Abstract
We evaluated the recently developed dipsticks for the rapid detection of Vibrio cholerae serotypes O1 and O139 from rectal swabs of hospitalized diarrheal patients after enrichment for 4 h in alkaline peptone water. The sensitivity and specificity of the dipsticks were above 92 and 91%, respectively. The dipsticks represent the first rapid test which has been successfully used to diagnose cholera from rectal swabs, and this would immensely improve surveillance for cholera, especially in remote settings.
In recent years, cholera has been becoming endemic in an increasing number of geographical areas, reflecting a failure of socioeconomic infrastructure and difficulties in implementation of control measures. Nearly 120 countries have reported indigenous cases of cholera since 1991; almost half of them have reported cases for at least 5 of the past 8 years (12). The current and ongoing seventh pandemic of cholera is caused by the El Tor biotype of Vibrio cholerae serogroup O1, which started in Indonesia in 1961 and reached Africa in the 1970s and South America in 1991 (7). Since 1992, V. cholerae O139, a variant of the El Tor biotype, has also spread to many parts of Asia, and, like the O1 strains, it has pandemic potential (4). The emergence of the O139 epidemic strain of V. cholerae is believed to have resulted from horizontal transfer of a fragment of DNA from an unknown donor into the region responsible for O-antigen biosynthesis of the seventh-pandemic V. cholerae O1 El Tor strain (3, 11). A conspicuous increase in the association of V. cholerae O139 with cholera outbreaks in India was recently reported (10). Likewise, a large outbreak of cholera caused predominantly by V. cholerae O139 occurred in Dhaka and adjoining areas of Bangladesh during March and April 2002, with an estimated 30,000 cases (5). Therefore, cholera continues to be a major problem in many developing countries, and both the O1 and the O139 serogroups are prevalent.
Cholera surveillance remains an important instrument for determining cholera trends in different regions of the world as well as within individual countries. The official annual World Health Organization figures on global incidence of cholera (13) underestimate the actual figures severalfold, since cholera is underreported and cholera surveillance figures represent only a small fraction of the actual number of people infected (9). Among the enteric pathogens, V. cholerae is perhaps the easiest to identify, but identification requires a laboratory infrastructure. Cholera is a disease of the poor, and outbreaks and epidemics of cholera usually occur in peripheral or war-ravaged areas where laboratory facilities are unavailable or are grossly inadequate. Consequently, efforts have been made to develop simple diagnostic tests which would allow the diagnosis of toxigenic strains of V. cholerae in the field itself. These rapid tests have had various degrees of success, but to date none have become widely available or are widely marketed. All the rapid tests for cholera developed so far require cholera stool samples, and none of the rapid tests have addressed the issue of rapid diagnosis of cholera from rectal swabs, which is usually how the specimen is received from the field or from remote settings.
Investigators at the Institute Pasteur, Paris, France, recently developed a one-step immunochromatographic dipstick test for rapid detection of V. cholerae O1 and O139 from stool samples. As reported previously, the detection thresholds with purified lipopolysaccharide (LPS) were 10 ng/ml for V. cholerae O1 and 50 ng/ml for V. cholerae O139 (8). In an evaluation of the dipstick assays in Madagascar and Bangladesh, two areas where cholera is endemic, the specificity of the O1 and O139 dipsticks ranged between 84 and 100% and the sensitivity ranged from 94.2 to 100% (8). The dipstick method requires minimal technical skill, and the test can be read in about 10 min. Additionally, the dipsticks can be stored at room temperature in a humidity-proof plastic bag, making them easily transportable (8). This prompted us to evaluate the efficacy of the dipsticks for the rapid detection of cholera from rectal swabs after a short enrichment.
