Abstract
NSP4-encoding genes of 78 human rotavirus strains of common or reassortant genotypes were characterized by reverse transcription-PCR followed by sequencing and phylogenetic analysis. It was found that all the human strains characterized clustered into only two of the five known NSP4 genotypes. Linkage between NSP4 genotypes and VP6 subgroups was 100%, NSP4 genotype A being linked to VP6 of subgroup I (SGI) and NSP4 of genotype B being linked to VP6 of SGII. The diversity among the NSP4- and VP6-encoding genes was significantly less than that among the VP7 and VP4 genes in cocirculating human rotavirus strains. Whereas G and P types appear to be shared among different animal species and humans, the NSP4- and VP6-encoding genes appear to segregate according to their host of origin, suggesting that these two proteins may be host restriction determinants. The NSP4-VP6 association may be structurally determined during rotavirus replication (morphogenesis).
Group A rotaviruses are the commonest etiologic agents of acute gastroenteritis in children and infants worldwide (22). They are ubiquitous viruses, also infecting the young of many animal species (22). The rotavirus genome consists of 11 segments of double-stranded RNA that encodes six structural and six nonstructural proteins. Group A rotaviruses are classified into subgroups, G types, and P types according to serological reactivities of the structural proteins VP6, VP7, and VP4, respectively, which constitute the middle and outer shells of the virions, or according to the nucleotide sequences of the genes encoding these proteins, gene segments 6, 9 (or 7 or 8, depending on the strain), and 4, respectively (12). Epidemiological studies of rotavirus infections in humans have revealed a great diversity of G and P types and combinations thereof cocirculating in different parts of the world (reviewed in reference 9). Rotaviruses readily reassort upon dual infection of a single cell, and this phenomenon, observed both in vitro and in vivo, is believed to be the major contributor to the natural evolution of rotaviruses (19).
The existence of genotypes is not, however, limited to the structural proteins. Studies of the genetic diversity of rotavirus gene segment 10, encoding nonstructural protein 4 (NSP4), have identified five genotypes (termed A to E) among human and animal rotaviruses of different G and P types (7, 8, 15, 28). NSP4 genotypes appear to cluster according to rotavirus host species, suggesting a conserved pattern of evolution within species (7).
NSP4 is a cotranslationally glycosylated 175-amino-acid (aa) protein that contains a noncleavable signal peptide (10, 11). The amino terminus of this protein interacts with the endoplasmic reticulum (ER) membrane and contains three hydrophobic regions; the glycosylation sites are located in the first one, and the second and third hydrophobic regions are membrane-associated and transmembrane domains, respectively (6). The hydrophilic carboxy terminus extends into the cytoplasm (6) and functions as an intracellular receptor for nascent double-layered particles (DLPs), mediating their budding into the ER lumen (1, 27, 36). The budding of DLPs involves specific interactions between the VP6, constituting the outer layer of the DLPs, and the cytoplasmic carboxy-terminal domain of the membrane-bound NSP4 (2, 27). However, membrane association is not an essential prerequisite for binding, and the soluble NSP4 carboxy-terminal domain retains the capacity to bind DLPs (36). Although the domain at aa 156 to 175 (2, 32) is critical for DLP binding, there is evidence that this binding domain is dependent on the conformational integrity of the carboxy terminus of NSP4 for efficient DLP binding (2).
NSP4 and peptides derived therefrom have been shown to induce diarrhea in mice in a specific and age-dependent manner (3). Differences between the NSP4 sequences from rotaviruses causing symptomatic and asymptomatic infections have been reported in some studies (24, 34, 39) but not in others (15, 32, 33, 38). Also, antibodies against NSP4 have been found to reduce the ability of NSP4 to induce diarrhea, and vaccination with NSP4 has been found to induce homotypic and heterotypic protection against rotavirus-induced diarrhea in mice (13). In humans, NSP4 has been found to induce humoral and cellular immune responses (20).
NSP4-encoding genes of human rotavirus strains of common or reassortant genotypes were characterized in order to determine the variability of NSP4 among rotaviruses circulating in diverse geographical areas during different rotavirus seasons and to investigate whether the gene encoding NSP4 segregates independently during reassortment, as do other rotavirus genes (16, 19).
