Abstract
An iCycler iQ real-time PCR assay targeting 18S rRNA Aspergillus-specific sequences was developed for the diagnosis of invasive pulmonary aspergillosis (IPA). Positive findings were obtained for 18 of 20 (90%) bronchoalveolar lavage (BAL) fluid specimens from patients with probable or confirmed IPA and were obtained for none of the 24 BAL samples from patients with no clinical evidence of aspergillosis. These results were concordant with those of a nested PCR assay, which detected 90% of the patients with IPA, while galactomannan ELISA revealed positivity for 100% of these patients, suggesting that combined use of methods might improve the diagnosis of IPA.
Invasive pulmonary aspergillosis (IPA) is a severe infection in immunocompromised hosts, such as those receiving cytoreductive or marrow-ablative therapy for hematological malignancies (7, 18, 24). Its high mortality rate is due in part to the rapid progression of the infection (i.e., 1 to 2 weeks from onset to death) and difficulties related to its diagnosis, particularly in the early stages (18). Although antifungal therapy is often initiated empirically, the drugs used against IPA have a high potential for toxicity. Definitive diagnosis requires histopathological evidence of deep-tissue invasion or a positive culture from normally sterile sites (7). However, invasive diagnostic procedures can be risky for debilitated neutropenic patients (18) and cultures for this fungus are characterized by low sensitivity (7, 19). Reliable noninvasive methods to diagnose IPA are thus essential for the optimal management of these high-risk patients.
Promising alternatives to culture or biopsy include enzyme-linked immunosorbent assay (ELISA) detection of the galactomannan (GM) antigen in serum and other biological fluids, e.g., bronchoalveolar lavage (BAL) fluid (2, 21), and PCR-based methods for the detection of Aspergillus-specific DNA (4, 5, 9, 10, 12, 14, 17, 26). New quantitative assays using real-time PCR have also been developed for the diagnosis of IPA (6, 13). In a recent study, however, Costa et al. (6) found that ELISA detection of GM in serum was superior to a LightCycler PCR assay in the diagnosis of invasive aspergillosis.
We developed a new real-time PCR assay for the diagnosis of IPA, based on iCycler iQ (Bio-Rad Laboratories, Hercules, Calif.) technology. In the present study, we evaluated its performance on BAL fluid specimens from hematology patients, comparing our results with those of a conventional PCR assay (14) and of ELISA detection of GM.
Specimen processing.
Forty-four BAL fluid specimens were obtained from 44 patients with hematological malignancies who were undergoing bronchoscopy in our center between January 2001 and July 2002 within a week after the diagnosis of lung infiltrates (Table 1). The specimens were collected in sterile vials without conservation medium and sent to the laboratory within 2 h. Upon receipt, each specimen was homogenized and divided into three parts: one was immediately processed for routine microbiological culture (23), the second was subjected to ELISA for GM, and the third was stored in a screw-cap tube at −20°C prior to PCR analyses, both conventional and real time (see below). To detect fungi, primary cultures for yeasts were done on Bacto Candida Growth agar (Kima, Padua, Italy) and held at 37°C for 6 days; those for filamentous fungi were performed on Sabouraud dextrose agar supplemented with chloramphenicol (40 μg/ml) and/or cycloheximide (0.05%) and incubated at 25°C for at least 2 weeks. All BAL specimens were simultaneously cultivated on these different media. When yeast growth was detected for a given specimen, incubation of the corresponding cultures for filamentous fungi was continued until the end point.
TABLE 1.
