and/or MEGA (version 2.1) (35), i.e., the Kimura 2-parameter neighbor-joining method, maximum likelihood, maximum parsimony, UPGMA (unweighted pair group method with arithmetic mean), and minimum evolution. Bootstrapping was done with 1,000 replicas where applicable. The minimum-evolution and maximum-parsimony algorithms in MEGA were used for phylogenetic analyses of the nagt sequences from 26 Leishmania genotypes together with four Trypanosoma and several other parasitic protozoa and also from 17 fungal species from GenBank (see below). For genealogical analyses, statistical parsimony in TCS (v. 1.21) (13) and median joining in Network (v. 4.1.1.2) software were used (3). Genetic distances were calculated using the Kimura 2-parameter model in MEGA. Phylogenetic analyses of other single-copy genes, i.e., fc, pgfs, p36, and dhfr-ts, were similarly carried out by using one or more of the above-mentioned programs.
Hypothetical model and motif predictions of Leishmania NAGT as an ER transmembrane protein.
This model was constructed by using five different topology prediction programs (DAS, HMMTOP, TMHMM, TMpred, and TOPpred) in ExPASy proteomics tools (http://ca.expasy.org/tools/). Topology of the Leishmania NAGT is defined by the ∼10 transmembrane hydrophobic domains and manually fine-tuned according to the hamster NAGT model (66). The protein families (Pfam) database (http://www.sanger.ac.uk/Software/Pfam/) was searched for consensus motifs of Leishmania NAGT, yielding 24 representative proteins of the eukaryotic glycosyltransferase 4 family. Conserved and nonconserved amino acid substitutions were determined using Blosum-80 or Blosum-62 for Leishmania NAGT proteins alone as a group or together with other NAGT proteins mentioned (see Fig. 5).
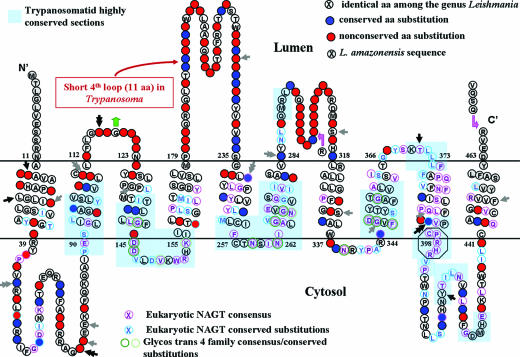
FIG. 5.
Phylogenetically useful residues in the hydrophilic loops predicted from a hypothetical model of Leishmania NAGT as a trans-ER membrane molecule. A Protein Family Database (Pfam) search places Leishmania NAGT in the glycosyltransferase 4 family. Circled letter, identical residues in NAGTs of all 26 Leishmania variants; solid blue/red circles, conserved/nonconserved aa substitutions among 26 Leishmania variants, respectively; black and gray arrows, heterozygous sites of the two L. tropica variants (I/II and IV/VI), resulting in three and four aa substitutions, respectively (double arrow); dark and light green circled residues, glycosyltransferase 4 family identical and conserved aa residues, respectively; gray circled letters, L. amazonensis NAGT sequences corresponding to the PCR primer sequences; octagonal sequence, NAGT consensus catalytic sites; letters circled pink/blue, eukaryotic NAGT consensus residues and conserved substitutions, respectively. Note that Trypanosoma as well as other eukaryotic NAGT-conserved residues are recognized in the cytosolic loops (third, fifth, and ninth) and the ninth transmembrane segment of the Leishmania sequence (blue-shaded and pink and blue circles). The motif of CPRHR (aa 396-400 in octagon) was experimentally proved to be crucial for enzymatic activity of the hamster NAGTs and conserved in all eukaryotic NAGTs examined. Absent from Leishmania NAGT is the potential dolichol recognition sequence [F-(I/V)-X-(F/Y)-X-X-I-P-F-X-(F/Y)] predicted for yeast, hamster, (66) and mammalian NAGTs as two separate sites in the second and seventh transmembrane regions (the second site only in mammalian enzymes). Leishmania NAGT dolichol phosphate binding sites appear to differ from the others.
GenBank accession numbers.
The 26 Leishmania nagt sequences are M96635 (36), AF205930-34 (1), AF291678, DQ836147-64 and LbrM35.3940 (L. braziliensis; GeneDB, http://www.genedb.org). Sequences of other Leishmania single-copy genes made available in this study for analyses include those for (i) 24 fc genes, DQ834284-301, DQ974212-3, EF088400-2, and LbrM17.1230; (ii) 13 pgfs genes, DQ834276-83, DQ981498-9, LmjF31.2150, LinJ31.2550, and LbrM31.2270; (iii) 16 p36 genes, L11705 (37), DQ834268-75, DQ974215-18, LmjF36.4170, LinJ36.6970, and LbrM35.3930; and (iv) 16 dhfr-ts genes, AF289072-3, AY122331, AY123971, DQ834262-6, DQ974214, M12734, X51733, X51735, LmjF06.0860, LinJ06.0890, and LbrM06.0760. The nagt sequences from four Trypanosoma and five other protozoa included for phylogenetic analyses are from the genome project (http://www.genedb.org) as follows: Trypanosoma cruzi (Tc00.1047053510283.140), T. brucei (Tb11.01.2220), T. gambiense (Tgamb.39611), T. vivax (tviv1098d08.q1k_5), Plasmodium berghei (PB000880.00.0), P. falciparum (PFC0935c), P. knowlesi (PK3_1860c), Entamoeba histolytica (142.m00140), and Theileria annulata (TA18965). The 17 fungal nagt sequences include five members of Euascomycota (AAL78196, EAA58397, EAA77141, EAL93419, CAF06073), seven members of Hemiascomycota (AAS53335, CAA68324, CAG58156, CAG83233, CAG86196, CAG98095, EAK97019), one member of Archeascomycota (AAA92799), three members of Basidiomycota (AAW46080, CAA67366, EAL17964), and one member of Ustilaginomycota (EAK84857).
RESULTS
PCR amplification of nagt genes from 193 independent isolates and other single-copy genes from selected samples. (i) nagt.
Using the L1/L4 primer set (Fig. 2A) for PCR, a single product of the expected size (∼1.4 kb) was amplified from all samples of the Leishmania subgenus examined (Fig. 2B), except for L. amazonensis variant 6 and variant 17 (Table 1, genotypes 2 and 3). Interestingly, the inability of the L1/L4 primer to PCR-amplify the nagt sequence from these two samples is not due to sequence heterogeneity in their 5′ and 3′ regions, corresponding to the primers used. Rather, it is apparently due to a diversion of the primers to spurious annealing sites in the non-nagt region. For these two samples, nagt was obtained with the alternative primers NGKF and NGKR, designed from the 5′ and 3′ flanking regions of the L. amazonensis nagt gene (Table 1, genotype 1, and Fig. 2A) (36). We focused on an ∼1.8-kb band among the multiple PCR products amplified with this set of primers (data not shown) because this size was expected for the specific product (Lnagt+; see Table S2 in the supplemental material) (36). Using this template for nested PCR amplification with the L1/L4 primers, the 1.4-kb nagt DNA fragment was obtained from both samples. The authenticity of the obtained products was verified by direct sequencing of the nested PCR product from L. amazonensis variant 17 and by sequencing of the ∼1.8-kb PCR product from L. amazonensis variant 6 after cloning into the pGEM-T vector. The latter thus provided a complete ORF of nagt plus its flanking regions. Notably, the 5′ and 3′ ends of this variant 6 nagt are almost identical in sequence to L1 and L4, except for a single base substitution at nt 1399 of the ORF (within the L4). In addition, the 5′ (250-bp) and 3′ (143-bp) flanking regions of the L. amazonensis and L. amazonensis variant 6 (Table 1, genotypes 1 and 2) were highly similar, except for two nucleotide deletions in the 5′ flank and a single base insertion in the 3′ flank.
