Abstract
Background
Modulation of Jak‐STAT signalling may provide an effective therapeutic strategy in inflammatory arthritis.
Objective
To document Jak‐STAT expression in a cohort of patients with active rheumatoid arthritis (RA), spondyloarthritis (SpA), and osteoarthritis (OA) and compare these subsets with normal synovial tissue.
Methods
Synovial tissue biopsy specimens from patients with RA, OA, and SpA and histologically normal tissue (n = 10 in each arthritis group) were examined for the presence of Jak3, STAT1, STAT4, and STAT6 expression using immunohistochemistry. Phenotyping was performed using immunohistochemistry and immunofluorescence. Clinical and serological characteristics of patients with RA expressing Jak3‐STAT4 were assessed.
Results
STAT1, STAT4, and Jak3 protein expression was generally increased in inflammatory arthritis. In contrast, STAT6 expression was relatively heterogeneous. A subpopulation of CD1a positive dendritic cells unique to seropositive patients with RA was detected. These cells showed intense protein expression for Jak3, STAT4, and STAT6.
Conclusion
CD1a positive dendritic cells intensely express Jak3, STAT4, and STAT6 in seropositive RA tissue and may be an alternative marker for dendritic cells in their early stages of activation as well as providing a tool for identifying RA at the level of the synovium. Jak3 inhibition may be a potential therapeutic target to prevent dendritic cell maturation in RA. STAT1 expression is increased in inflammatory arthritis, suggesting that its pro‐apoptotic and anti‐inflammatory effects cannot effectively counteract inflammation. STAT6 expression is heterogeneous in synovium, suggesting a possible homoeostatic role in addition to any anti‐inflammatory effects.
Keywords: dendritic cells, rheumatoid arthritis, transcription factors
Rheumatoid arthritis (RA) is a chronic inflammatory disease, primarily of synovium.1 Cytokines, particularly macrophage derived tumour necrosis factor α (TNFα) and interleukin (IL) 1, have important roles in perpetuating inflammation. Antibodies against TNFα and the use of IL1 receptor antagonist have successfully suppressed inflammation in many patients with RA, but only 60% of patients obtain a partial response and a minority experience no benefit.2
A study of transcription factor pathways may explain the variable responses to IL1 and TNFα antagonists. The smaller molecules in these pathways make an attractive alternative therapeutic target because pharmaceutical agents can be given orally. In addition, only a few inducible factors seem to have a pivotal role in the regulation of inflammatory genes (for example, activator protein‐1 (AP‐1), CCAT/enhancer‐binding proteins (C/EBPs), STATs, NF‐AT (nuclear factor of activated T cells), and NF‐κΒ (nuclear factor‐κΒ).3
The janus kinase and Signal Transducer and Activator of Transcription (Jak‐STAT) pathway is the signalling target of a multitude of cytokines, including interferon (IFN)γ, IL2, IL4, IL6, IL7, IL10, IL12, and IL15, all of which are thought to have biologically significant roles in rheumatoid synovial inflammation.4,5 Seven STATs have been identified and preliminary work in human synovial tissue suggests that:
STAT1 expression and activity are increased in RA synovium, at least in those with active disease6,7
STAT3 promotes survival of RA synovial fibroblasts8
STAT4 is expressed in RA synovium9
IL4 STAT mRNA (IL4 primarily signals through STAT6) is increased in RA tissue.10
In addition, murine arthritis knockout models suggest that IL12, signalling through a STAT4 pathway, is required for the development of proteoglycan‐induced inflammatory arthritis, whereas IL4, signalling through STAT6, has an anti‐inflammatory effect.11
Generally STATs confer signalling specificity, whereas Jaks are more widely expressed, but Jak3 expression is mostly limited to haematopoietic cell lines and as such, it is a potential therapeutic target in patients with autoimmune disease. As far as we know, no published work has investigated Jak3 expression in human synovial tissue, although it has been identified as a potential therapeutic target in autoimmune disease. The immunomodulatory effects of Jak3 extend beyond its role in STAT activation as it is (a) phosphorylated during dendritic cell activation through the CD40‐CD154 ligand12 and (b) phosphorylated upon T cell receptor stimulation in murine models, and therefore may have a direct role in T cell activation.13
With the exception of STAT1, most published research investigating Jak‐STAT in inflammatory arthritis has been limited to work in synovial fibroblasts or tissue obtained at the time of joint replacement surgery, often without detailed information about clinical activity. Therefore, results must be interpreted with caution because the clinical and histological features of RA alter significantly depending on disease activity.14 In addition, work in synovial fibroblasts cannot be extrapolated to macrophages or lymphocytes, and both of these cell populations are likely to have important pathogenic roles in RA.
We proposed the hypothesis that Jak‐STAT would be differentially expressed in RA compared with other forms of arthritis. We now report on the protein expression of STAT1, STAT4, STAT6, and Jak3 in synovial tissue taken from a series of patients with clinically active RA, spondyloarthritis (SpA), and osteoarthritis (OA), and normal controls.