Results of real-time PCR, conventional PCR, and GM ELISA of BAL fluid specimens from patients with IPA
| Patient no. | Sex, age (yr) | Primary diseasea | Outcome | IPA diagnosis (EORTC criteriab) | BAL fungal culture | GM ELISA | BAL PCR result
|
|
|---|---|---|---|---|---|---|---|---|
| Real time (copies/ml) | Conventional | |||||||
| 1 | F, 47 | NHL | Survival | Probable | C. albicans | Positive | Negative | Negative |
| 2 | F, 72 | NHL | Death | Probable | C. glabrata | Positive | 3,000 | Positive |
| 3 | F, 65 | ML | Death | Probable | Negative | Positive | 4,500 | Positive |
| 4 | M, 41 | AML | Survival | Probable | Negative | Positive | 8,000 | Positive |
| 5 | M, 50 | ALL | Death | Proven | A. fumigatus | Positive | 60,000 | Positive |
| 6 | M, 65 | SAA | Death | Proven | A. flavus | Positive | 45,000 | Positive |
| 7 | M, 68 | AML | Survival | Probable | Negative | Positive | 4,200 | Positive |
| 8 | M, 48 | SAA | Death | Proven | A. fumigatus | Positive | 75,000 | Positive |
| 9 | F, 72 | ML | Survival | Probable | C. albicans | Positive | 2,000 | Positive |
| 10 | M, 65 | AML | Death | Probable | Negative | Positive | 3,900 | Positive |
| 11 | M, 60 | AML | Death | Probable | Negative | Positive | 9,800 | Positive |
| 12 | F, 60 | AML | Death | Probable | Negative | Positive | 10,500 | Positive |
| 13 | M, 67 | ALL | Survival | Probable | Negative | Positive | 8,700 | Positive |
| 14 | F, 42 | NHL | Death | Proven | A. fumigatus | Positive | 85,000 | Positive |
| 15 | M, 77 | ALL | Death | Probable | Negative | Positive | 5,400 | Positive |
| 16 | M, 70 | AML | Death | Probable | A. fumigatus | Positive | 23,000 | Positive |
| 17 | M, 70 | NHL | Death | Proven | A. flavus | Positive | 100,000 | Positive |
| 18 | M, 74 | AML | Death | Probable | Negative | Positive | 7,600 | Positive |
| 19 | F, 39 | AML | Survival | Probable | C. albicans | Positive | Negative | Negative |
| 20 | M, 54 | ALL | Survival | Probable | C. albicans | Positive | 3,200 | Positive |
NHL, non-Hodgkin's lymphoma; ML, malignant lymphoma; AML, acute myeloblastic leukemia; ALL, acute lymphoblastic leukemia; SAA, severe aplastic anemia.
EORTC, European Organization for Research and Treatment of Cancer.
ELISA for GM was performed with a commercial kit (Platelia Aspergillus; Bio-Rad) according to the manufacturer's instructions. Assays were classified as positive when the optical density ratio was ⩾1.5; ratios between 1 and 1.5 were classified as doubtful. For PCR assays, DNA was extracted from 1.5 ml of each BAL fluid specimen with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), suspended in 100 μl of distilled water, and stored at −20°C until used. One aliquot (10 μl) of this DNA solution was subjected to the conventional nested PCR developed by Kawamura et al. (14). Briefly, in the first step, 30 pmol of each outer primer (i.e., M5c and M6b) was added to the reaction mixture, and PCR was performed as follows: denaturation at 94°C for 45 s, annealing at 57°C for 1 min, and extension at 72°C for 1 min 30 s for 40 cycles. In the next step, 10 μl of the product obtained from the first amplification was added to a new reaction mixture containing 30 pmol of each inner primer (i.e., M5cN and M6bN), and the process was performed under the following conditions: denaturation at 94°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 60 s for a total of 30 cycles (14). Amplification was carried out with appropriate precautions to avoid cross contamination (15). The PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide and examined under UV light to detect the 136-bp band (14). The second 10-μl aliquot of extracted DNA was used for iCycler real-time PCR.
iCycler-based Aspergillus PCR assay.
The iCycler iQ real-time detection system (Bio-Rad) was used to develop a quantitative PCR assay for Aspergillus spp. DNA. Primers consisted of Asp-fwd (5′-CCG ATT ACG TCC CTG CCC TT) and Asp-rev (5′-TTG ACC AAC TTT CCG GCT CTG), which amplify a 124-bp sequence in a conserved region of the 18S rRNA genes of several Aspergillus species (i.e., A. fumigatus, A. flavus, A. glaucus, A. niger, and A. terreus) as deposited in GenBank. A TaqMan probe that hybridizes to the region between primers (5′-TCG GCC CTT AAA TAG CCC GGT CCG C) was labeled at the 5′ end with FAM (6-carboxy-fluorescein) as the reporter dye and at the 3′ end with TAMRA (6-carboxy-teremethyl-rhodamine) as the quencher. Primers and probe were designed with Beacon Designer 2 version 2.06 software (Premier Biosoft International, Palo Alto, Calif.) and synthesized by MWG Biotech (Florence, Italy). For quantification, DNA from the A. fumigatus strain H11-20 (16) was extracted as described above, amplified with the same primers, and cloned into pCR2 TA cloning vector (Invitrogen, San Diego, Calif.). Serially diluted samples of this plasmid ranging from 10 to 106 copies/reaction were used as external standards in each run. The iCycler PCRs were carried out in a 50-μl volume containing 10 μl of DNA sample, both primers (each at 5 μM), TaqMan probe (5 μM), and 25 μl of the Platinum-qPCR Supermix-UDG (Invitrogen), which contained 1.5 U of heat-activable Taq polymerase and 1 U of UDG. After one 5-min step at 50°C to allow UDG to act and a second one at 94°C for 4 min to inactivate the UDG and activate the Taq polymerase, samples were subjected to 40 cycles of 15 s at 94°C and 1 min at 59°C. Fluorescent data were collected during the 59°C annealing and extension step and were analyzed with iCycler iQ software. The detection threshold was set at 10 times the mean standard deviation above the baseline fluorescence, which was calculated from cycles 1 through 15. Each sample was tested in triplicate, and the results were averaged to obtain the final threshold cycle (Ct), i.e., the point at which sample fluorescence rises above the background level. A standard curve constructed by plotting the log of starting plasmid concentration versus the Ct was used to determine Aspergillus DNA concentrations in specimens.