PCR amplification of nagt genes from the members of the Viannia group was successful only with the L1b/L4 primer set but not with the L1/L4 primer set (Fig. 2A and see Vnagt and Lnagt in Table S2 in the supplemental material;). This is expected, since L1 and L1b differ in 7 out of 20 nucleotide positions. These primers serve for subgenus-specific PCR amplification of nagt genes from the L. (Leishmania) subgenus and the L. (Viannia) subgenus groups, respectively. The L1/L4 primer set successfully PCR-amplified nagt from L. enriettii, classified with neither of these two subgenera. By using either of the two primer sets, we were unsuccessful in attempts to PCR-amplify the nagt sequence from lizard Leishmania (L. tarentolae) (data not shown). The L1/L4 primer set was originally designed from the nagt sequence of L. amazonensis of Brazilian origin (36). Its ability to PCR-amplify nagt genes from both New World and Old World members of L. (Leishmania) but not those of L. (Viannia) is consistent with the taxonomic grouping of L. amazonensis-L. mexicana into the former subgenus, even though it coexists with Viannia in Neotropics.
(ii) Other single-copy genes.
A total of 27 ferrochelatase (encoded by the fc gene), 8 dihydrofolate reductase-thymidylate synthase (encoded by the dhfr-ts gene), 18 zeta-crystalline/NADPH oxidoreductase (encoded by the p36 gene), and 10 prostaglandin F2 alpha-synthase (encoded by the pgfs gene) genes were PCR-amplified from representatives of the nagt variants. These single-copy genes could not be PCR-amplified as readily as nagt. Different primers and PCR conditions required to complete the list were not pursued further.
The genes under study are single copied per haploid genome.
This was done to provide crucial evidence for the lack of paralogy among the investigated genes. The status of nagt as a single-copy gene has been previously established for L. amazonensis (LV78; genotype 1 in Table 1) (32, 36) and L. major (Friedlin), used for the genome project. Genomic DNA from selected samples was digested with the restriction enzyme SacII, which cuts once at nt position 1219 in all nagt sequences of the Leishmania subgenus examined (Fig. 3A). Hybridization of SacII digests of the genomic DNAs with the labeled ∼1.4-kb L. amazonensis nagt gene probe revealed no more than two bands with expected relative intensity (Fig. 3B), indicative of nagt as a single-copy gene per haploid genome in all representative samples examined. Analyses of additional restriction enzyme digests supported this conclusion (data not shown). Those from L. tropica (I/II) (Fig. 3B, lanes 8 and 9), L. donovani-L. infantum (Fig. 3B, lanes 1 to 3 and 10) and L. major (Fig. 3B, lanes 4 to 6) produced one heavy band (∼4 kb, ∼7 kb, and 1.5 kb, respectively) and one light band (∼3 kb, ∼1.3 kb, ∼6 kb, respectively; visible after prolonged exposure), as expected from the location of the SacII site 182 bp upstream of the stop codon of the nagt ORF (Fig. 3A). The fragments observed for L. major and L. infantum are consistent in size with those expected from their genomic database sequences (LmjF 36.4180 and LinJ 36_20005090_v20, respectively). Two fragments (∼3 kb and ∼0.9 kb) were observed for the genomic DNA of L. amazonensis (Table 1, genotype 1 and Fig. 3B, lane 7) and (∼1.2 kb and ∼0.2 kb, respectively) for the PCR-amplified nagt DNA fragment from this species (Fig. 3B, lane 13). These results are also consistent with sequence data published previously (36). L. amazonensis variants 6 and 17 are different from L. amazonensis, giving a single band of ∼8 kb (Fig. 3B, lane 11 to 12 versus lane 7). SacII digestion of these two samples was complete, and no additional signals emerged after overexposure of the blot. There is no evidence for the presence of an additional copy of nagt in these or in any other Leishmania samples examined. The single-copy status was similarly verified for the fc and pgfs genes (data not shown). The well-characterized genes dhfr-ts and p36 are known to be single-copy genes (30, 37).
RFLP and sequence analyses of PCR-amplified nagt recognize 26 genotypes.
Construction of restriction maps began with six original nagt sequences published earlier (1, 36). The L. braziliensis sequence from GenBank was subsequently included. The analysis was continued for 193 PCR-amplified 1.4-kb nagt DNA samples. The RFLP patterns obtained allowed grouping of the isolates as follows: 91 isolates of L. donovani-L. infantum species complex, 48 of L. tropica, 23 of L. major, 21 of L. turanica-L. gerbilli, 5 of L. amazonensis-L. mexicana, 2 of L. braziliensis, 2 of L. panamensis, and 1 of L. enriettii (Table 1 and see Table S1 in the supplemental material). Twenty six nagt variants were established by restriction mapping as distinct genotypes (Fig. 4A and B). Their differences were verified by sequencing. Also sequenced were many additional samples from isolates that appeared identical by RFLP but differed in their origins (geographic areas, animal sources, and years in which they were isolated) and/or their cutaneous leishmaniasis (CL) and visceral leishmaniasis (VL) disease phenotypes. Based on these analyses, a sequence database of nagt genotypes from 77 isolates was built (65 of the 77 isolates are listed in Table 1).
Sixteen of the 26 nagt genotypes (Table 1, genotypes 4 to 6, 8 to 12, 14 to 18, 20, 22, and 25) were obtained from multiple independent isolates (the actual number of isolates is indicated in parentheses after each genotype listed in Table 1). All isolates within each of these 16 showed identical nagt RFLP patterns, and their identities were further verified by sequencing analysis of 2 to 9 isolates from each genotype. Each of the remaining 10 variants was represented by a single isolate. These were validated rigorously by PCR and sequencing of different batches of the same isolates from the original sites or donors (especially for genotypes 2 and 3, as shown in Table 1). Genotypes represented by single isolates were further validated by the following criteria: (i) agreement with donors' original assignments based on different typing methods (Table 1, genotypes 1, 7, 19, 21, 24, and 26) and (ii) sequence divergence and phylogenetic analyses of additional single-copy genes (fc, dhfr, p36, and pgfs) (Table 1, genotypes 1, 13, 19, 21, and 26; and cf. Fig. 6C and Fig. S1 in the supplemental material).
FIG. 6.