Patients and methods
Patients
Synovial tissue was obtained from 38 subjects, including 10 patients with RA, 10 patients with SpA, 10 patients with osteoarthritis OA, and 8 control subjects. Control specimens were obtained from patients undergoing knee or ankle joint arthroscopy for undiagnosed joint pain, who were found to have non‐inflamed synovium at the time of arthroscopy and on subsequent histological analysis. Patients with SpA comprised five patients with undifferentiated SpA, four with psoriatic arthritis, and one with ankylosing spondylitis. The diagnoses of RA and OA conformed to 1987 American College of Rheumatology criteria.15,16Tables 1 and 2 summarise clinical details of individual patients with RA and SpA, and provide details of the whole group of patients with OA and normal control patients. All studies were approved by the ethics committee of the Repatriation General Hospital.
Table 1 Clinical characteristics of study groups.
| Characteristics | Rheumatoid arthritis | Spondylo‐ arthritis | Osteoarthritis | Normal subjects |
|---|---|---|---|---|
| (n = 10) | (n = 10) | (n = 10) | (n = 8) | |
| Age (years), mean (SD) | 61.0 (15.5) | 49.1 (17.5) | 66 (10.4) | 34.4 (10.4)* |
| Male:female ratio | 4:6 | 5:5 | 6:4 | 6:2 |
*p<0.05.
Table 2 Clinical and serological characteristics of patients with RA and SpA.
| Patients | Age | Sex | DMARD | Disease duration | CRP | RF | HLA‐B27 |
|---|---|---|---|---|---|---|---|
| (months) | (mg/l )* | (IU/ml)* | |||||
| 1RA | 60 | M | SSZ, Pred, | 132 | 17 | 248 | N/A |
| 2RA | 74 | M | IM gold, MTX | >12 | 5 | 101 | N/A |
| 3RA | 36 | F | CyA, Pred, NSAID | 156 | 3 | 80 | N/A |
| 4RA | 63 | F | NSAID | <12 | <1 | <20 | N/A |
| 5RA | 71 | M | Nil | 3 | 120 | 1220 | N/A |
| 6RA | 59 | F | MTX, SSZ | 216 | 35 | 531 | N/A |
| 7RA | 41 | F | Nil | 6 | <1 | 26 | N/A |
| 8RA | 74 | F | Nil | 24 | 9 | 1552 | N/A |
| 9RA | 48 | F | Nil | <12 | 8 | 335 | N/A |
| 10RA | 84 | M | MTX, Pred | 84 | 90 | <20 | Negative |
| 11uSpA | 27 | F | Depomedrol | 12 | 6 | N/A | Negative |
| 12PsA | 59 | F | SSZ (2 days), Pred | 96 | 164 | N/A | Negative |
| 13uSpA | 42 | F | Nil | >60 | 16 | N/A | Negative |
| 14PsA | 49 | F | Allopurinol, NSAID | 60 | 45 | N/A | Negative |
| 15PsA | 64 | M | MTX | >12 | 48 | N/A | Negative |
| 16AS | 50 | M | SSZ, NSAID | 384 | 34 | N/A | positive |
| 17PsA | 58 | M | NSAID | >12 | 6 | N/A | Negative |
| 18uSpA | 33 | M | NSAID | >12 | 26 | N/A | Positive |
| 19uSpA | 27 | F | NSAID | 24 | 3 | N/A | Positive |
| 20uSpA | 82 | M | NSAID | 6 | 384 | N/A | Positive |
DMARD, disease modifying antirheumatic drug; SSZ, sulfasalazine; Pred, prednisolone; IM gold, intramuscular gold; MTX, methotrexate; CyA, ciclosporin A; NSAID, non‐steroidal anti‐inflammatory drug; uSpA, undifferentiated spondyloarthritis; PsA, psoriatic arthritis; AS, ankylosing spondyloarthritis; N/A, not applicable.
*CRP, normal <60 mg/l; RF, normal <20 IU/ml.
Arthroscopic biopsies
Synovial membrane samples were obtained from affected joints (knee, wrist or ankle) of patients with RA, SpA, and OA under direct vision using a 2.7 mm miniarthroscope (Dyonics, Andover, MA, USA) and standard approaches as previously described.17 Normal specimens were obtained from patients undergoing arthroscopy for undiagnosed joint pain at a sports medicine facility who were found to have non‐inflamed synovium at the time of arthroscopy and on subsequent histological analysis. Samples were frozen in Tissue Tek OCT compound (Miles Diagnostics) and stored at −80°C.
Immunohistochemistry
Table 3 list the antibodies used. Cryosections (4 μm) were fixed in ice cold acetone for 4 minutes. Sections were brought to room temperature, washed in phosphate buffered saline (PBS), and endogenous peroxidase activity was inhibited using 1%H2O2 and 0.1% sodium azide in PBS. Primary antibody was applied for 2 hours (1.5 hours for STAT1) at room temperature. (Control sections were incubated with diluent PBS/1% bovine serum albumin.) Horseradish peroxidase conjugated goat antirabbit antibody was applied for 30 minutes, followed by horseradish peroxidase conjugated swine antigoat for 30 minutes. A signal was developed using 3‐amino‐9‐ethylcarbazole (Sigma, St Louis, MO) and hydrogen peroxide for 20 minutes in the dark. Sections were counterstained with haematoxylin and mounted (Gurr, BDH). Where Jak‐STAT activity was compared across arthritis groups, each antibody was tested in all tissues at one time to exclude run to run variability and allow direct comparison of staining between the tissues. Sequential sections were performed to look for colocalisation of Jak3, STAT4, and STAT6 compared with STAT1.