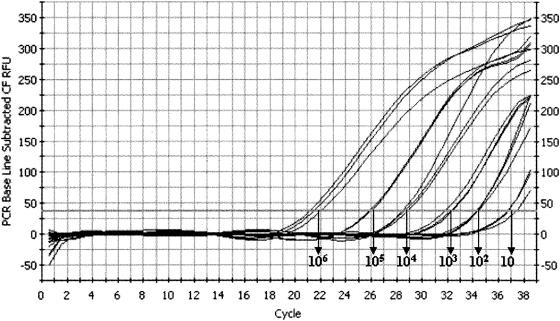
The dynamic range of the real-time PCR assay (determined in preliminary studies) was 10 to 106 plasmid copies. Figure 1 shows amplification results with the TaqMan probe for the standard DNA dilution series. When these samples were tested in five independent runs to determine assay reproducibility, the coefficient of variation was ≤1.7%. DNAs extracted from A. fumigatus, A. flavus, A. glaucus, A. niger, A. terreus and from Candida albicans, Candida glabrata, Candida tropicalis, Saccharomyces cerevisiae, Cryptococcus neoformans, Fusarium solani, Fusarium oxysporum, and Mucor spp. were tested to evaluate assay specificity. As expected, positive results were obtained only with DNA from the Aspergillus species.
FIG. 1.
Real-time quantitative amplification results of standard Aspergillus fumigatus DNA dilution series with a TaqMan probe. The dynamic range covers 10 to 106 copies/well of the target.
All 44 BAL fluid specimens were tested with real-time and conventional PCRs and ELISA. Diagnoses of IPA were based on culture, histopathological, clinical, and radiological data and classified according to the European Organization for Research and Treatment of Cancer criteria (1). Proven IPA was defined as histopathologic or cytopathologic demonstration of septate branched hyphae from deep-tissue biopsy and/or autopsy specimens and compatible clinical symptoms. Probable IPA was defined as the presence of at least one host factor criterion (e.g., neutropenia or persistent fever refractory to broad-spectrum antibacterial therapy) and one major clinical criterion (e.g., cavitary lesion or halo sign on computerized tomography imaging) or two minor clinical criteria (e.g., symptoms of lower respiratory tract infection, pleural effusion, or any new infiltrate not fulfilling major criteria) from abnormal site consistent with infection and a positive culture for Aspergillus from a respiratory specimen (sputum or BAL) or at least two blood samples positive for Aspergillus antigen. Twenty-four of the patients showed no evidence of aspergillosis, and specimens from these patients yielded negative results in all three assays and in cultures. Table 1 shows the test results and clinical data for the remaining 20 patients with proven (n = 5) or probable (n = 15) IPA. All 20 of these specimens were ELISA positive, and 18 of 20 were also positive in both PCR assays. The sensitivity, specificity, and positive and negative predictive values of the iCycler and nested PCR assays for diagnosing IPA were 90, 100, 100, and 92%, respectively. Of the 18 patients with positive PCR results, 6 had culture-proven aspergillosis (A. fumigatus and A. flavus cultured from 4 and 2 patients, respectively); BAL fluid cultures from the two patients with negative PCR results yielded Candida albicans (Table 1).