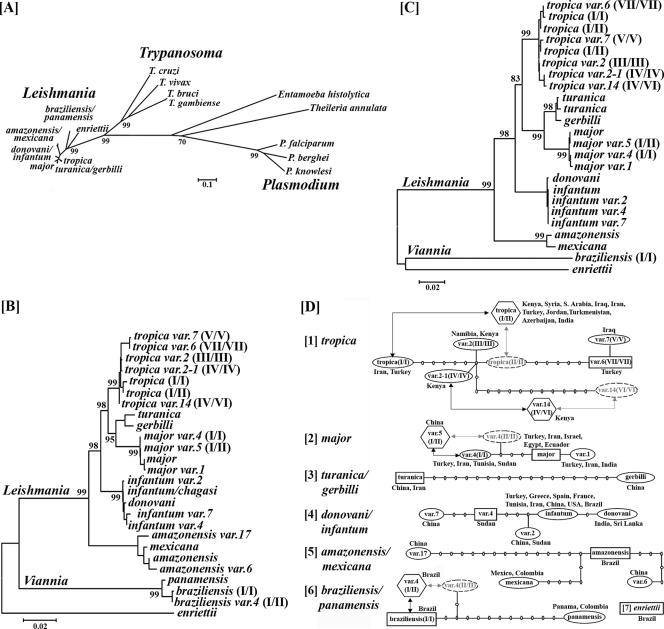
Phylogenetic analyses. The trees shown were constructed from 26 Leishmania nagt gene sequences (each from nt 37 to 1,360 of a 1,401-bp ORF) together with (A) and without (B) the nagt genes from the following parasitic protozoa: (i) four Trypanosoma spp. (T. cruzi, 1,236 bp; T. brucei, 1,182 bp; T. gambiense, 1,182 bp; T. vivax, 1,185 bp); (ii) three Plasmodium spp. (P. berghei, 1,203 bp; P. falciparum, 1,194; P. knowlesi, 1,212 bp); and one each from Entamoeba histolytica (1,098 bp) and Theileria annulata (1,179 bp). (C) Phylogenetic trees constructed from 24 Leishmania fc gene sequences. All programs mentioned in Materials and Methods produced trees of essentially the same topology. Trees presented are from analyses by the minimum evolution method with Kimura's 2 parameter by pair-wise comparison with bootstrapping of 1,000 replicas (MEGA). Note the clustering of all Leishmania spp. separated from Trypanosoma spp. (panel A) and the clustering of Leishmania spp. into five clades/subclades (≥95% bootstrap values) separated from Viannia subgenus (L. braziliensis and L. panamensis) and the unclassified L. enriettii. The topology of the tree remains essentially the same when constructed from amino acid sequences of the nagt gene (cf. Fig. S2, maximum parsimony bootstrap tree, in the supplemental material). Similar results were also obtained by such analyses of four additional single-copy, conserved gene sequences (C) for the fc gene (also see Fig. S1, genes dhfr-ts, p36, and pgfs, in the supplemental material). (D) Genealogical analyses: 26 Leishmania nagt allelic haplotype sequences grouped by using statistical parsimony with TCS (version 1.21) at a 93% confidence level, indicative of relatedness among subspecies variants and predictive of a hypothetical ancestor for each network (square box). Hexagonal box, allelically heterogeneous variants observed. The numbers of lines between nodes denote evolutionary events of substitutions between named genotypes. Faded letters in dotted oval represent hypothetical parental homozygotes.
Sequence divergence of the Leishmania nagt and other single-copy genes.
Alignment of the 26 nagt gene sequences revealed no gaps, except for a codon deletion of G-118 in L. turanica and a codon insertion of R after S-302 in L. enriettii (Fig. 5, green and pink arrows). In addition, a point mutation was observed in the C-terminal region of the L. amazonensis variant 6 gene corresponding to the stop codon in other isolates, resulting in a C-terminal tetrapeptide (QSQV) extension.
Pair-wise comparisons of the 26 nagt nt and aa sequences reveal their divergences according to species complexes based on the percentage of nt identity as well as the values of their genetic distances (see Table S4 in the supplemental material). Similar results were obtained from such analyses of four additional single-copy genes available, i.e., 24 fc, 16 dhfr-ts, 13 pgfs, and 16 p36 (data not shown).
Subspecies divergences of nagt genes vary significantly with different species complexes (Table 2). The nucleotide polymorphism seen among the subspecies variants within each complex is as follows: L. amazonensis-L. mexicana and L. tropica (29 sites) > L. braziliensis-L. panamensis and L. turanica-L. gerbilli (15 sites) > L. donovani-L. infantum and L. major (6 to 8 sites) (Table 2). A similar intraspecies divergence within each complex is noted also from the values of their genetic distances, as follows: L. tropica > L. amazonensis-L. mexicana > L. turanica-L. gerbilli > L. braziliensis-L. panamensis > L. donovani-L. infantum > L. major (Table 2). Intraspecies divergence of the fc gene sequences measured by distance analyses also showed the highest and the lowest values for L. tropica and L. donovani-L. infantum, respectively (data not shown). The intraspecies nucleotide polymorphisms resulted largely in silent mutations, especially in the case of L. major (Table 2).
TABLE 2.
Leishmania nagt intraspecies diversity
| Species complex | No. of variants/total | No. nt polymorphic sites | No. amino acid substitutionsa
|
Intraspecies distance (nt 37 to 1360)c | ||
|---|---|---|---|---|---|---|
| Silent | Conserved | Nonconserved | ||||
| L. major | 4/23 | 6 | 6 | 0 | 0 | 0.001-0.004 |
| L. turanica-L. gerbillib | 2/21 | 15 | 7 | 3 | 5 | 0.011 |
| L. tropica | 7/48 | 29 | 18 | 8 | 3 | 0.001-0.016 |
| L. donovani-L. infantum | 5/91 | 8 | 3 | 1 | 3 | 0.002-0.005 |
| L. amazonensis-L. mexicana | 4/5 | 29 | 16 | 5 | 8 | 0.005-0.016 |
| L. braziliensis-L. panamensis | 3/4 | 15 | 12 | 2 | 1 | 0.003-0.009 |
Scored by using Blosum-80 for conserved or nonconserved amino acid substitutions.
Excluding the codon deletion (nt 353 to 355) in L. turanica.
Values were determined by pair-wise comparisons among different genotypes within each species complex (see Materials and Methods). Compare the scale of genetic distances with that in Fig. 6B.
Allelic sequence heterogeneity (heterozygosity) of nagt is prominent and apparently genome wide in the L. tropica complex.
Direct PCR cycle sequencing of nagt from 20 isolates classified as L. tropica revealed seven diplotype variants (Table 1, genotypes 16 to 22). In two cases, i.e., L. tropica (I/II) and variant 14 (IV/VI), the DNA sequence traces contained 7 and 14 overlapping nucleotide positions, respectively (see Table S5 in the supplemental material, allele combinations are shown as I/II and IV/VI). These double peaks were subsequently found to persist in samples from cloned cell populations of L. tropica (I/II). The seven heterozygous sites are thus present in the same individual cells but not in different cells of a mixed population. Since nagt is present as a single-copy gene per haploid genome and Leishmania are diploid, the double peaks represent allelic differences (heterozygosity). No overlapping peaks appeared in the remaining five diplotypes, hence indicative of their nagt homozygosity, i.e., L. tropica I/I, III/III, IV/IV, V/V, and VII/VII (Table 1, genotypes 17 to 21). The findings of homozygous isolates, i.e., L. tropica I/I and IV/IV, allow us to infer the sequences of alleles II and VI in the heterozygous allelic “recombinants” of I/II and IV/VI. It is also possible to predict the potential existence of homozygous “recombinants” of II/II and VI/VI, although they have not been encountered so far. Hypothetical combinations of all seven haplotypes increase the total to 28 diplotypes in this species complex. The 7 and 14 heterozygous sites in L. tropica (I/II) and variant 14 (IV/VI), respectively (see Table S5 in the supplemental material), result in three and four aa substitutions, respectively, between the alleles in these genotypes (Fig. 5, double black and gray arrows). Allelic nucleotide differences were also observed in two to four sites of nagt sequences in L. major variant 5 (I/II) and L. braziliensis variant 4 (I/II) (data not shown and cf. Fig. 6D). Heterozygosity was also noted in sequences of dhfr-ts and fc from L. tropica (I/II) and variant 14 (IV/VI) (not shown), indicating that it is not limited to nagt, at least in this species complex.