Table 3 Affinity purified primary antibodies and secondary antibodies.
| Antibody | Type | Dilution | Source |
|---|---|---|---|
| STAT1 | Rabbit polyclonal | 1/900 | Santa Cruz (CA, USA) |
| STAT4 | Rabbit polyclonal | 1/200 | Santa Cruz (CA, USA) |
| STAT6 | Rabbit polyclonal | 1/700 | Santa Cruz (CA, USA) |
| Jak3 | Rabbit polyclonal | 1/600 | Santa Cruz (CA, USA) |
| CD68 | Mouse monoclonal | 1/400 | DAKO (Australia) |
| CD22 | Mouse monoclonal | 1/50 | Serotec (Oxford, UK) |
| CD55 | Mouse monoclonal | 1/7000 | Serotec (Oxford, UK) |
| CD1a | Mouse monoclonal | 1/50 | Serotec (Oxford, UK) |
| CD3 | Mouse monoclonal | 1/50 | Becton Dickinson (NJ, USA) |
| CD45 | Mouse monoclonal | 1/100 | D Beroukas (Dept Immunology, Flinders Medical Centre, South Australia) |
| Goat anti rabbit | HRP conjugated | 1/100 | DAKO (Australia) |
| Swine antigoat | HRP conjugated | 1/200 | BioSource (CA, USA) |
| Donkey antirabbit | Cy3 | 1/100 | Jackson ImmunoResearch (PA, USA) |
| Donkey antimouse | FITC | 1/100 | Jackson ImmunoResearch (PA, USA) |
HRP, horseradish peroxidase.
For double immunohistochemistry, sections were incubated with STAT4, followed by a secondary and tertiary antibody. Subsequently, tissue was blocked with 0.1 M Tris 0.02 M glycine for 60 minutes at room temperature. A 20% normal donkey serum block was applied for 60 minutes and the second primary antibody (CD68, CD55, CD3, CD22) was added overnight at 4°C in a humidified chamber. Biotinylated donkey antimouse was added for 40 minutes followed by APAAP (DAKO, Australia) 1:50 for 60 minutes at room temperature. Signal was detected with fast blue substrate. A counterstain was not performed on double stained sections.
Further immunohistochemistry using the previously described method with the addition of a 20% normal human serum block was performed upon one patient with seropositive RA to ensure that cell bound rheumatoid factor (RF) was not cross reacting with Jak3 or STAT4 antibodies.
Immunofluorescence
Cryosections (4 μm) were cut from synovial tissue, fixed, and blocked for endogenous peroxidases as described previously. Specimens were then incubated overnight at 4°C with primary antibody to STAT1, STAT4, STAT6, Jak3, CD55, CD45, CD68, and CD1a. Subsequently, specimens were incubated with an antirabbit fluorescein conjugate and an antimouse cyanine 3 conjugate, mounted with DPX (dibutyl phthalate with xylene) and analysed using confocal microscopy for the presence or absence of Jak‐STAT colocalisation with cell lineage markers.
Semiquantitative analysis
Synovial tissue in the lining and sublining region was evaluated by two independent observers using a semiquantitative scoring system as previously described18 where 0, <5% staining; 1, 5–25% staining; 2, 26–50% staining; 3, 51–75% staining; and 4, >75% staining. Scores were compared and where differences occurred, a consensus was reached.
Statistical analysis
Results are given as mean (SD). Interobserver variability was assessed using Spearman's rank correlation and the κ statistic. Occasionally, tissue was inadequate (for technical reasons, lifting of tissue, inadequate synovium), and these samples were excluded from the final analysis. Details are included in the “Results” section.
Comparison between patient groups was performed using the Kruskal‐Wallis test. Post hoc analyses were performed with Student's t test or the Mann‐Whitney U test, where appropriate. For statistical analysis, RF <40 IU/ml was recorded as negative. To account for multiple comparisons, p values ⩽0.01 were considered significant.
Results
Interobserver variability
Variability between observers was assessed for Jak‐STAT protein expression lining and sublining scores. Spearman's rank correlation ranged between 0.785 and 0.914 (p<0.01), indicating excellent correlation. The κ statistic was assessed for all groups except STAT6 lining (owing to insufficient patient data in the RA group) and ranged between 0.266 and 0.708 (p⩽0.002), indicating fair to substantial agreement.
To evaluate expression of Jak3, STAT1, STAT4, and STAT6 in differing forms of arthritis, we compared tissue from those with osteoarthritis, SpA, and RA against normal synovial tissue and then analysed differences in Jak‐STAT expression between the arthritis groups. Results describe protein expression and so do not differentiate between activated and quiescent Jak‐STAT.