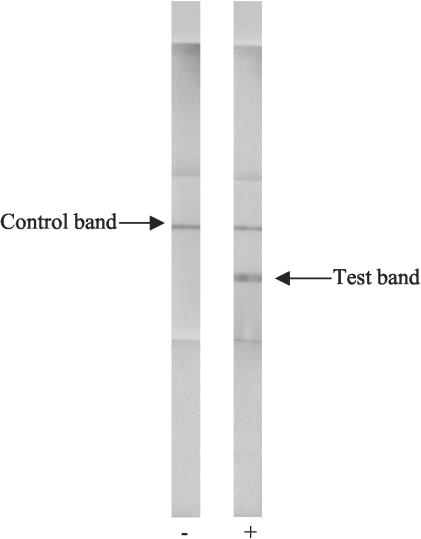
Stool specimens were collected from patients enrolled in the 2% systematic routine surveillance system at the Clinical Research and Service Centre of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B). In this surveillance system, every 50th patient attending the hospital is screened for major enteric pathogens. The Dhaka hospital of the ICDDR,B identifies cases of cholera throughout the year and treats patients during large outbreaks of the disease. For this study, single rectal swabs were obtained from 134 patients with severe watery diarrhea who were seen at the ICDDR,B hospital in Dhaka between August and October 2002. Rectal swabs were immediately placed into Cary Blair medium (in grams per liter: sodium thioglycolate, 1.5 g; disodium phosphate, 1.1.g; calcium chloride, 0.1 g; sodium chloride, 5 g; agar, 5 g; pH 8.4) and transported to the laboratory within 30 min. In the laboratory, the rectal swabs were placed in alkaline peptone water (APW; in grams per liter: Bacto Peptone, 10 g; sodium chloride, 10 g; pH 8.8) and incubated at 37°C for 4 h. The dipstick test was performed by simultaneously introducing both the O1 and O139 dipsticks into 200 μl of the 4-h-enriched broth contained in a fresh tube such that the end containing the absorbent pad was partially in the enriched broth sample. A positive result appeared as two pink lines (upper control line and lower LPS-positive line), and a negative result was a single upper pink control line; the results became discernible within 10 min (Fig. 1). Dipsticks that did not give a positive test result in 10 min were kept in the enriched stool sample for an additional 30 min to see if they would yield delayed positive results. The dipsticks were made available from the Institute Pasteur and were sent to the ICDDR,B laboratory at an ambient temperature in grip seal bags by Federal Express. The rectal swabs as well as the 4-h-enriched broth were cultured by inoculation onto taurocholate tellurite gelatin agar (in grams per liter: Trypticase, 10 g; sodium chloride, 10 g; sodium taurocholate, 5 g; sodium carbonate, 1 g; gelatin, 30 g; agar, 15 g; pH 8.5; after autoclaving, the agar is cooled to 50°C, 5 ml of 0.1% potassium tellurite solution is added and mixed well, and the agar is poured into plates). Suspected colonies resembling V. cholerae were tested by slide agglutination with polyvalent anti-O1 and anti-O139 sera. Samples that were positive by either the O1 or O139 dipstick but negative by culture were stored at −20°C and later examined by a multiplex PCR for concurrent detection of wbe and wbf sequences specific for O1 and the O139 serogroups of V. cholerae, respectively, and for ctxA-specific sequences (6). Toxigenic V. cholerae O1 (strain MAK 757 El Tor) and O139 (strain AI1852) were used as positive controls for the multiplex PCR.
FIG. 1.
Two dipsticks showing typical negative and positive results after being kept for 10 min in secretory diarrhea stool samples.
The clinical characteristics of all 134 patients from whom the rectal swabs were taken for this evaluation study are shown in Table 1. For the O1 dipstick evaluation, 65 of the 134 rectal swabs after enrichment were both dipstick and culture positive, 5 were dipstick positive culture negative, 3 were dipstick negative and culture positive, and 61 were negative by both tests (Table 2). We further analyzed the five samples that were O1 dipstick positive but culture negative by the multiplex PCR and found that all five were negative by PCR for the O1-specific 192-bp amplicon and for the 308-bp ctxA amplicon, indicating that the five dipstick-positive results were false-positives. The sensitivity of the O1 dipstick on enriched rectal swabs compared to culture was 96%, with a specificity of 92% and positive predictive value of 93%.
TABLE 1.