Our iCycler PCR assay is one of a number of PCR-based methods developed to improve the diagnosis of invasive aspergillosis (25). Methods of this type have been used successfully to detect Aspergillus DNA in serum (4, 14, 30), whole blood (8, 17, 26), and BAL fluid (3, 12, 22, 28-29) from patients with IPA. Detection of circulating Aspergillus GM antigen has also proved useful for early diagnosis of IPA. Three methods are currently used: the latex agglutination test, measurement of the plasma (1→3)-β-d-glucan concentration, and ELISA (18). The last approach appears to be the most sensitive (27), but for most patients with IPA this assay is positive only at advanced stages of infection (13). This is a serious drawback because recent studies have shown that, whereas IPA can be successfully treated with amphotericin B, the drug must be started early, i.e., before signs and symptoms of infection become apparent (18).
PCR-based methods have proved to be more sensitive (with detection limits of ≤10 fg of Aspergillus DNA) than antigen detection methods, particularly for patients with IPA (5, 14). The nested PCR assay developed by Kawamura et al. (14) was successfully used in serum samples from 44 patients with pulmonary aspergillosis, although only four had invasive disease. Unlike ELISA GM assays, however, conventional PCR methods cannot be used to monitor antigen titers during antifungal treatment. This limitation has been overcome by the introduction of real-time PCR assays (6, 11, 13, 20). In a recent study by Kami et al. (13), maximum levels of fungal copies revealed in blood samples by real-time PCR showed good correlation with those obtained with ELISA, and the molecular method yielded positive results earlier. These findings encouraged us to develop a quantitative PCR assay based on iCycler technology and to compare its performance in the diagnosis of IPA with those of Kawamura's nested PCR assay and ELISA detection of GM.
The new assay proved to be highly sensitive, specific, and accurate. It displayed full concordance with the results of nested PCR and was only slightly less sensitive than the ELISA method. The excellent positive predictive value (100%) of our assay is consistent with those of some conventional PCR methods (12, 22), but much lower values have emerged from other studies (3, 5, 10, 26, 29). False-positive results may be caused by accidental contamination of BAL fluid specimens by ubiquitous Aspergillus conidia. Hayette et al. (10) have also suggested that PCR positivity might reflect a transient presence of aspergilli in the respiratory tract rather than true infection. In this respect, real-time PCR can be more helpful than conventional PCR because it provides quantitative information on the fungal burden that can be used to distinguish between infection and simple colonization. The estimated DNA load in our positive specimens, for example, ranged from 2,000 to 100,000 copies/ml, and in those with the highest levels (i.e., ⩾23,000 copies/ml) the PCR result was also confirmed by culture findings. In contrast, those with the lowest fungal loads grew only C. albicans (two samples) or C. glabrata (one sample). In addition, the two specimens that produced negative results in both of the PCR assays also grew C. albicans, which suggested that the presence of DNA of other fungal species might somehow affect the PCR. Furthermore, the negative PCR results for these specimens may be the result of infections with Aspergillus species that were not detected by primers used in our assay (only five Aspergillus species are recognized). Alternatively, the ELISA positivity for these specimens could be due to the presence of another mold (e.g., the Penicillium species) that cross-reacts in the GM assay (21). Although possible, the second hypothesis appears less convincing.
In conclusion, use of this new real-time PCR assay with BAL fluid specimens seems to be a promising method for diagnosis of IPA, especially when used in association with the GM detection test. As other authors have shown (5, 19), PCR analysis of these specimens is particularly suited to early detection of IPA, and while BAL is not a noninvasive procedure, it is certainly less traumatic than lung biopsy, which can be an important consideration for critically ill patients such as the ones included in our study. Prospective studies are under way in our lab to determine the actual diagnostic value of the new assay.
Acknowledgments
This study was supported by a grant from Istituto Superiore di Sanità (3rd National Program of Research on AIDS, grant number 50 C.9).