Expression of Jak‐STAT in inflammatory arthritis and OA compared with normal synovium
STAT1, STAT4, and Jak3 expression was increased in RA compared with normal tissue (p⩽0.001, table 4). STAT1 and Jak3 lining and sublining expression and STAT4 sublining expression were increased in SpA compared with normal tissue (p⩽0.01), while there was a trend towards increased STAT4 lining expression (p = 0.024). Expression of Jak3 in lining and sublining areas was increased in OA in comparison with normal synovium (p⩽0.01), and there was a trend towards increased STAT4 and STAT6 sublining expression (p = 0.038, p = 0.016). Subsequent results focus on differences between inflammatory and degenerative arthritis and differences within inflammatory arthritis groups.
Table 4 Summary of the differential expression of Jak/STAT in the four study groups.
| Comparison | RA v normal | RA v OA | RA v SpA | SpA v normal | SpA v OA | OA v normal |
|---|---|---|---|---|---|---|
| STAT1 lining | ↑ p<0.001 | NS | NS | ↑ p = 0.001 | ↑ p = 0.013 | NS |
| STAT1 sublining | ↑ p<0.001 | ↑ p = 0.001 | NS | ↑ p<0.001 | ↑ p = 0.004 | NS |
| STAT6 lining | NS | NS | NS | NS | NS | NS |
| STAT6 sublining | NS | NS | NS | NS | NS | NS |
| STAT4 lining | ↑ p<0.001 | NS | ↑ p = 0.012 | NS | NS | ↑ p = 0.002 |
| STAT4 sublining | ↑ p = 0.001 | ↑ p = 0.011 | NS | ↑ p = 0.011 | NS | NS |
| Jak3 lining | ↑ p<0.001 | NS | NS | ↑ p = 0.011 | NS | ↑ p = 0.001 |
| Jak3 sublining | ↑ p = 0.001 | ↑ p = 0.008 | NS | ↑ p = 0.002 | NS | ↑ p = 0.010 |
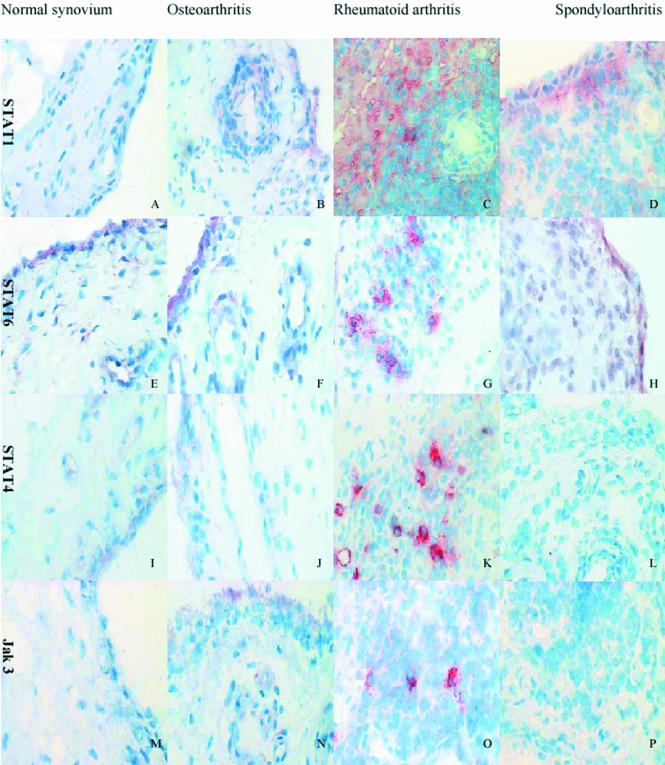
STAT1 protein expression is increased in inflammatory arthritis compared with OA
STAT1 lining and sublining expression in SpA and STAT1 sublining expression in RA were significantly increased compared with OA (table 4, fig 1). There was a trend towards increased expression of STAT1 in RA lining when compared with OA (p⩽0.015). This was the only STAT to be increased in all inflammatory arthritis when compared with OA synovium. In contrast, in RA tissues, Jak3 and STAT4 sublining expression was also increased (p⩽0.01). STAT4 expression in one patient with OA and Jak3 expression in two patients with OA were excluded from the final analysis owing to poor quality tissue.
Figure 1 Representative examples of staining for Jak‐STAT in patients' tissues. Sections taken at ×400 magnification and counterstained with haematoxylin. Note the prominent staining with Jak3/STAT4/STAT6 in cells with dendritic morphology in the rheumatoid synovium (panels G, K, O).
STAT6 protein expression is heterogeneous across arthritis groups
There was no significant difference in STAT6 protein expression between the different patient groups (p>0.01, table 4, fig 1). Even in normal synovium, there was a moderate baseline expression of STAT6, with only a trend towards increased expression recorded in the sublining of RA (p = 0.02) and OA tissue (p = 0.02). Occasional intense staining cells were seen in the sublining tissue, particularly in RA synovium, but these were difficult to interpret given the overall high level of STAT6 protein expression. Scores from two patients with RA had to be excluded from the final analysis owing to poor tissue quality. The high baseline level of STAT6 suggests that it may have important homoeostatic functions within the joint.