MATERIALS AND METHODS
A total of 78 human rotavirus strains of common or reassortant G and P genotype combinations were selected for NSP4 characterization by reverse transcription (RT)-PCR and sequence analysis. Thirty-nine of these strains were collected in the United Kingdom between 1995 and 2002 (17), and the remaining 39 were collected between 1995 and 1999 in Vellore, India (21). The G and P types of these strains were determined by RT-PCR as previously described (18). The subgroups of 58 of these strains (34 from the United Kingdom and 24 from Vellore) were determined by RT-PCR and sequence analysis of a 378-bp VP6 amplicon as previously described (16). The subgroups and G and P types of these strains are listed in Table 1
TABLE 1.
G types, P types, and subgroup specificities of 58 human rotaviruses selected for NSP4 characterization
| Strains | G type | P type | Sub- group | No. of strains analyzed |
|---|---|---|---|---|
| Common human rotavirus strains | G1 | P[8] | II | 14 |
| G2 | P[4] | I | 5 | |
| G3 | P[8] | II | 4 | |
| G4 | P[8] | II | 8 | |
| Human rotavirus reassortant strains | G1 | P[4] | I | 3 |
| G1 | P[4] | II | 4 | |
| G3 | P[8] | I | 1 | |
| G4 | P[4] | I | 1 | |
| G4 | P[4] | II | 3 | |
| G4 | P[8] | I | 1 | |
| G9 | P[6] | I | 2 | |
| G9 | P[8] | I | 1 | |
| G9 | P[8] | II | 4 | |
| Strains not fully characterized | G1 | ND | I | 1 |
| G1 | ND | II | 1 | |
| G8 | ND | I | 1 | |
| G8 | ND | II | 1 | |
| G9 | ND | II | 1 | |
| ND | P[8] | II | 1 | |
| ND | ND | I | 1 |
Nucleic acid was extracted from 200 μl of 10% rotavirus-positive fecal suspensions by the guanidinium isothiocyanate method (5), and RT was performed with random hexamers (18). The resulting cDNA was used for the VP6-, VP7-, VP4-, and NSP4-specific PCRs by using oligonucleotide primers and methods previously described (7, 16, 18). G and P types were determined by seminested PCR with type-specific primers (18). VP6 and NSP4 genotypes were determined by direct sequencing of the 379-bp and full-length amplicons of the VP6- and NSP4-encoding genes, respectively, with an automated sequencer (Beckman Coulter) followed by phylogenetic analysis with the Bionumerics software package (Applied Maths, Kortrij, Belgium). The NSP4 sequences obtained were aligned and compared to other NSP4 sequences of rotaviruses of human or animal origin available in GenBank/EMBL. Dendrograms were confirmed by two methods, neighbor joining and maximum parsimony. Genotypes and genetic lineages within genotypes were confirmed by bootstrap values of >90%.
The NSP4 sequences described in this work are freely available from the corresponding author upon request.
RESULTS
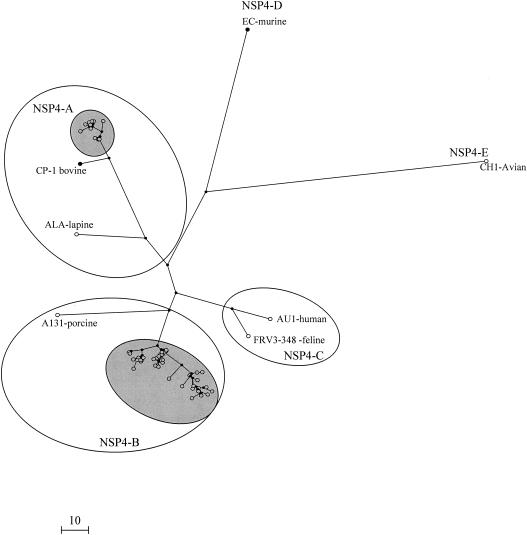
The 78 NSP4 sequences analyzed clustered in two groups (Fig. 1) Of these, 57 sequences, 26 from rotavirus strains isolated in India and 31 United Kingdom strains, clustered within genotype B. Twenty-one sequences, 13 and 8 from strains isolated in India and the United Kingdom, respectively, clustered within genotype A (Fig. 1).
FIG. 1.
Unrooted dendrogram constructed by using rotavirus NSP4 nucleotide sequences (coding region) and the maximum parsimony method. The NSP4 cDNA sequences of the 78 human rotavirus strains characterized in this study are highlighted. Sequences representative of the different existing NSP4 genotypes were included for comparison (accession numbers: EC, U96337; A131, AF144798; ALA, AF144793; CP-1, AF448854; CH1, AB065287; AU1, D89873; FR3-348, AB 048203). The calibration bar indicates the number of base conversions.