Possible chimeric nagt in L. amazonensis variant 17.
In this isolate, the bulk of the 5′ region of the nagt gene (1,132 bp from nt position 37 to nt position 1178, excluding the PCR primer-corresponding region) is most similar to other nagt genes in the L. amazonensis-L. mexicana complex, with only 5 to 10 nucleotide substitutions among them. In contrast, the last 182 bp extending from nt position 1179 to nt position 1360 are completely identical to the corresponding 3′-end region of the nagt gene in three variants of the L. donovani-L. infantum complex and have only two to three substitutions in the remaining two variants of this species complex. As expected, this variant 17 nagt is segregated into the clades of L. amazonensis-L. mexicana (not shown, but see Fig. 6B) and L. donovani-L. infantum (see Fig. S3 in the supplemental material), respectively, when subjected to phylogenetic analyses of the respective 5′ and 3′ regions in question, together with the corresponding regions of the other 25 nagt genotypes. Sample mix ups and technical errors were ruled out as factors accounting for the emergence of such an apparently chimeric sequence. The chimeric nagt cannot possibly be created by PCR or by cycle-sequencing errors with the L1/L4 primers, judging from their positions (Fig. 2A and see Table S2 and S3 in the supplemental material) relative to the “crossover site” of the chimeric sequence. In addition, had the variant 17 DNA been mixed with other samples, PCR with the L1/L4 primer set would have produced the 1.4-kb nagt. Most importantly, the results were verified by repeated PCR for DNA analyses from different batches of the same isolate reacquired from the original donor.
Phylogenetic analyses of the 26 nagt sequences allow grouping of the variants according to their hierarchical taxonomic levels and evaluating subspecies divergence.
Phylogenetic analyses of the Leishmania and Trypanosoma nagt genes show that the 26 Leishmania sequences are clustered together as a group well separated from Trypanosoma (Fig. 6A and see Fig. S2 in the supplemental material), as reported previously, by using different markers (25, 28, 56). The topology of the trypanosomatid trees remains essentially unchanged when subjected to further analyses together with the nagt genes from other parasitic protozoa, such as Plasmodium spp. (Fig. 6A and see Fig. S2 in the supplemental material). The nagt sequences are thus sufficiently divergent to distinguish the two genera of trypanosomatids. The 26 Leishmania nagt sequences formed three distinct groups according to their subgeneric status, i.e., L. (Leishmania), L. (Viannia), and the branch of L. enriettii which does not belong to either subgenus (Fig. 6B). Those in the subgenus L. (Leishmania) are grouped into five known species complexes, i.e., L. (L.) amazonensis-L. mexicana, L. (L.) donovani-L. infantum, L. (L.) tropica, L. (L.) major, and L. (L.) turanica-L. gerbilli (Fig. 6B). Analyses of additional single-copy genes available, i.e., dhfr-ts, fc, p36, and pgfs produced trees with similar topology at the species level (see Fig. 6C for fc and see Fig. S1 in the supplemental material for the rest). The separation of the Leishmania and Viannia groups, as well as the species complexes therein, agrees completely with the data from similar analyses of other protein-coding genes (14, 27, 39, 44) and internal-transcribed-spacer (16) sequences.
Incongruence between the Leishmania nagt gene-based grouping and the CL/VL disease phenotype of the isolates is noted, i.e., the coexistence of subspecies variants with different disease phenotypes in the same species complex and, more strikingly, the same genotype from different disease phenotypes (Table 1). In the L. donovani-L. infantum complex, for example, we observed a nagt genotype of L. infantum with both CL and VL phenotypes, isolated from southern Turkey (Table 1, genotype 5, strains OG-VL and Adana #7) (54). Another one is L. infantum variant 7, which shows CL exclusively in western China (38) but VL in eastern China (Table 1, genotype 8, strains KXG-LIU and Shandong).
Phylogenetically useful sites of Leishmania nagt sequences.
These are unevenly distributed, as noted from amino acid substitutions in the NAGT polypeptide chain presented as a putative ER transmembrane model (Fig. 5). Of the ∼130 aa substitutions present among the 26 sequences (Fig. 5, solid circle), a majority, that is, two-thirds (∼93), appears to occur in the hydrophilic loops, especially the longest fourth and the adjacent one. The fourth loop is unique to Leishmania, since it is truncated in the NAGT of Trypanosoma (Fig. 5) and other eukaryotes (not shown). The remaining one-third of the substitutions (∼37) are dispersed throughout the hydrophobic domains. In addition, nonconserved substitutions are almost three times more frequent in the hydrophilic loops (∼66) than in the transmembrane segments (∼24) (Fig. 5, solid red circle).
Leishmania intraspecies divergence.
The 26 nagt haplotype sequences were further subjected to genealogy analyses by statistical parsimony (13) (Fig. 6D) and median-joining analyses (3) (see Fig. S4 in the supplemental material). Both algorithms sorted the 26 sequences into independent networks of their respective species complexes (Fig. 6D and see Fig. S4 in the supplemental material). This result provides further confirmation of the phylogenetic grouping of the 26 variants presented in Fig. 6A through C. There are four heterozygous genotypes or “recombinants” (Fig. 6D, subspecies [1], [2], and [6], and see Fig. S4, [1] and [2], hexagon, in the supplemental material), i.e., L. tropica (I/II), L. tropica variant 14 (IV/VI), L. major variant 5 (I/II), and L. braziliensis variant 4 (I/II). Homozygous genotypes with some of the above-mentioned allele-specific sequences were found, including L. tropica (I/I), L. tropica variant 2-1 (IV/IV), L. major variant 4 (I/I), and L. braziliensis (I/I) (Fig. 6D and see Fig. S4 in the supplemental material), but not those with the remaining alleles, such as putative L. tropica (II/II), L. tropica variant 14 (VI/VI), L. major variant 4 (II/II), and L. braziliensis variant 4 (II/II) (Fig. 6D, dotted oval, and see Fig. S4, faded letters, in the supplemental material). The presumed “parental” variants for the “recombinants” (lined arrows) are not necessarily close to each other evolutionarily (Fig. 6D and see Fig. S4 in the supplemental material), but they colocalize geographically, as shown by the coexistence of the L. tropica variant 2-1 (IV/IV) and variant 14 (IV/VI) from Kenya and by L. tropica (I/I) and (I/II) from Asia and the Middle East (Fig. 6D) (1). Preliminary genealogical analyses of the eight fc sequences available for the most divergent L. tropica complex yielded results which supported the observation of a distant relationship between the “parental” variants, although the genealogy of the subspecies variants is not entirely identical in topology to that derived from the nagt sequences (not shown). This situation was noted previously in similar analyses of five different single-copy genes for the subspecies genealogy of L. donovani-L. infantum (41). In the present study, while the levels of intraspecies divergence are clearly reflected in the number of polymorphic sites and the values of genetic distance observed in the investigated species complexes, the conclusions about evolutionary relationships of genotypes within each complex remain tentative, pending further analyses of additional sequences for more informative sites.