Clinical characteristics of patients
| Characteristic | No. of patients (%)
|
|||
|---|---|---|---|---|
| With V. cholerae
|
Others (n = 39) | All patients (n = 134) | ||
| O1 (n = 68) | O139 (n = 27) | |||
| Age (yr) | ||||
| <5 | 19 (27.9) | 1 (3.7) | 2 (5.1) | 22 (16.4) |
| 6-14 | 17 (25.0) | 2 (7.4) | 5 (12.8) | 24 (17.9) |
| 15-45 | 27 (39.7) | 20 (74.1) | 20 (51.3) | 67 (50.0) |
| 45 | 5 (7.4) | 4 (14.8) | 12 (30.8) | 21 (15.7) |
| Mean ± SD | 15.9 ± 14.7 | 29.1 ± 14.3 | 32.8 ± 19.2 | 23.5 ± 17.8 |
| Median | 10.0 | 26.0 | 30.0 | 20.0 |
| Range | 0.2-60.0 | 3.0-55.0 | 1.1-70.0 | 0.2-70.0 |
| Watery stool | 68 (100) | 27 (100) | 39 (100) | 134 (100) |
| Vomiting | 68 (100) | 27 (100) | 37 (94.9) | 132 (98.5) |
| Abdominal pain | 34 (50.0) | 12 (44.4) | 24 (61.5) | 70 (52.2) |
| Duration of diarrhea (days) | ||||
| <1 | 62 (91.2) | 26 (96.3) | 34 (87.2) | 122 (91.0) |
| 1-3 | 5 (7.4) | 1 (3.7) | 5 (12.8) | 11 (8.2) |
| 4-6 | 1 (1.5) | 1 (0.7) | ||
| Dehydration status | ||||
| None | 1 (1.5) | 1 (0.7) | ||
| Some | 10 (14.7) | 1 (3.7) | 4 (10.3) | 15 (11.2) |
| Severe | 57 (83.8) | 26 (96.3) | 35 (89.7) | 118 (88.1) |
| Duration of stay at hospital (h) | 59 | 27 | 35 | 121 |
| 0-11 | 10 (16.9) | 8 (29.6) | 18 (51.4) | 36 (29.8) |
| 12-23 | 8 (13.6) | 8 (29.6) | 6 (17.1) | 22 (18.2) |
| 24-47 | 29 (49.2) | 10 (37.0) | 11 (31.4) | 50 (41.3) |
| 48-95 | 11 (18.6) | 1 (3.7) | 12 (9.9) | |
| 96+ | 1 (1.7) | 1 (0.8) | ||
TABLE 2.
Sensitivity and specificity of the O1 dipstick test in comparison with culture of V. cholerae O1
| Culture result (24 h) | No. of samples with dipstick result (4 h of APW culture)a
|
Total | |
|---|---|---|---|
| + | − | ||
| + | 65 | 3 | 68 |
| − | 5 | 61 | 66 |
| Total | 70 | 64 | 134 |
Sensitivity of the O1 dipstick test was 96%, specificity was 92%, and positive predictive value was 93%.
For the O139 dipstick evaluation, of the 134 stool samples, 25 were both dipstick and culture positive, 2 were dipstick positive but culture negative, 2 were dipstick negative but culture positive, and 105 were negative by both dipstick and culture (Table 3). In one case, the dipstick was positive for both O1 and O139, which was subsequently confirmed by culture. The two samples which were positive by the O139 dipstick but negative by culture were examined by the multiplex PCR. Both samples were negative for the 449-bp O139-specific band and for the 308-bp ctxA-specific band, indicating that in these samples the dipstick gave a false-positive result. The sensitivity of the O139 dipstick on enriched rectal swabs compared to culture was 93%, with a specificity of 98% and a positive predictive value of 93%.
TABLE 3.
Sensitivity and specificity of the O139 dipstick test in comparison with culture of V. cholerae O139
| Culture result (24 h) | No. of samples with dipstick result (4 h of culture)a
|
Total | |
|---|---|---|---|
| + | − | ||
| + | 25 | 2 | 27 |
| − | 2 | 105 | 107 |
| Total | 27 | 107 | 134 |
Sensitivity of the O139 dipstick test was 93%, specificity was 98%, and positive predictive value was 93%.
Overall, the sensitivity and specificity of the dipstick tests for detection of V. cholerae O1 or O139 from rectal swabs were excellent. All samples which were negative for test results after 10 min remained negative after the dipsticks were kept for an additional 30 min in the stool sample. Compared to our previous evaluation of the dipsticks directly on stool samples (8), the dipsticks were more efficient for detection of cholera from enriched rectal swabs than directly from stool samples. Although we did encounter false positives and false negatives with both the O1 and O139 dipsticks, such results were few. At this point, we do not have an explanation for the false positives. In the case of V. cholerae O139, at least, previous studies have shown that other bacteria within the family Vibrionaceae and also strains of Aeromonas trota share somatic antigens with O139 (1, 2). The false negatives may relate to the fact that the numbers of V. cholerae present in the broth after 4 h of enrichment were below the detection thresholds of the O1 and O139 dipsticks. The detection thresholds with purified LPS are 10 ng/ml for V. cholerae O1 and 50 ng/ml for V. cholerae O139 (8). However, this clearly was the exception rather than the rule. The prior use of antibiotics by some patients may have lessened the excretion of viable V. cholerae in the rectal swabs.