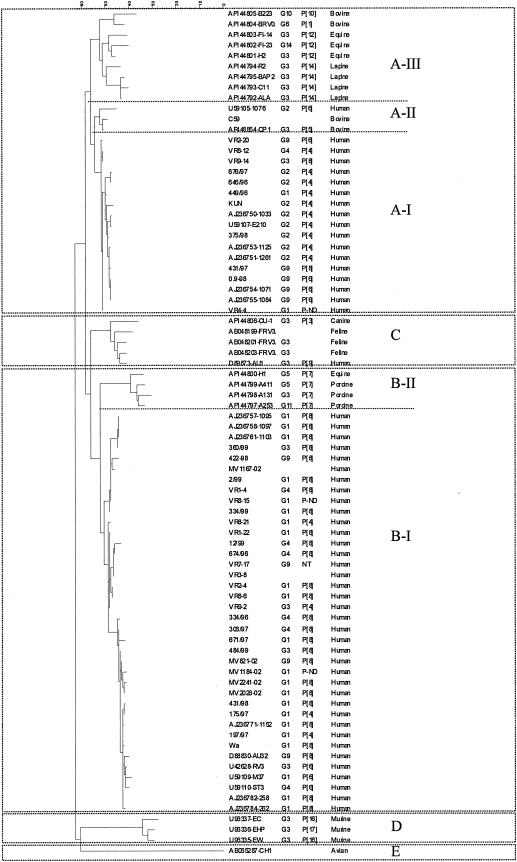
Genetic lineages were distinguished among the sequences in genotypes A and B when known sequences of animal and human rotavirus strains (available from GenBank/EMBL) were included in the phylogenetic analysis (Fig. 2) Three clusters, or genetic lineages, were identified within genotype A. Lineage A-I uniquely comprised sequences from human strains, whereas the other two, lineages A-II and A-III, comprised sequences derived from animal rotavirus strains (bovine, equine, and lapine). A single human strain isolated in Australia clustered in lineage A-II (Fig. 2). Prototype human rotavirus strain AU1 (D89843), isolated in Japan, is the only human strain with an NSP4 genotype other than A or B and clustered with feline and canine strains of NSP4 genotype C. Three genetic lineages were distinguished in genotype B. Lineage B-I consisted uniquely of sequences derived from human strains, and lineage B-II comprised sequences of animal (equine or porcine) rotaviruses (Fig. 2). Genotype D was composed of sequences derived from murine rotavirus strains, and genotype E was defined by an NSP4 sequence derived from an avian rotavirus (Fig. 2).
FIG. 2.
Phylogenetic tree of rotavirus NSP4 sequences constructed by the maximum parsimony method. The genotype-defining clusters were confirmed by bootstrap values of >90% (not shown). Sequences available from GenBank/EMBL from human and animal rotavirus strains (accession numbers and strain identifiers are in the first column) were included alongside representative NSP4 nucleotide sequences of human strains from this study for comparison. G and P type (where available) and host of origin are indicated after each strain identifier or accession number.
A 100% linkage between subgroup and NSP4 genotype was identified among the human common and reassortant strains for which G, P, and subgroup data were available: Of the 58 strains for which G and P types, subgroups, and NSP4 genotypes were known, all 17 strains of NSP4 genotype A had a VP6 of subgroup I (SGI), whereas all 41 strains of NSP4 genotype B had a VP6 of SGII. By contrast, no associations were found between the NSP4 genotype and G or P types (Fig. 3)
FIG. 3.
Phylogenetic tree of rotavirus NSP4 sequences (maximum parsimony method) derived from human strains of SGI or SGII; G and P type specificities, where available, are indicated. Sequences available at GenBank/EMBL were included for comparison. Lineages identified within NSP4 genotypes A or B, confirmed by bootstrap values of >90% (not shown), are indicated with horizontal lines. The NSP4 gene sequence of the human strain AU1 (accession number D89873) was included as an outlier.
When 17 NSP4 sequences of prototype human rotavirus strains or other rotavirus strains isolated in the United States (25) (available in GenBank/EMBL; accession numbers are given in Fig. 3) were included in the comparison, the segregation of NSP4 genotype with subgroup specificity remained 100%: NSP4 type A segregated with SGI, and NSP4 type B segregated with SGII (Fig. 3).