There is discordance between the subspecies divergence of nagt sequences within the species complexes (Table 2 and Fig. 6D, number of nodes) and the extent of their geographic distribution. Data from analyses of 24 fc sequences (not shown) support this observation from nagt data discussed below. The L. donovani-L. infantum complex is geographically very wide spread (Fig. 1), but it is relatively homogeneous genetically, with the range of intraspecies genetic distances from 0.002 to 0.005 and only eight polymorphic sites (Table 2) found among five homozygous genotypes (see Table 1, Fig. 6D, [4], and Fig. S4, [3], in the supplemental material). In contrast, the L. tropica complex is geographically more restricted (Fig. 1), but it is the most divergent, with intraspecies genetic distances varying from 0.001 to 0.016 and 29 polymorphic sites (Table 2) among two “recombinants” (see Table 1, Fig. 6D, [1], and Fig. S4 [1], hexagon, in the supplemental material) and five homozygous genotypes. The remaining species complexes present a picture intermediate between these two species complexes. The L. major complex is nearly as widely spread (Fig. 1) as L. donovani-L. infantum but is less divergent (Fig. 6D, [2], and Fig. S4 [2], with its four variants including one “recombinant” [hexagon], in the supplemental material). Closely related to L. major is the L. turanica-L. gerbilli complex, which contains nonpathogenic parasites of the great gerbil (Rhombomys opimus) (Fig. 6B). This complex is rather restricted in distribution (Fig. 1) and consists of two quite divergent members (Fig. 6D, [3]). Restricted to Neotropics is the L. braziliensis-L. panamensis complex (Fig. 1), which includes both types of genotypes (Fig. 6D, [6]). Genetic diversity of the Viannia group is underscored by a high level of sequence heterogeneity obvious by examining only a few isolates. Most interesting is the finding that the L. amazonensis-L. mexicana complex has two genotypes from Asia (Fig. 1 and Table 1, genotypes 2 and 3), including what appears to be a chimeric hybrid with L. donovani-L. infantum, in addition to the two genotypes from Neotropics (Table 1, genotypes 1 and 4) (Fig. 6D, [5], and Fig. S4, [4], in the supplemental material). It should be emphatically stressed that exceptional caution was exercised to exclude any technical or human errors, such as sample mix up, to ascertain that the Asian origin of the two L. amazonensis variants was real. In fact, this finding should not be that surprising, except for the nomenclature of this species complex, considering the phylogenetic grouping of L. amazonensis-L. mexicana with other species complexes of the Old World (Leishmania).
DISCUSSION
Leishmania nagt sequence database for phylogenetic analyses.
We have identified 26 nagt genotypes (19 from this work) by sequencing 77 PCR-amplified nagt genes selected from a total of 238 independent isolates (193 from this study). These genotypes represent true nagt gene divergence of the samples examined, since data collection and analyses are greatly facilitated by several favorable factors, i.e., the ease of nagt PCR amplification (Fig. 2), the absence of paralogous genes (Fig. 3), the precision of sequence-based RFLP analyses of the PCR products (Fig. 4), and the unambiguous alignment of nagt sequences (Fig. 5). Although the sample size examined here is not small, sample collection is inevitably biased by its accessibility or availability, which favors Asian isolates from the Old World, in this case. Thus, analyses of additional samples will undoubtedly uncover new genotypes, especially among those that are under-represented from the Viannia group in the present study. The database presented for the Old World Leishmania subgenus is sufficiently robust, especially in conjunction with those from additional single-copy genes.
Genes encoding transmembrane molecules, such as nagt, are useful, albeit previously untapped phylogenetic markers.
Consistent with previous findings based on other protein-coding (39) and nonprotein-coding sequences (16, 47, 64), phylogenetic analyses of the 26 nagt sequences used segregate the isolates into expected genera and species complexes (Fig. 6A and B and see Fig. S2 in the supplemental material). The phylogenetic trees obtained are identical in topology to those obtained from additional single-copy genes from selected samples in the present study, i.e., dhfr-ts, fc, p36, and pgfs (Fig. 6C and Fig. S1 in the supplemental material). The location of these additional genes on different chromosomes (see Table S2 in the supplemental material) strongly implies that the phylogenetic relationship of the genotypes deduced is genome wide but not limited to the nagt site.
The phylogenetically useful sites of Leishmania nagt are localized mostly in a ∼500-bp section of this ∼1.4-kb gene, corresponding to the putative fourth and sixth hydrophilic loops of the trans-ER membrane enzyme (Fig. 5). Divergent sites of the nagt sequences in other single-cell eukaryotes, such as fungi, are also informative for phylogenetic analyses (see Fig. S5 in the supplemental material). In other genes encoding cytosolic enzymes, e.g., dhfr-ts, p36, fc, and pgfs, the phylogenetically useful sites are dispersed randomly throughout the ORF. The advantage of the Leishmania nagt gene is, thus, its utility for PCR amplification of only a small section of this sequence (∼500 bp) to yield the maximal number of informative sites. This is of practical importance in avoiding in vitro isolation and cultivation of parasites, which is cumbersome, not always successful, and potentially selective for cultivable stocks. It is possible to envision the use of a battery of genes encoding similar transmembrane molecules in this way for phylogenetic analyses of single-cell eukaryotes toward the goal of genome taxonomy.
Leishmania subspecies divergence and evolutionary adaptation.
Within the Leishmania subgenus, there is a lack of correlation between intraspecies divergence and geographic distribution of the species complexes. Namely, the less divergent L. donovani-L. infantum and L. major complexes are widely distributed, whereas the more divergent L. tropica and L. turanica-L. gerbilli complexes are geographically restricted (see Fig. 1, Fig. 6D, and Fig. S4 in the supplemental material; also see Table 2). The latter scenario appears true for the Neotropic-restricted L. (Viannia) subgenus on account of the great intraspecies divergence seen among the few samples examined in this study. The observation may have relevance to the evolutionary adaptability of Leishmania to a new environment subsequent to the organism's dispersal but not to the dispersal mechanisms per se. Since Leishmania are vector-borne obligate endoparasites, their clonal evolution may be driven by two sets of direct pressures: (i) primarily, the internal microenvironments of their long-term residence (months to years) intraphagolysosomally in the macrophages of the mammalian hosts/reservoirs; and (ii) secondarily, their short-term residence (up to several weeks) in the fly gut. Vector- or reservoir-parasite specificity has long been considered to be the apparent factor limiting the distributions of leishmaniasis. The external macroenvironments are relevant to the evolution of reservoirs and vectors but exert no direct pressures on Leishmania, especially when they are in homeothermal mammalian hosts. While the precise nature of the host-dependent selective pressures remains to be defined, reservoirs are most pertinent to Leishmania evolution, since leishmaniasis is fundamentally a zoonotic disease. Reported cases of anthroponosis are based on the negative finding of animal reservoirs and are limited to few specific geographic sites, e.g., kala-azar, caused by Indian L. donovani, and CL by L. tropica in Asia but not in Africa. Genetic changes of isolates due to geographic isolation or genetic drift are demonstrable more readily in the fast-evolving sequences, e.g., microsatellite DNA markers (45, 52).
Contributions of genetic recombination to Leishmania evolution?