Signal transduction expression in RA
We were interested in identifying Jak‐STAT protein expression unique to RA compared with other forms of inflammatory arthritis to (a) demonstrate examples of differences in pathogenesis at the level of signal transduction, (b) identify any clinically useful measure for differentiating between RA and other inflammatory arthritis at the level of the synovium. When RA Jak‐STAT protein expression was compared with expression in SpA synovium, the only significant difference using semiquantitative analysis was STAT4 protein expression. This was increased at the lining level in RA tissue (p = 0.012, table 4). More interesting, however, was the staining pattern for Jak3 and STAT4 in 7/10 patients with RA. These cases exhibited isolated, “brightly staining cells” (BCs) in the sublining region that appeared to have dendritic morphology (fig 1). These cells were not seen in any other arthritis groups. There was no difference in age, sex, disease duration, disease modifying antirheumatic drug use, or C reactive protein in the BC group compared with other patients with RA (p>0.05). However, the presence of a positive RF was strongly associated with these cells (p = 0.008). To exclude the possibility of the BCs being the result of non‐specific binding to cell bound RF, we added a 1 hour serum block with 20% normal human serum to one of the tissues known to exhibit BC staining for Jak3 and STAT4. Bright cells were still seen in the presence of a normal human serum block.
Cells staining brightly for Jak3 also express intense STAT4 and STAT6 protein expression or closely interdigitate with these cells
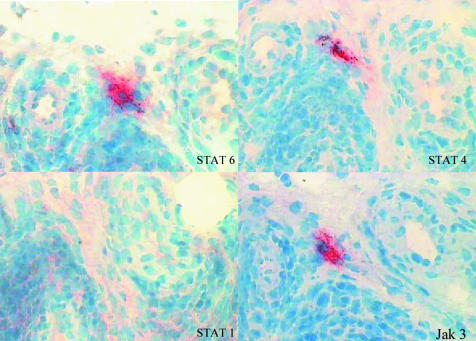
Murine models have shown that Jak3 signals through STAT6, but our immunohistochemistry work suggested that STAT4 expression more closely follows Jak3 expression. To determine whether cells staining brightly for Jak3 express STAT4 or STAT6, or both, sequential sections in one patient with RA were stained. We were able to locate cells prominently expressing Jak3 which also appeared to express prominently, or were extremely closely associated with cells prominently expressing, STAT4 and STAT6. In contrast, no staining was seen for STAT1 in this region (fig 2). In summary, intense Jak3 expression in RA tissue is closely linked with intense STAT4 and STAT6 protein expression.
Figure 2 Sequential staining of Jak‐STAT showing close association of Jak3/STAT4/STAT6 staining. Synovium was obtained from a patient with RA. Sections were taken at ×400 magnification and counterstained with haematoxylin.
Phenotype of BCs in RA tissue
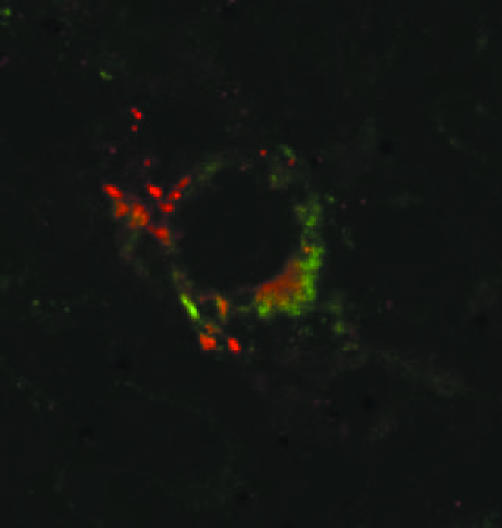
Colocalisation studies were performed to phenotype BCs found in RA tissue. No colocalisation was observed with CD3 (T cell marker), CD22 (B cell marker), CD55 (lining fibroblast marker) or CD68 (macrophage marker). There was intermittent colocalisation using confocal microscopy with CD1a, a marker for immature dendritic cells (fig 3).
Figure 3 Colocalisation of STAT4 (Cy3, red) with CD1a (FITC, green) as seen in synovium from a patient with RA. Magnification ×600 digitally enhanced to ×1800.
Confocal studies identified cell types expressing Jak‐STAT in RA
Confocal microscopy was performed on one RA tissue to look for colocalisation with CD45 (a pan lymphocyte marker), CD55, and CD68. Colocalisation was observed with STAT6 for all cellular markers. STAT1 colocalised predominantly with CD55, but CD68 and CD45 colocalisation was also seen. STAT4 and Jak3 were found occasionally to colocalise with CD68 and CD45, but these were not specifically bright cells.
Discussion
The Jak‐STAT pathway is the primary signal transduction path for a number of cytokines known to have important roles in inflammatory arthritis. IFNγ and IL6 signal through STAT1, IL4 (thought to have an anti‐inflammatory role) signals through STAT6, and IL12 (critical in Th1 mediated inflammation) and IFNα signal through STAT4.5,19 To date, much of the work on these signalling pathways has been based on murine and in vitro models. We have documented Jak and STAT protein expression in vivo in RA, SpA, and OA and compared their expression with that in a normal cohort.