We thank David Perlin (Public Health Research Institute, Newark, N.J.) for the A. fumigatus H11-20 strain.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. J., S. de Marie, D. Willemse, H. A. Verbrugh, and I. A. Bakker-Woudenberg. 2000. Quantitative galactomannan detection is superior to PCR in diagnosing and monitoring invasive pulmonary aspergillosis in an experimental rat model. J. Clin. Microbiol. 38:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretagne, S., J. M. Costa, A. Marmorat-Khuong, F. Poron, C. Cordonnier, M. Vidaud, and J. Fleury-Feith. 1995. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J. Clin. Microbiol. 33:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretagne, S., J. M. Costa, E. Bart-Delabesse, N. Dhedin, C. Rieux, and C. Cordonnier. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., C. Baust, H. Skladny, J. Ritter, T. Suedhoff, M. Baldus, W. Seifarth, C. Leib-Moesch, and R. Hehlmann. 2001. Detection of Aspergillus species in blood and bronchoalveolar lavage samples from immunocompromised patients by means of 2-step polymerase chain reaction: clinical results. Clin. Infect. Dis. 33:428-435. [DOI] [PubMed] [Google Scholar]
- 6.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 8.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferns, R. B., H. Fletcher, S. Bradley, S. Mackinnon, C. Hunt, and R. S. Tedder. 2002. The prospective evaluation of a nested polymerase chain reaction assay for the early detection of Aspergillus infection in patients with leukaemia or undergoing allograft treatment. Br. J. Haematol. 119:720-725. [DOI] [PubMed] [Google Scholar]
- 10.Hayette, M. P., D. Vaira, F. Susin, P. Boland, G. Christiaens, P. Melin, and P. De Mol. 2001. Detection of Aspergillus species DNA by PCR in bronchoalveolar lavage fluid. J. Clin. Microbiol. 39:2338-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 12.Jones, M. E., A. J. Fox, A. J. Barnes, B. A. Oppenheim, P. Balagopal, G. R. Morgenstern, and J. H. Scarffe. 1998. PCR-ELISA for the early diagnosis of invasive pulmonary Aspergillus infection in neutropenic patients. J. Clin. Pathol. 51:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura, S., S. Maesaki, T. Noda, Y. Hirakata, K. Tomono, T. Tashiro, and S. Kohno. 1999. Comparison between PCR and detection of antigen in sera for diagnosis of pulmonary aspergillosis. J. Clin. Microbiol. 37:218-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz, M. B., E. M. Bernard, F. F. Edwards, J. A. Marrinan, J. Dropinski, C. M. Douglas, and D. Armstrong. 1995. Aerosol and parenteral pneumocandins are effective in a rat model of pulmonary aspergillosis. Antimicrob. Agents Chemother. 39:1784-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lass-Flörl, C., J. Aigner, E. Gunsilius, A. Petzer, D. Nachbaur, G. Gastl, H. Einsele, J. Loffler, M. P. Dierich, and R. Wurzner. 2001. Screening for Aspergillus spp. using polymerase chain reaction of whole-blood samples from patients with haematological malignancies. Br. J. Haematol. 113:180-184. [DOI] [PubMed] [Google Scholar]
- 18.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy, H., D. A. Horak, B. R. Tegtmeier, S. B. Yokota, and S. J. Forman. 1992. The value of bronchoalveolar lavage and bronchial washings in the diagnosis of invasive pulmonary aspergillosis. Respir. Med. 86:243-248. [DOI] [PubMed] [Google Scholar]
- 20.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the Light Cycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maertens, J., J. Verhaegen, K. Lagrou, J. Van Eldere, and M. Boogaerts. 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood 97:1604-1610. [DOI] [PubMed] [Google Scholar]
- 22.Melchers, W. J., P. E. Verweij, P. van den Hurk, A. van Belkum, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 24.Pagano, L., C. Girmenia, L. Mele, P. Ricci, M. E. Tosti, A. M. Nosari, M. Buelli, M. Picardi, B. Allione, L. Corvatta, D. D'Antonio, M. Montillo, L. Melillo, A. Chierichini, A. R. Cenacchi, A. Tonso, L. Cudillo, A. Candoni, C. Savignano, A. Bonini, P. Martino, and A. Del Favero. 2001. Infections caused by filamentous fungi in patients with hematologic malignancies—a report of 391 cases by GIMEMA Infection Program. Haematologica 86:862-870. [PubMed] [Google Scholar]
- 25.Perea, S., and T. F. Patterson. 2002. Invasive Aspergillus infections in hematologic malignancy patients. Semin. Respir. Infect. 17:99-105. [DOI] [PubMed] [Google Scholar]
- 26.Skladny, H., D. Buchheidt, C. Baust, F. Krieg-Schneider, W. Seifarth, C. Leib-Mosch, and R. Hehlmann. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stynen, D., A. Goris, J. Sarfati, and J. P. Latgé. 1995. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J. Clin. Microbiol. 33:497-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang, C. M., D. W. Holden, A. Aufauvre-Brown, and J. Cohen. 1993. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am. Rev. Respir. Dis. 148:1313-1317. [DOI] [PubMed] [Google Scholar]
- 29.Verweij, P. E., J. P. Latgé, A. J. Rijs, W. J. Melchers, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1995. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J. Clin. Microbiol. 33:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamakami, Y., A. Hashimoto, I. Tokimatsu, and M. Nasu. 1996. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J. Clin. Microbiol. 34:2464-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]