Three genetic lineages were distinguished among the sequences derived from human rotavirus strains in genotype B-I, which did not correlate with any temporal or geographical pattern of virus isolation (Fig. 3).
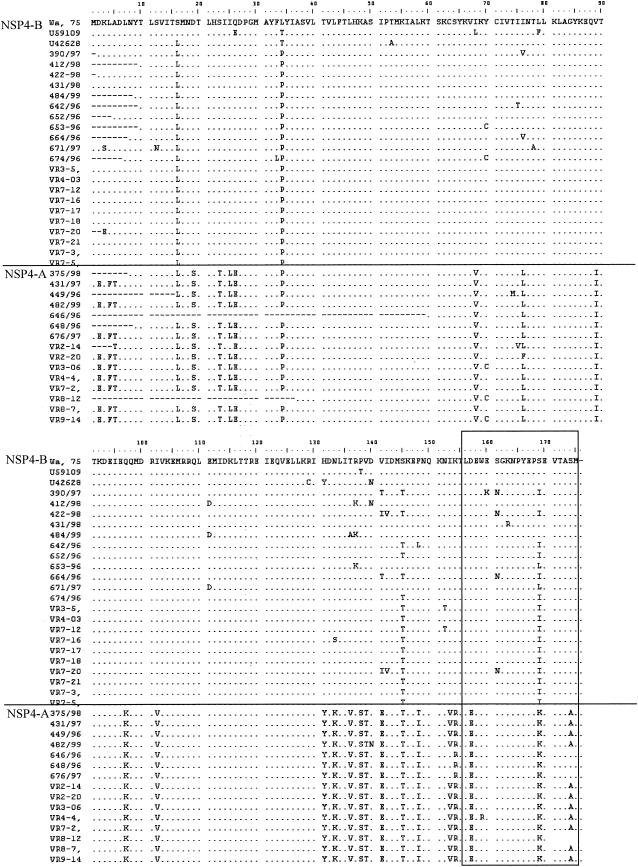
Alignment of the deduced amino acid sequences of NSP4 derived from human strains showed that a number of significant amino acid changes between the sequences of genotypes A and B were concentrated mainly in the cytoplasmic domain of the protein, within the DLP binding region (aa 156 to 175) (31), or in the region immediately upstream (aa 97 to 148) (Fig. 4)
FIG. 4.
Alignment of the deduced amino acid sequences of the genes encoding NSP4 genogroups A and B of human rotavirus strains. The DLP binding domain is boxed.
DISCUSSION
Although five rotavirus NSP4 genotypes have been identified to date (7, 8, 15, 28), the diversity of the NSP4-encoding genes among human rotaviruses is restricted to genotypes A or B. Previously, Ciarlet et al. (7) found that NSP4 clustering correlated with rotavirus host species. NSP4 sequences from animal rotaviruses which had previously been genotyped within NSP4 A or B according to their deduced amino acid sequences form clusters that are distinct from the human rotaviruses of NSP4 genotype A or B (Fig. 1 and 2). The only exception identified to date is the human strain AU1, which possesses an NSP4 C genotype, typical of feline and canine rotavirus strains. There is evidence through total genome hybridization and through sequence comparison of the VP4-encoding gene that this strain was of animal origin, most likely feline (30). The NSP4 sequence supports this further, strongly suggesting direct zoonotic transmission from a feline host. Interestingly, this strain does not appear to have spread widely in the human population.
A great variety of G and P types and combinations thereof have been identified in rotaviruses infecting humans, many of which are shared with rotaviruses found in different animal species (reviewed in reference 9). However, much less diversity has been found among the VP6-encoding genes of human rotaviruses. The molecular characterization of a region of rotavirus VP6 associated with subgroup specificity has shown that SGI and SGII are the only two VP6 subgroups occurring in human rotaviruses (16). Similarly, it has now emerged that the diversity of the NSP4-encoding genes among human rotavirus strains is restricted to two of the five genotypes that have been described to date.