In real-life situations, most specimens for diagnosis of cholera from the field or from remote areas come as rectal swabs. Both the O1 and O139 dipsticks performed well with rectal swabs enriched for 4 h in APW. The dipsticks was also useful in identifying a case of mixed infection caused by both O1 and O139, which would not have been recognized in bacteriological culture, because usually only one or two colonies are picked for serotyping. The introduction of dipsticks will help identify cholera cases in the most remotely affected regions because the test is so simple and does not require any special infrastructure. The dipstick method in its current format cannot specifically detect toxigenic V. cholerae O1 or O139 strains, and therefore an additional step would be required to detect cholera toxin production. However, more than 95% of O1 and O139 strains are toxigenic (4). Efforts to integrate both O1 and O139 and the ability to detect cholera toxin production into one dipstick as opposed to two or more dipsticks are ongoing. This would simplify the test further and make it more informative.
Acknowledgments
Research work in the ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States.
REFERENCES
- 1.Albert, M. J., M. Ansaruzzaman, T. Shimada, A. Rahman, N. A. Bhuiyan, S. Nahar, F. Qadri, and M. S. Islam. 1995. Characterization of Aeromonas trota strains that cross-react with Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 33:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaruzzaman, M., T. Shimada, N. A. Bhuiyan, S. Nahar, M. S. Islam, and M. J. Albert. 1999. Cross-reaction between a strain of Vibrio mimicus and V. cholerae O139 Bengal. J. Med. Microbiol. 48:873-877. [DOI] [PubMed] [Google Scholar]
- 3.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, Q. S. Ahmad, A. S. G. Faruque, M. A. Salam, T. Ramamurthy, G. B. Nair, A. Weintraub, and D. A. Sack. 2002. Analysis of reemergent Vibrio cholerae O139 strains associated with an epidemic of cholera in Bangladesh, p. 11-15. In Proceedings of the 37th US-Japan Joint Conference on Cholera and Other Bacterial Enteric Infections Panel, December 17-19, 2002, Naha, Okinawa, Japan. US-Japan Cooperative Medical Science Program, Bethesda, Md.
- 6.Hoshino, K., S. Yamasaki, A. K. Mukhopadhyay, S. Chakraborty, A. Basu, S. K. Bhattacharya, G. B. Nair, T. Shimada, and Y. Takeda. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol. Med. Microbiol. 20:201-207. [DOI] [PubMed] [Google Scholar]
- 7.Lacey, W. L. 1995. Cholera: calamitous past, ominous future. Clin. Infect. Dis. 20:1409-1419. [DOI] [PubMed] [Google Scholar]
- 8.Nato, F., A. Boutonnier, M. Rajerison, P. Grosjean, S. Dartevelle, A. Guenole, N. A. Bhuiyan, D. A. Sack, G. B. Nair, J. M. Fournier, and S. Chanteau. 2003. One-step immunochromatographic dipstick tests for rapid detection of Vibrio cholerae O1 and O139 in stool samples. Clin. Diagn. Lab. Immunol. 10:476-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sack, D. A. 1995. Underestimating the cholera problem and the potential for vaccination---a case for accelerating the use of cholera vaccines. Bull. Inst. Pasteur 93:229-235. [Google Scholar]
- 10.Sinha, S., R. Chakraborty, K. De, A. Khan, S. Datta, T. Ramamurthy, S. K. Bhattacharya, Y. Takeda, and G. B. Nair. 2002. Escalating association of Vibrio cholerae O139 with cholera outbreaks in India. J. Clin. Microbiol. 40:2635-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stroeher, U. H., K. E. Jedani, and P. A. Manning. 1998. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene 223:269-282. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. 2001. Cholera vaccines. WHO position paper. Wkly. Epidemiol. Rec. 76:117-124.11338983 [Google Scholar]
- 13.World Health Organization. 2002. Cholera 2001. Wkly. Epidemiol. Rec. 77:257-268. [PubMed] [Google Scholar]