There is a notable disparity of different species complexes in the frequency of the heterozygosity encountered. Heterozygosity is more prominent among the extant genotypes of L. tropica, as observed here and previously (51, 53), than among those of L. donovani-L. infantum (41). This may result from different timelines for the event of “recombination” to occur in different species. Genetic recombination is thought to occur at a low rate in Leishmania (48), as found experimentally in Trypanosoma spp. (19, 20). Leishmania sequence analyses show hybrid and nonhybrid genotypes reminiscent of those seen in T. cruzi (40, 63), suggestive of “genetic exchange.” Its contribution to Leishmania evolution may be suggested, as heterozygosity results in amino acid substitutions in multiple genes of functionally important enzymes, as shown here at least for L. tropica (Fig. 5).
Interspecies recombination is hinted at by the observation of a chimeric nagt sequence in a single isolate as a hybrid of L. amazonensis-L. mexicana and L. donovani-L. infantum, i.e., L. amazonensis variant 17 (Fig. 6B and see Fig. S3 in the supplemental material). Heterologous species hybrids have also been reported to occur between L. major and L. arabica (33) and between L. braziliensis and other Viannia species (4, 6, 8, 61). Such hybrids could result in a chimeric sequence by anomalous chromosomal breakage/joining (8). Meaningful genetic implications of such interspecies hybridization and chimeras await further investigation.
Incongruity of Leishmania speciation and disease phenotypes.
This observation has long been reported for the Leishmania subgenus (1, 2, 5, 22, 31, 34, 46). Although host genetics may be involved (9, 43), the inherent differences of the causative agents remain to be significant contributing factors. Differential acquisition of “pathogenic islands” laterally by the same genotype is a possibility, but there is no evidence for this. Although no genetic marker is available to discriminate CL- from VL-specific isolates (60), A2 (65) and K39 (21) repeats have been related to “visceralization” of Leishmania relevant to immunopathology (11). Of interest is the elucidation of the mechanism by which the expression of these and other phenotype-specific genes is differentially regulated in VL and CL isolates of an identical genotype apparently independent of Leishmania speciation.
We demonstrate here for the first time the potential utility of single-copy genes encoding transmembrane versus soluble enzymes for phylogenetic analyses of eukaryotic protists. Such analyses of trans-ER nagt genes from >200 Leishmania samples readily group them into taxa of an expected hierarchy, revealing considerable variations in subspecies divergence and evidence of heterozygosity in L. tropica, resulting in changes at the protein level. Further analyses of this and additional sequence databases will bear on Leishmania evolution and mechanisms of their recombination crucial for elucidating the clinical epidemiology of leishmaniasis and the control measures against these wide-spread diseases.
ADDENDUM
Two articles, which came to our attention after the acceptance of this report, contain information of relevance to the present discussion. One article describes the L. major-L. infantum hybrid strains isolated originally from human immunodeficiency virus (HIV)-positive patients in Portugal (49a). This finding is supportive of our proposed existence of putative L. infantum-L. amazonensis hybrids, although our cases involve different parental species and have no record of coinfection with HIV. The very low subspecies divergence among different genotypes in the L. donovani-L. infantum complex that we noted is consistent with data presented in another article based on the examination of a different set of protein-coding gene sequences (64a). In that article, subspecies variants were found geographically segregated into Indian, African, and Mediterranean groups within this species complex (64a). This conclusion is not inconsistent with our data presented in Fig. 6D, [4], by excluding some isolates from China, Brazil, and the United States.
Supplementary Material
Acknowledgments
This work is partially supported by NIH AI-20486 to K.P.C.
We thank Leyla (Akman) Anderson for initiating the hypothetical model presented in Fig. 5 and colleagues who generously provided Leishmania isolates or their DNA samples (see a complete list at http://66.99.255.20/cms/micro/sample%20list.pdf).
Footnotes
Published ahead of print on 1 December 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akman, L., H. S. Aksu, R. Q. Wang, S. Ozensoy, Y. Ozbel, Z. Alkan, M. A. Ozcel, G. Culha, K. Ozcan, S. Uzun, H. R. Memisoglu, and K. P. Chang. 2000. Multi-site DNA polymorphism analyses of Leishmania isolates define their genotypes predicting clinical epidemiology of leishmaniasis in a specific region. J. Eukaryot. Microbiol. 47:545-554. [DOI] [PubMed] [Google Scholar]
- 2.Alborzi, A., M. Rasouli, and A. Shamsizadeh. 2006. Leishmania tropica-isolated patient with visceral leishmaniasis in southern Iran. Am. J. Trop. Med. Hyg. 74:306-307. [PubMed] [Google Scholar]
- 3.Bandelt, H. J., P. Forster, and A. Rohl. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37-48. [DOI] [PubMed] [Google Scholar]
- 4.Banuls, A. L., F. Guerrini, F. Le Pont, C. Barrera, I. Espinel, R. Guderian, R. Echeverria, and M. Tibayrenc. 1997. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J. Eukaryot. Microbiol. 44:408-411. [DOI] [PubMed] [Google Scholar]
- 5.Barral, A., R. Badaro, M. Barral-Netto, G. Grimaldi, Jr., H. Momem, and E. M. Carvalho. 1986. Isolation of Leishmania mexicana amazonensis from the bone marrow in a case of American visceral leishmaniasis. Am. J. Trop. Med. Hyg. 35:732-734. [DOI] [PubMed] [Google Scholar]
- 6.Belli, A. A., M. A. Miles, and J. M. Kelly. 1994. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology 109:435-442. [DOI] [PubMed] [Google Scholar]
- 7.Beverley, S. M., R. B. Ismach, and D. M. Pratt. 1987. Evolution of the genus Leishmania as revealed by comparisons of nuclear DNA restriction fragment patterns. Proc. Natl. Acad. Sci. USA 84:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britto, C., C. Ravel, P. Bastien, C. Blaineau, M. Pages, J. P. Dedet, and P. Wincker. 1998. Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New World Leishmania genomes. Gene 222:107-117. [DOI] [PubMed] [Google Scholar]
- 9.Bucheton, B., L. Abel, M. M. Kheir, A. Mirgani, S. H. El-Safi, C. Chevillard, and A. Dessein. 2003. Genetic control of visceral leishmaniasis in a Sudanese population: candidate gene testing indicates a linkage to the NRAMP1 region. Genes. Immun. 4:104-109. [DOI] [PubMed] [Google Scholar]