It has become increasingly apparent that mixed infections and subsequent reassortment of rotaviruses occur relatively frequently in humans and that this is the principal mechanism generating rotavirus diversity (19). Gene biases during reassortment have been identified both in vitro and in vivo (14, 29), and particular associations of VP7, VP4, and VP6 appear to be more frequent than others. In general, G1, G3, and G4 strains appear to be more frequently associated with P[8] types and are mostly of SGII, and G2 strains are commonly associated with P[4] and SGI. However, other subgroup and G and P type combinations, resulting from reassortment upon dual infection, have been found in epidemiological studies (9, 17, 19). Recently evidence for the independent segregation of VP4, VP6, and VP7 was obtained through sequence analysis of the genes encoding these three rotavirus proteins from human rotavirus strains of common and of reassortant genotypes (16, 19).
The most significant finding of the work presented here is the 100% linkage between NSP4 genotype and VP6 subgroup in human rotaviruses, with NSP4 type A being associated always with SGI and NSP4 type B being associated always with SGII. Kirkwood et al. (23) reported a possible association between NSP4 genotype and subgroup through Northern hybridization studies of genomes of common human rotavirus strains with probes complementary to NSP4 genotypes A or B. No evidence for reassortment between genes 6 and 10 was found. The work presented here confirms this association and lack of reassortment between the genes encoding VP6 and NSP4 of particular subgroups or genotypes in common and reassortant human rotavirus strains, confirmed by sequence analysis of the genes encoding NSP4, VP4, VP6, and VP7. We have found no exception to this rule, even in reassortant strains in which VP6, VP7, and VP4 had segregated independently. This is the more remarkable as the human rotavirus strains investigated originated from very different population backgrounds.
This linkage may be a consequence of NSP4 acting as a receptor for VP6 during viral morphogenesis and may suggest coevolution of the genes encoding these two proteins. The segregation of both NSP4 genotypes and VP6 subgroups according to the rotavirus host species (7, 16) suggests that either of these two proteins or both may be important host restriction determinants.
There are at present no experimental data to indicate at which step of the replicative cycle this linkage may occur. One possibility is that conformational restrictions to interactions of particular NSP4 and VP6 molecules may occur during morphogenesis. Virus maturation is dependent on the internalization of the nascent DLP into the ER lumen, which is mediated by direct interaction between the ER membrane-bound NSP4 and the VP6 on the DLPs (1, 2, 4, 6, 27, 37). It is remarkable that the highest sequence diversity between NSP4 genotypes is located in the cytoplasmic domain of NSP4 (Fig. 4), which is responsible for interactions with VP6 (13). Several significant amino acid changes were observed between the cytoplasmic domains of NSP4s of genogroup A or B at aa 133, 136, 138, 139, 141, and 148 and also within the previously identified DLP binding domain (2, 32) at aa 170 (Fig. 4). It is therefore possible that only particular NSP4-VP6 interactions lead to internalization and maturation of the viral progeny. This may explain why even in reassortant strains the genes encoding NSP4 and VP6 cosegregate. This process may or may not be host specific for particular NSP4 and VP6 alleles.
The NSP4-VP6 linkage as well as other gene biases observed in reassortant viruses may be the result of the ability of the reassortant progeny to replicate efficiently in a given host. Further work is necessary to elucidate the mechanism by which NSP4 and VP6 interact to form this exclusive linkage.
Analysis of the NSP4- and VP6-encoding genes of animal rotavirus strains will reveal whether linkages also exist among the NSP4 and VP6 alleles not found in human rotavirus strains. Binding of SA11 DLPs to membranes expressing SA11-derived NSP4 was abolished by incubation of the DLPs with an SGI-specific monoclonal antibody (27), which recognizes aa 259 to 296 and 305 (26, 35). However, the blocking induced by SGI-specific antibody may be the result of steric hindrance, and there are no experimental data indicating whether this or any other subgroup-specific domain is implicated in NSP4 binding. Identification of the exact domain of VP6 responsible for binding to NSP4 and investigation of whether differences exist between the binding domains of the NSP4 and or VP6 proteins of different genotypes or subgroups, respectively, will contribute to the understanding of rotavirus morphogenesis. Such studies may also lead to a better understanding of interspecies transmission of rotaviruses and whether the successful introduction of novel rotaviruses into the human population through the acquisition of genes derived from animal strains depends on coinfection with human strains and the generation of novel strains with NSP4-VP6 genes characteristic of human rotavirus strains. Also, NSP4 characterization may be used as a tool for monitoring interspecies transmission of rotaviruses in epidemiological studies.