STAT1 expression was increased in inflammatory arthritis in comparison with OA and normal controls. Cells expressing STAT1 were predominantly fibroblasts (CD55+) but colocalisation with leucocytes (CD45+) and macrophages (CD68+) was also found.
The primary activator of STAT1 is thought to be IFNγ but IL6, IL10, and IFNα/β may also contribute to its activation.19 Yokota et al found that IL6 was the primary activator of STAT1 in synovial fluid cells consisting predominantly of neutrophils, suggesting the presence of cell lineage specific activation pathways.20 We have reviewed the current understanding of the physiological role of STAT1 in arthritis elsewhere,21 but to summarise, human tissue and rodent arthritis models suggest that its action is predominantly anti‐inflammatory and that modulation of STAT1 function, either directly or through suppression of cytokine signalling (SOCS‐1), may have therapeutic applications.
Two published reports have described STAT1 expression in synovial tissue. Our findings concur with those of van der Pouw Kraan et al.6 They used microarray techniques and real time polymerase chain reaction to demonstrate increased expression of STAT1 and genes known to be regulated by STAT1 in active RA. Kasperkovitz et al reported increased STAT1 expression in the synovium in 12 patients with RA compared with OA and reactive arthritis (ReA).7 Furthermore, they showed that the ratio of STAT1 to pSTAT1 expression in the RA group remained the same, suggesting that STAT1 activation may increase concomitantly with STAT1 expression. In contrast, we found that STAT1 expression was significantly increased in both SpA and RA. However, no mention of disease activity in the ReA group was made and in most cases the entire control group was used for the comparison with the RA group, rather than considering ReA and OA separately.
IL4, a prototypical Th2 cytokine, is the primary activator of STAT6 and it has been suggested that altering the Th1/Th2 balance in inflammatory arthritis through STAT6 may modulate disease expression.22 In support of this, Müller‐Ladner et al found IL4 STAT (IL4 primarily signals through STAT6) expression in 8/10 RA tissues in both early (<12 months) and late (>2 years) disease.10 IL4 STAT was found in macrophages, fibroblasts, and also in follicular infiltrates. In contrast, very weak expression was seen in 3/3 patients with OA.10 Using a proteoglycan‐induced murine arthritis model and knockout models, Finnegan et al found that IL4, signalling through STAT6, was necessary to attenuate the severity of joint inflammation. Its anti‐inflammatory role appeared to function through the regulation of IL12 production.11
Among our patient cohort, STAT6 protein was widely expressed, with representation in all tested cell lineages (fibroblast, macrophage, and lymphocyte). The only statistically significant findings were an increase in sublining expression in RA and OA in comparison with normal tissue. Our cellular distribution concurs with that of Müller‐Ladner's group,10 but we did not note any marked difference in expression between arthritis types. Although STAT6 may have an anti‐inflammatory role, the widespread expression we observed suggests that it may not be an ideal immunomodulatory target. However, we have only observed STAT6 protein expression. STAT6 phosphorylation may be widely skewed between arthritis groups, supporting a role for STAT6 modulation as a therapeutic target.
Our most intriguing finding is the demonstration of multiple cells of a dendritic phenotype (CD1a+, CD22−, CD68−, CD3−, CD55−) in rheumatoid synovial tissue, expressing large amounts of STAT4, STAT6, and Jak3. This staining pattern was unique to patients with seropositive RA, suggesting a differing pathogenic process than for SpA and the seronegative RA group. In addition, the location of these cells was consistent with previous work documenting the distribution of CD1a+ cells in the rheumatoid synovium.23
Dendritic cells are the major antigen presenting cells in rheumatoid synovium.24 The specific pattern of Jak3, STAT4, and STAT6 expression gives an indication of the stage of differentiation of these cells. Recent work on purified dendritic cell lines has shown that Jak3 is phosphorylated in response to CD40 ligation by CD154 on activated T cells, a critical component in dendritic cell maturation. Therefore, dendritic cells in their earlier stages of activation might be expected to express large amounts of Jak3 and its downstream transcription factors, STAT6 (and possibly, STAT5a/b). Monocyte derived dendritic cells triggering through CD40 also show expression of costimulatory molecules and production of IL12, which acts in a paracrine and autocrine fashion.12,25 IL12 signals through STAT4 and so dual cellular expression of Jak3, STAT6, and STAT4 is consistent with dendritic cells that have recently undergone activation. Although the presence of costimulatory molecules (such as CD80 and CD86) would support this hypothesis, their absence may only mean that the changes in Jak3, STAT6, and STAT4 expression occur at an early stage, before the production of other costimulatory molecules.
Overall expression of Jak3 and STAT4 was increased in the sublining but not the lining regions of the inflammatory arthritis groups when compared with OA. The absence of any significant difference in lining expression of Jak3 and STAT4 between the inflammatory and OA groups is somewhat surprising. Although lining tissue from patients with RA and SpA was more hypertrophied, the percentage of lining cells expressing STAT4 and Jak3 in the OA group was similar, possibly reflecting low grade inflammatory change in the OA synovial lining.26
There is little published work documenting the expression of Jak3 or STAT4 in arthritis and there are no studies documenting synovial distribution in a series of arthritides. Only one study has reported the presence of Jak3 in inflammatory arthritis and they used microarray techniques on peripheral blood monocytes.27 Selected genes were further assessed by reverse transcription‐polymerase chain reaction assay. Among other changes, Jak3 expression was increased in patients with psoriasis but not other inflammatory arthritis groups. Although these findings are intriguing, they cannot be extrapolated to changes at the synovial level.