- 10.Chance, M. L., W. Peters, and L. Shnur. 1974. Biochemical taxonomy of Leishmania. I. Observations on DNA. Ann. Trop. Med. Parasitol. 68:307-316. [PubMed] [Google Scholar]
- 11.Chang, K. P., S. G. Reed, B. S. McGwire, and L. Soong. 2003. Leishmania model for microbial virulence: the relevance of parasite multiplication and pathoantigenicity. Acta Trop. 85:375-390. [DOI] [PubMed] [Google Scholar]
- 12.Chen, D. Q., H. Lu, and K. P. Chang. 1999. Replacement of Leishmania N-acetylglucosamine-1-phosphate transferase gene requires episomal rescue. Mol. Biochem. Parasitol. 100:223-227. [DOI] [PubMed] [Google Scholar]
- 13.Clement, M., D. Posada, and K. A. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657-1659. [DOI] [PubMed] [Google Scholar]
- 14.Croan, D. G., D. A. Morrison, and J. T. Ellis. 1997. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 89:149-159. [DOI] [PubMed] [Google Scholar]
- 15.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50:296-311. [DOI] [PubMed] [Google Scholar]
- 16.Davila, A. M., and H. Momen. 2000. Internal-transcribed-spacer (ITS) sequences used to explore phylogenetic relationships within Leishmania. Ann. Trop. Med. Parasitol. 94:651-654. [DOI] [PubMed] [Google Scholar]
- 17.Du, Y., and K. P. Chang. 1994. Phylogenetic heterogeneity of three Crithidia spp. vs. Crithidia fasciculata. Mol. Biochem. Parasitol. 66:171-174. [DOI] [PubMed] [Google Scholar]
- 18.Du, Y., D. A. Maslov, and K. P. Chang. 1994. Monophyletic origin of beta-division proteobacterial endosymbionts and their coevolution with insect trypanosomatid protozoa Blastocrithidia culicis and Crithidia spp. Proc. Natl. Acad. Sci. USA 91:8437-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaunt, M. W., M. Yeo, I. A. Frame, J. R. Stothard, H. J. Carrasco, M. C. Taylor, S. S. Mena, P. Veazey, G. A. Miles, N. Acosta, A. R. de Arias, and M. A. Miles. 2003. Mechanism of genetic exchange in American trypanosomes. Nature 421:936-939. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, W., and J. Stevens. 1999. Genetic exchange in the trypanosomatidae. Adv. Parasitol. 43:1-46. [DOI] [PubMed] [Google Scholar]
- 21.Goto, Y., R. N. Coler, J. Guderian, R. Mohamath, and S. G. Reed. 2006. Cloning, characterization, and serodiagnostic evaluation of Leishmania infantum tandem repeat proteins. Infect. Immun. 74:3939-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gramiccia, M., L. Gradoni, and E. Pozio. 1987. Leishmania infantum sensu lato as an agent of cutaneous leishmaniasis in Abruzzi region (Italy). Trans. R. Soc. Trop. Med. Hyg. 81:235-237. [DOI] [PubMed] [Google Scholar]
- 23.Grimaldi, G., and D. McMahon-Pratt. 1996. Monoclonal antibodies for the identification of New World Leishmania species. Mem. Inst. Oswaldo Cruz 91:37-42. [DOI] [PubMed] [Google Scholar]
- 24.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41:95-98. [Google Scholar]
- 25.Hamilton, P. B., J. R. Stevens, M. W. Gaunt, J. Gidley, and W. C. Gibson. 2004. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int. J. Parasitol. 34:1393-1404. [DOI] [PubMed] [Google Scholar]
- 26.Hanafi, R., M. Barhoumi, S. B. Ali, and I. Guizani. 2001. Molecular analyses of Old World Leishmania RAPD markers and development of a PCR assay selective for parasites of the L. donovani species complex. Exp. Parasitol. 98:90-99. [DOI] [PubMed] [Google Scholar]
- 27.Hannaert, V., F. R. Opperdoes, and P. A. Michels. 1998. Comparison and evolutionary analysis of the glycosomal glyceraldehyde-3-phosphate dehydrogenase from different Kinetoplastida. J. Mol. Evol. 47:728-738. [DOI] [PubMed] [Google Scholar]
- 28.Hughes, A. L., and H. Piontkivska. 2003. Phylogeny of Trypanosomatidae and Bodonidae (Kinetoplastida) based on 18S rRNA: evidence for paraphyly of Trypanosoma and six other genera. Mol. Biol. Evol. 20:644-652. [DOI] [PubMed] [Google Scholar]
- 29.Jamjoom, M. B., R. W. Ashford, P. A. Bates, S. J. Kemp, and H. A. Noyes. 2002. Towards a standard battery of microsatellite markers for the analysis of the Leishmania donovani complex. Ann. Trop. Med. Parasitol. 96:265-270. [DOI] [PubMed] [Google Scholar]
- 30.Kapler, G. M., K. Zhang, and S. M. Beverley. 1990. Nuclease mapping and DNA sequence analysis of transcripts from the dihydrofolate reductase-thymidylate synthase (R) region of Leishmania major. Nucleic. Acids Res. 18:6399-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karunaweera, N. D., F. Pratlong, H. V. Siriwardane, R. L. Ihalamulla, and J. P. Dedet. 2003. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme MON-37. Trans. R. Soc. Trop. Med. Hyg. 97:380-381. [DOI] [PubMed] [Google Scholar]
- 32.Kawazu, S., H. G. Lu, and K. P. Chang. 1997. Stage-independent splicing of transcripts from two heterogeneous neighboring genes in Leishmania amazonensis. Gene 196:49-59. [DOI] [PubMed] [Google Scholar]
- 33.Kelly, J. M., J. M. Law, C. J. Chapman, G. J. Van Eys, and D. A. Evans. 1991. Evidence of genetic recombination in Leishmania. Mol. Biochem. Parasitol. 46:253-263. [DOI] [PubMed] [Google Scholar]
- 34.Kreutzer, R. D., M. Grogl, F. A. Neva, D. J. Fryauff, A. J. Magill, and M. M. Aleman-Munoz. 1993. Identification and genetic comparison of leishmanial parasites causing viscerotropic and cutaneous disease in soldiers returning from Operation Desert Storm. Am. J. Trop. Med. Hyg. 49:357-363. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 36.Liu, X., and K. P. Chang. 1992. The 63-kilobase circular amplicon of tunicamycin-resistant Leishmania amazonensis contains a functional N-acetylglucosamine-1-phosphate transferase gene that can be used as a dominant selectable marker in transfection. Mol. Cell. Biol. 12:4112-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, X., and K. P. Chang. 1994. Identification by extrachromosomal amplification and overexpression of a zeta-crystallin/NADPH-oxidoreductase homologue constitutively expressed in Leishmania spp. Mol. Biochem. Parasitol. 66:201-210. [DOI] [PubMed] [Google Scholar]
- 38.Lu, H. G., L. Zhong, L. R. Guan, J. Q. Qu, X. S. Hu, J. J. Chai, Z. B. Xu, C. T. Wang, and K. P. Chang. 1994. Separation of Chinese Leishmania isolates into five genotypes by kinetoplast and chromosomal DNA heterogeneity. Am. J. Trop. Med. Hyg. 50:763-770. [DOI] [PubMed] [Google Scholar]
- 39.Luyo-Acero, G. E., H. Uezato, M. Oshiro, K. Takei, K. Kariya, K. Katakura, E. Gomez-Landires, Y. Hashiguchi, and S. Nonaka. 2004. Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology 128:483-491. [DOI] [PubMed] [Google Scholar]
- 40.Machado, C. A., and F. J. Ayala. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98:7396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauricio, I. L., M. Yeo, M. Baghaei, D. Doto, F. Pratlong, E. Zemanova, J. P. Dedet, J. Lukes, and M. A. Miles. 2006. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int. J. Parasitol. 36:757-769. [DOI] [PubMed] [Google Scholar]