Early rotavirus studies using electropherotyping as a means of strain characterization are suggestive of a linkage between VP6 and NSP5, as SGI strains were always found in association with short electropherotypes and SGII was associated with long electropherotypes. Also, the study of rotavirus strain diversity using RNA-RNA hybridization and the proposed classification of rotaviruses into genogroups (reviewed in reference 31) may suggest the existence of linkages between other rotavirus gene segments, based on the scarce cross-hybridization observed between genogroups. However, these studies have also provided evidence of interspecies transmission of rotaviruses. Only detailed sequence analysis of whole genomes of a significant number of common and unusual rotavirus strains will determine with confidence the degree of diversity of all rotavirus gene segments and whether they can segregate independently during reassortment.
Acknowledgments
Support for this work was obtained through an MRC Research Training Fellowship awarded to Miren Iturriza-Gòmara and from a Wellcome Trilateral Infectious Disease Initiative grant (no. 063144).
We thank Max Ciarlet for helpful discussions and Annabel Guun for technical assistance.
REFERENCES
- 1.Au, K. S., W. K. Chan, J. W. Burns, and M. K. Estes. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au, K. S., N. M. Mattion, and M. K. Estes. 1993. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology 194:665-673. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. M., P. Tian, C. Q. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101-104. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, C. C., D. Maass, M. S. Poruchynsky, P. H. Atkinson, and A. R. Bellamy. 1989. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 8:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. A. Sol, M. M. M. Salismans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van den Noordaa. 1990. Rapid and simple method for the purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, W. K., K. S. Au, and M. K. Estes. 1988. Topography of the simian rotavirus nonstructural glycoprotein (NS28) in the endoplasmic reticulum membrane. Virology 164:435-442. [DOI] [PubMed] [Google Scholar]
- 7.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145:371-383. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe, N. A., P. A. Woods, J. P. Leite, B. K. Das, M. Ramachandran, M. K. Bhan, C. A. Hart, R. I. Glass, and J. R. Gentsch. 1997. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J. Med. Virol. 53:41-50. [PubMed] [Google Scholar]
- 9.Desselberger, U., M. Iturriza-Gòmara, and J. Gray. 2001. Rotavirus epidemiology and surveillance. Novartis Found Symp. 238:125-147. [DOI] [PubMed] [Google Scholar]
- 10.Ericson, B. L., D. Y. Graham, B. B. Mason, H. H. Hanssen, and M. K. Estes. 1983. Two types of glycoprotein precursors are produced by the simian rotavirus SA11. Virology 127:320-332. [DOI] [PubMed] [Google Scholar]
- 11.Ericson, B. L., B. L. Petrie, D. Y. Graham, B. B. Mason, and M. K. Estes. 1983. Rotaviruses code for two types of glycoprotein precursors. J. Cell. Biochem. 22:151-160. [DOI] [PubMed] [Google Scholar]
- 12.Estes, M. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Estes, M., G. Kang, C. Zeng, S. Crawford, and M. Ciarlet. 2001. Pathogenesis of rotavirus gastroenteritis. Novartis Found Symp. 238:83-96. [DOI] [PubMed] [Google Scholar]
- 14.Gombold, J. L., and R. F. Ramig. 1986. Analysis of reassortment of genome segments in mice mixedly infected with rotaviruses SA11 and RRV. J. Virol. 57:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie, Y., O. Masamune, and O. Nakagomi. 1997. Three major alleles of rotavirus NSP4 proteins identified by sequence analysis. J. Gen. Virol. 78:2341-2346. [DOI] [PubMed] [Google Scholar]
- 16.Iturriza-Gòmara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 76:6596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iturriza-Gòmara, M., J. Green, D. Brown, M. Ramsay, U. Desselberger, and J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iturriza-Gòmara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 19.Iturriza-Gòmara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen, K., J. Hinkula, F. Espinoza, M. Levi, C. Zeng, U. Ruden, T. Vesikari, M. Estes, and L. Svensson. 1999. Humoral and cell-mediated immune responses in humans to the NSP4 enterotoxin of rotavirus. J. Med. Virol. 59:369-377. [PubMed] [Google Scholar]