STAT4 has been demonstrated in type A synovial macrophages in rheumatoid tissue taken at the time of joint replacement surgery,9 but the expression of STAT4 in early and active human inflammatory arthritis has not, to our knowledge, been documented. Studies of proteoglycan‐induced arthritis in STAT4 knockout mice have demonstrated an attenuated form of arthritis, suggesting that STAT4 is important in perpetuating inflammation.11
We postulate that STAT4/Jak3/STAT6 dendritic cell expression in seropositive RA reflects a unique pathogenic process, and Jak3/STAT4 expression may be a useful marker for identifying RA at the synovial level. Further large scale studies would explain whether the characteristic histological picture seen in these patients has any clinical application.
Our findings identify alternative pathways to modulate dendritic cell maturation in RA. Jak3, in particular, is an attractive target. Its expression is largely limited to haematopoietic cell lines and in addition to its roles in dendritic cell activation, it is also responsible for signalling of multiple cytokines important in lymphocyte activation through haematopoietin receptors (namely, IL2, IL7, IL9, and IL15)5. Jak3 inhibition would have the potential to alter dendritic cell maturation at a time when the inflammatory process is still evolving and therefore may provide long term disease modulating activity. An orally bioavailable Jak3 antagonist has been found to reduce transplant rejection in two animal models.28
In conclusion, we have documented the differential expression of Jak‐STAT signalling pathways in four groups of synovial tissue. STAT1 expression was up regulated in inflammatory arthritis, consistent with its role in IFNγ and IL6 signalling. Our preliminary work shows that Jak3 and STAT4 are uniquely and prominently expressed by dendritic cells in RF positive patients with RA. These findings suggest a pathogenic process unique to seropositive RA. Intense Jak3, STAT6, and STAT4 protein expression is likely to correspond with an early stage of dendritic cell activation. Their characteristic staining profile raises the possibility of an alternative marker which could be used to identify dendritic cells in frozen synovial tissue and may also provide a method for identifying RA at the level of the synovium. Our findings suggest that Jak3 inhibition may be an appropriate therapeutic target in inflammatory arthritis and especially seropositive RA. Animal model studies may further define any therapeutic roles of a Jak3‐specific inhibitor in the treatment of RA.
Acknowledgements
We thank Dr Toby Coates for assistance with morphology studies.
Supported by the Daw Park Research Foundation. Dr Walker's work is supported by the National Health and Medical Research Council of Australia and the Arthritis Foundation of Australia.
Abbreviations
BCs - brightly staining cells
IFN - interferon
IL - interleukin
Jak‐STAT - janus kinase and Signal Transducer and Activator of Transcription
OA - osteoarthritis
PBS - phosphate buffered saline
SpA - spondyloarthritis
RA - rheumatoid arthritis
ReA - reactive arthritis
RF - rheumatoid factor
TNFα - tumour necrosis factor α
References
- 1.Weyand C M, Goronzy J J, Takemura S, Kurtin P J. Cell‐cell interaction in synovitis. Interactions between T cells and B cells in rheumatoid arthritis. Arthritis Res 20002457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firestein G. Evolving concepts of rheumatoid arthritis. Nature 2003423356–361. [DOI] [PubMed] [Google Scholar]
- 3.Makarov S S, NF‐κΒ in rheumatoid arthritis: a pivotal regulator of inflammation, hyperplasia and tissue destruction Arthritis Res. 2001;3:200–206. doi: 10.1186/ar300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinarello C A, Moldawer L L.Proinflammatory and anti‐inflammatory cytokines in rheumatoid arthritis. A primer for clinicians. Amgen 2002
- 5.Gadina M, Hilton D, Johnston J A, Morinobu A, Lighvani A, Zhou Y.et al Signaling by type I and II cytokine receptors: ten years after. Curr Opinion Immunol 200113363–373. [DOI] [PubMed] [Google Scholar]
- 6.van der Pouw Kraan T C, van Gaalen F A, Verbeet N, Smeets T, Kraan M.et al Rheumatoid arthritis is a heterogeneous disease. Evidence for differences in the activation of the STAT‐1 pathway between tissues. Arthritis Rheum 2003482132–2145. [DOI] [PubMed] [Google Scholar]