- 42.McGwire, B. S., and K. P. Chang. 1996. Posttranslational regulation of a Leishmania HEXXH metalloprotease (gp63). The effects of site-specific mutagenesis of catalytic, zinc binding, N-glycosylation, and glycosyl phosphatidylinositol addition sites on N-terminal end cleavage, intracellular stability, and extracellular exit. J. Biol. Chem. 271:7903-7909. [DOI] [PubMed] [Google Scholar]
- 43.Mohamed, H. S., M. E. Ibrahim, E. N. Miller, J. K. White, H. J. Cordell, J. M. Howson, C. S. Peacock, E. A. Khalil, A. M. El Hassan, and J. M. Blackwell. 2004. SLC11A1 (formerly NRAMP1) and susceptibility to visceral leishmaniasis in The Sudan. Eur. J. Hum. Genet. 12:66-74. [DOI] [PubMed] [Google Scholar]
- 44.Noyes, H. A., D. A. Morrison, M. L. Chance, and J. T. Ellis. 2000. Evidence for a neotropical origin of Leishmania. Mem. Inst. Oswaldo Cruz 95:575-578. [DOI] [PubMed] [Google Scholar]
- 45.Ochsenreither, S., K. Kuhls, M. Schaar, W. Presber, and G. Schönian. 2006. Multilocus microsatellite typing as a new tool for discrimination of Leishmania infantum MON-1 strains. J. Clin. Microbiol. 44:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveira Neto, M. P., G. Grimaldi, Jr., H. Momen, R. S. Pacheco, M. C. Marzochi, and D. McMahon Pratt. 1986. Active cutaneous leishmaniasis in Brazil, induced by Leishmania donovani chagasi. Mem. Inst. Oswaldo Cruz 81:303-309. [DOI] [PubMed] [Google Scholar]
- 47.Orlando, T. C., M. A. Rubio, N. R. Sturm, D. A. Campbell, and L. M. Floeter-Winter. 2002. Intergenic and external transcribed spacers of ribosomal RNA genes in lizard-infecting Leishmania: molecular structure and phylogenetic relationship to mammal-infecting Leishmania in the subgenus Leishmania (Leishmania). Mem. Inst. Oswaldo Cruz 97:695-701. [DOI] [PubMed] [Google Scholar]
- 48.Panton, L. J., R. B. Tesh, K. C. Nadeau, and S. M. Beverley. 1991. A test for genetic exchange in mixed infections of Leishmania major in the sand fly Phlebotomus papatasi. J. Protozool. 38:224-228. [DOI] [PubMed] [Google Scholar]
- 49.Pratlong, F., J.-A. Rioux, P. Marty, F. Faraut-Gambarelli, J. Dereure, G. Lanotte, and J.-P. Dedet. 2004. Isoenzymatic analysis of 712 strains of Leishmania infantum in the south of France and relationship of enzymatic polymorphism to clinical and epidemiological features. J. Clin. Microbiol. 42:4077-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Ravel, C., S. Cortes, F. Pratlong, F. Morio, J. P. Dedet, and L. Campino. 2006. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int. J. Parasitol. 36:1383-1388. [DOI] [PubMed] [Google Scholar]
- 50.Rosypal, A. C., G. C. Troy, A. M. Zajac, R. B. Duncan, Jr., K. Waki, K. P. Chang, and D. S. Lindsay. 2003. Emergence of zoonotic canine leishmaniasis in the United States: isolation and immunohistochemical detection of Leishmania infantum from foxhounds from Virginia. J. Eukaryot. Microbiol. 50:691-693. [DOI] [PubMed] [Google Scholar]
- 51.Schonian, G., M. El Fari, S. Lewin, C. Schweynoch, and W. Presber. 2001. Molecular epidemiology and population genetics in Leishmania. Med. Microbiol. Immunol. (Berlin) 190:61-63. [DOI] [PubMed] [Google Scholar]
- 52.Schwenkenbecher, J. M., C. Frohlich, F. Gehre, L. F. Schnur, and G. Schonian. 2004. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect. Genet. Evol. 4:99-105. [DOI] [PubMed] [Google Scholar]
- 53.Schwenkenbecher, J. M., T. Wirth, L. F. Schnur, C. L. Jaffe, H. Schallig, A. Al-Jawabreh, O. Hamarsheh, K. Azmi, F. Pratlong, and G. Schonian. 2006. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int. J. Parasitol. 36:237-246. [DOI] [PubMed] [Google Scholar]
- 54.Serin, M. S., K. Daglioglu, M. Bagirova, A. Allahverdiyev, S. Uzun, Z. Vural, B. Kayar, S. Tezcan, M. Yetkin, G. Aslan, G. Emekdas, and F. Koksal. 2005. Rapid diagnosis and genotyping of Leishmania isolates from cutaneous and visceral leishmaniasis by microcapillary cultivation and polymerase chain reaction-restriction fragment length polymorphism of miniexon region. Diagn. Microbiol. Infect. Dis. 53:209-214. [DOI] [PubMed] [Google Scholar]
- 55.Shaw, J. J. 2002. New world Leishmaniasis: the ecology of leishmaniasis and the diversity of leishmanial species in Central and South America. p. 11-32. In J. P. Farrell, (ed.), World class parasites, vol. 4, Leishmania. Kluwer Academic Publishers, Boston, MA. [Google Scholar]
- 56.Simpson, A. G., J. R. Stevens, and J. Lukes. 2006. The evolution and diversity of kinetoplastid flagellates. Trends. Parasitol. 22:168-174. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tibayrenc, M., and F. J. Ayala. 1999. Evolutionary genetics of Trypanosoma and Leishmania. Microbes. Infect. 1:465-472. [DOI] [PubMed] [Google Scholar]
- 59.Tibayrenc, M., F. Kjellberg, and F. J. Ayala. 1990. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. USA 87:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toledo, A., J. Martin-Sanchez, B. Pesson, C. Sanchiz-Marin, and F. Morillas-Marquez. 2002. Genetic variability within the species Leishmania infantum by RAPD. A lack of correlation with zymodeme structure. Mol. Biochem. Parasitol. 119:257-264. [DOI] [PubMed] [Google Scholar]
- 61.Torrico, M. C., S. De Doncker, J. Arevalo, D. Le Ray, and J. C. Dujardin. 1999. In vitro promastigote fitness of putative Leishmania (Viannia) braziliensis/Leishmania (Viannia) peruviana hybrids. Acta Trop. 72:99-110. [DOI] [PubMed] [Google Scholar]
- 62.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Westenberger, S. J., C. Barnabe, D. A. Campbell, and N. R. Sturm. 2005. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zelazny, A. M., D. P. Fedorko, L. Li, F. A. Neva, and S. H. Fischer. 2004. Evaluation of 7SL RNA gene sequences for the identification of Leishmania spp. Am. J. Trop. Med. Hyg. 72:415-420. [PubMed] [Google Scholar]
- 64a.Zemanova, E., M. Jirku, I. L. Mauricio, A. Horak, M. A. Miles, and J. Lukes. 18 Sept. 2006, posting date. The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, PI, G6PDH, and FH), new targets for multilocus sequence typing. Int. J. Parasitol. [Epub ahead of print.] doi: 10.1016/j.ijpara.2006.08.008. [DOI] [PubMed]
- 65.Zhang, W. W., and G. Matlashewski. 2001. Characterization of the A2-A2rel gene cluster in Leishmania donovani: involvement of A2 in visceralization during infection. Mol. Microbiol. 39:935-948. [DOI] [PubMed] [Google Scholar]
- 66.Zhu, X. Y., and M. A. Lehrman. 1990. Cloning, sequence, and expression of a cDNA encoding hamster UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase. J. Biol. Chem. 265:14250-14255. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.