- 21.Kang, G., J. Green, C. I. Gallimore, and D. W. Brown. 2002. Molecular epidemiology of rotaviral infection in South Indian children with acute diarrhea from 1995-1996 to 1998-1999. J. Med. Virol. 67:101-105. [DOI] [PubMed] [Google Scholar]
- 22.Kapikian, A., Y. Hoshino, and R. Chanock. 2001. Rotaviruses, p. 1787-1834. In D. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Kirkwood, C., J. Gentsch, and R. Glass. 1999. Sequence analysis of the NSP4 gene from human rotavirus strains isolated in the United States. Virus Genes 19:113-122. [DOI] [PubMed] [Google Scholar]
- 24.Kirkwood, C. D., B. S. Coulson, and R. F. Bishop. 1996. G3P2 rotaviruses causing diarrhoeal disease in neonates differ in VP4, VP7 and NSP4 sequence from G3P2 strains causing asymptomatic neonatal infection. Arch. Virol. 141:1661-1676. [DOI] [PubMed] [Google Scholar]
- 25.Kirkwood, C. D., and E. A. Palombo. 1997. Genetic characterization of the rotavirus nonstructural protein, NSP4. Virology 236:258-265. [DOI] [PubMed] [Google Scholar]
- 26.Lòpez, S., R. Espinosa, H. B. Greenberg, and C. F. Arias. 1994. Mapping the subgroup epitopes of rotavirus protein VP6. Virology 204:153-162. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, J. C., C. C. Bergmann, and A. R. Bellamy. 1989. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology 171:98-107. [DOI] [PubMed] [Google Scholar]
- 28.Mori, Y., M. A. Borgan, N. Ito, M. Sugiyama, and N. Minamoto. 2002. Diarrhea-inducing activity of avian rotavirus NSP4 glycoproteins, which differ greatly from mammalian rotavirus NSP4 glycoproteins in deduced amino acid sequence in suckling mice. J. Virol. 76:5829-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossel, E. C., and R. F. Ramig. 2002. Rotavirus genome segment 7 (NSP3) is a determinant of extraintestinal spread in the neonatal mouse. J. Virol. 76:6502-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagomi, T., and O. Nakagomi. 1989. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J. Virol. 63:1431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagomi, O., and T. Nakagomi. 2002. Genomic relationships among rotaviruses recovered from various animal species as revealed by RNA-RNA hybridisation assays. Res. Vet. Sci. 73:207-214. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, J. A., J. A. Taylor, and A. R. Bellamy. 2000. Probing the structure of rotavirus NSP4: a short sequence at the extreme C terminus mediates binding to the inner capsid particle. J. Virol. 74:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka, T., T. Nakagomi, and O. Nakagomi. 2001. A lack of consistent amino acid substitutions in NSP4 between rotaviruses derived from diarrheal and asymptomatically-infected kittens. Microbiol. Immunol. 45:173-177. [DOI] [PubMed] [Google Scholar]
- 34.Pager, C. T., J. J. Alexander, and A. D. Steele. 2000. South African G4P[6] asymptomatic and symptomatic neonatal rotavirus strains differ in their NSP4, VP8*, and VP7 genes. J. Med. Virol. 62:208-216. [DOI] [PubMed] [Google Scholar]
- 35.Tang, B., J. M. Gilbert, S. M. Matsui, and H. B. Greenberg. 1997. Comparison of the rotavirus gene 6 from different species by sequence analysis and localization of subgroup-specific epitopes using site-directed mutagenesis. Virology 237:89-96. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, J. A., J. A. O'Brien, V. J. Lord, J. C. Meyer, and A. R. Bellamy. 1993. The RER-localized rotavirus intracellular receptor: a truncated purified soluble form is multivalent and binds virus particles. Virology 194:807-814. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, J. A., J. A. O'Brien, and M. Yeager. 1996. The cytoplasmic tail of NSP4, the endoplasmic reticulum-localized non-structural glycoprotein of rotavirus, contains distinct virus binding and coiled coil domains. EMBO J. 15:4469-4476. [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, R. L., B. B. Mason, D. I. Bernstein, D. S. Sander, V. E. Smith, G. A. Zandle, and R. S. Rappaport. 1997. Attenuation of a human rotavirus vaccine candidate did not correlate with mutations in the NSP4 protein gene. J. Virol. 71:6267-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, M., C. Q. Zeng, Y. Dong, J. M. Ball, L. J. Saif, A. P. Morris, and M. K. Estes. 1998. Mutations in rotavirus nonstructural glycoprotein NSP4 are associated with altered virus virulence. J. Virol. 72:3666-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]