- 7.Kasperkovitz P, Verbeet N, Smeets T, van Rietschoten J G, Kraan M C, van der Pouw Kraan T C.et al Activation of the STAT1 pathway in rheumatoid arthritis. Ann Rheum Dis 200463233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause A, Scaletta N, Ji J, Ivashkiv L B. Rheumatoid arthritis synoviocyte survival is dependent on Stat3. J Immunol 20021696610–6616. [DOI] [PubMed] [Google Scholar]
- 9.Frucht D M, Aringer M, Galon J, Danning C, Brown M, Fan S.et al Stat4 is expressed in activated peripheral blood monocytes, dendritic cells and macrophages at sites of Th1‐mediated inflammation. J Immunol 20001644659–4664. [DOI] [PubMed] [Google Scholar]
- 10.Müller‐Ladner U, Judex M, Ballhorn W, Kullmann F, Distler O, Schlottmann K.et al Activation of the IL‐4 STAT pathway in rheumatoid synovium. J Immunol 20001643894–3901. [DOI] [PubMed] [Google Scholar]
- 11.Finnegan A, Grusby M J, Kaplan C D, O'Neill S K, Eibel H, Koreny T.et al IL‐4 and IL‐12 regulate proteoglycan‐induced arthritis through Stat‐dependent mechanisms. J Immunol 20021693345–3352. [DOI] [PubMed] [Google Scholar]
- 12.Saemann M D, Diakos C, Keleman P, Kriehuber E, Zeyda M, Bohmig G A.et al Prevention of CD40‐triggered dendritic cell maturation and induction of T cell hyporeactivity by targeting of janus kinase 3. Am J Transplant 200331341–1349. [DOI] [PubMed] [Google Scholar]
- 13.Tomita K, Saijo K, Yamasaki S, Iida T, Nakatsu F, Arase H.et al Cytokine‐independent Jak3 activation upon T cell Receptor (TCR) stimulation through direct association of Jak3 and the TCR complex. J Biol Chem 200127625378–25385. [DOI] [PubMed] [Google Scholar]
- 14.Smith M D, Kraan M C, Slavotinek J, Au V, Weedon H, Parker A.et al Treatment‐induced remission in rheumatoid arthritis patients is characterised by a reduction in macrophage content of synovial biopsies. Rheumatology (Oxford) 200140367–374. [DOI] [PubMed] [Google Scholar]
- 15.Arnett F, Edworthy S, Bloch D, McShane D, Fries J, Cooper N.et al The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 198831315–324. [DOI] [PubMed] [Google Scholar]
- 16.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K.et al Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum 1986291039–1049. [DOI] [PubMed] [Google Scholar]
- 17.Smith M, Chandran G, Youssef P, Darby T, Ahern M. Day case knee arthroscopy under regional anaesthesia performed by rheumatologists. Aust NZ J Med 199626108–109. [DOI] [PubMed] [Google Scholar]
- 18.Tak P P, van der Lubbe P A, Cauli A, Daha M R, Smeets T J, Kluin P M.et al Reduction of synovial inflammation after anti‐CD4 monoclonal antibody treatment in early rheumatoid arthritis. Arthritis Rheum 1995381457–1465. [DOI] [PubMed] [Google Scholar]
- 19.Ivashkiv L, Xiaoyu H. The JAK/STAT pathway in rheumatoid arthritis: pathogenic or protective? Arthritis Rheum 2003482092–2096. [DOI] [PubMed] [Google Scholar]
- 20.Yokota A N M, Shima Y, Murata N, Tanaka T, Suemura M, Yoshizaki K.et al Preferential and persistent activation of the STAT1 pathway in rheumatoid synovial fluid cells. J Rheumatol 2001281952–1959. [PubMed] [Google Scholar]
- 21.Walker J G, Smith M D. The Jak‐STAT pathway in rheumatoid arthritis. J Rheumatol 2005321650–1653. [PubMed] [Google Scholar]
- 22.O'Shea J, Visconti R, Cheng T P, Gadina M. Jaks and Stats as therapeutic targets. Ann Rheum Dis 200059(suppl I)i115–i118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page G, Lebecque S, Miossec P. Anatomic localization of immature and mature dendritic cells in an ectopic lymphoid organ: correlation with selective chemokine expression in rheumatoid synovium. J Immunol 20021685333–5341. [DOI] [PubMed] [Google Scholar]
- 24.Thomas R, Lipsky P. Presentation of self peptides by dendritic cells: possible implications for the pathogenesis of rheumatoid arthritis. Arthritis Rheum 199639183–190. [DOI] [PubMed] [Google Scholar]
- 25.Fukao T, Frucht D M, Yap G, Gadina M, O'Shea J J, Koyasu S. Inducible expression of Stat4 in dendritic cells and macrophages and its critical role in innate and adaptive immune responses. J Immunol 20011664446–4455. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier J P, Martel‐Pelletier J, Abramson S B. Osteoarthritis, an inflammatory disease. Potential implication for the selection of new therapeutic targets. Arthritis Rheum 2001441237–1247. [DOI] [PubMed] [Google Scholar]
- 27.Gu J, Marker‐Hermann E, Baeten D, Tsai W C, Gladman D, Xiong M.et al A 588‐gene microarray analysis of the peripheral blood mononuclear cells of spondyloarthropathy patients. Rheumatology (Oxford) 200241759–766. [DOI] [PubMed] [Google Scholar]
- 28.O'Shea J J. Targeting the Jak/STAT pathway for immunosuppression. Ann Rheum Dis 200463(suppl II)ii67–ii71. [DOI] [PMC free article] [PubMed] [Google Scholar]