Abstract
Background
Tumour necrosis factor α (TNFα) is a cytokine of critical importance in psoriatic arthritis.
Objectives
(1) To examine the association between TNFα promoter gene polymorphisms and psoriatic arthritis in two well characterised Canadian populations with the disease; (2) to carry out a meta‐analysis of all TNFα association studies in white psoriatic arthritis populations.
Methods
DNA samples were genotyped for five TNF variants by time of flight mass spectrometry using the Sequenom platform. All five single nucleotide polymorphisms were in the 5′ flanking region of TNFα gene at the following positions: −1031 (T→C), −863 (C→A), −857 (C→T), −308 (G→A), and −238 (G→A). Primary analyses were based on logistic regression. Summary estimates of disease/genotype relations from several studies were derived from random effects meta‐analyses.
Results
237 psoriatic arthritis subjects and 103 controls from Newfoundland and 203 psoriatic arthritis subjects and 101 controls from Toronto were studied. A combined analysis of data from both populations, showed a significant association between disease status and the −238(A) variant (p = 0.01). The meta‐analysis estimate for the −238(A) TNFα variant in eight psoriatic arthritis populations was also significant (odds ratio = 2.29 (95% confidence interval, 1.48 to 3.55)).
Conclusions
Analysis of TNFα variants in psoriatic arthritis populations shows that the −238 (A) variant is a significant risk factor for this disease.
Keywords: psoriatic arthritis, TNFα, meta‐analysis, genetic association studies
Psoriatic arthritis is a complex immunologically mediated disorder that results from interplay between multiple genetic and environmental factors.1 Although the pathogenesis of psoriatic arthritis is still unclear, there is a substantial contribution of genetic factors to the aetiology of the disease.2 The MHC region has long been considered to harbour the underlying psoriatic arthritis susceptibility gene or genes and it has been estimated that at least one third of the genetic contribution of psoriatic arthritis resides within this region.2 The tumour necrosis factor α (TNFα) gene, which is located 250 kb centromeric from HLA‐B, has been proposed as a high priority candidate gene in psoriatic arthritis.3 This premise is supported by studies noting significantly higher levels of serum, synovial fluid, and synovial membrane levels of TNFα in patients with psoriatic arthritis than in those with osteoarthritis or healthy controls.4,5 The importance of TNFα in psoriatic arthritis is further strengthened by the marked clinical response of TNFα blockade.6 Thus TNFα appears to be a cytokine of critical importance in this disease.
However, genetic studies of TNFα polymorphisms and psoriatic arthritis have produced conflicting results.7,8,9,10,11,12,13,14 Certain studies have noted a significant association of TNFα polymorphisms in psoriatic arthritis,7,8 while others have failed to detect any such association.9,10,11,12 The inconsistencies in these results may reflect the small effect size of TNFα, insufficient sample size of subjects and controls, differences in populations, the presence of linkage disequilibrium, or multiple testing. In view of the location and proposed biological effect of TNFα, we felt it was prudent to evaluate further the relation between TNFα promoter polymorphisms and psoriatic arthritis in two well characterised Canadian psoriatic arthritis populations—a founder population from Newfoundland and a heterogeneous population from Toronto, Ontario. This was followed by a meta‐analysis of the TNFα association studies in psoriatic arthritis populations.
Methods
Patients
This study was approved by the local ethics committees at Memorial University of Newfoundland and the University of Toronto. Informed consent was obtained from all patients. Psoriatic arthritis was diagnosed as an inflammatory arthritis in patients with psoriasis, in the absence of other causes of inflammatory arthritis. All psoriatic arthritis probands were white. Information was collected systematically and included age at onset of psoriasis and psoriatic arthritis, and the disease pattern. The control subjects were of similar ethnicity to the cases. Controls for the Newfoundland population were volunteers from Newfoundland who participated in our study as a result of a local campaign seeking population based controls for genetic studies. The Toronto controls were ascertained from the local HLA laboratory DNA bank which includes healthy volunteers and organ donors.
Laboratory methods
Blood samples were collected from patient volunteers with psoriatic arthritis and healthy controls in EDTA anticoagulant. DNA was extracted from peripheral blood lymphocytes using the Wizard Genomic DNA purification kit from Promega (Madison, Wisconsin, USA). DNA samples were genotyped for five TNF variants by time of flight mass spectrometry using the Sequenom platform. All five single nucleotide polymorphisms (SNPs) were in the 5′ flanking region of TNFα gene at the following positions: −1031 (T→C), −863 (C→A), −857 (C→T), −308 (G→A), and −238 (G→A). Primer sequences were determined using Sequenom SpectroDESIGNER software v1.3.4 (table 1).
Table 1 Primer sequences for TNFα single nucleotide polymorphisms.
| SNP ID | rs No | Forward primer | Reverse primer | Mass extend primer | ||
|---|---|---|---|---|---|---|
| −1031 | rs1799964 | 5′‐ACG TTG GAT GGG GAA GCA AAG GAG AAG CTG‐3′ | 5′‐ACG TTG GAT GTA CAT GTG GCC ATA TCT CCC‐3′ | 5′‐GAC CCT GAC TTT TCC TTC‐3′ | ||
| −857 | rs1799724 | 5′‐ACG TTG GAT GCT ATG GAA GTC GAG TAT GGG‐3′ | 5′‐ACG TTG GAT GTA TTC CAT ACC TGG AGG TCC‐3′ | 5′‐CCT CTA CAT GGC CCT GTC TTC‐3′ | ||
| −238 | rs361525 | 5′‐ACG TTG GAT GAC ACA AAT CAG TCA GTG GCC‐3′ | 5′‐ACG TTG GAT GAT CAA GGA TAC CCC TCA CAC‐3′ | 5′‐AGA AGA CCC CCC TCG GAA TC‐3′ | ||
| −308 | rs1800629 | 5′‐ACG TTG GAT GGG TCC CCA AAA GAA ATG GAG‐3′ | 5′‐ACG TTG GAT GGA TTT GTG TGT AGG ACC CTG‐3′ | 5′‐GAG GCT GAA CCC CGT CC‐3′ | ||
| −863 | rs1800630 | 5′‐ACG TTG GAT GCT ATG GAA GTC GAG TAT GGG‐3′ | 5′‐ACG TTG GAT GTA TTC CAT ACC TGG AGG TCC‐3′ | 5′‐CGA GTA TGG GGA CCC CC‐3′ |
TNFα, tumour necrosis factor α.
For each sample, 2.5 ng of genomic DNA were amplified under standard conditions using the forward and reverse primer pairs. After DNA amplification, all unincorporated nucleotides in the polymerase chain reaction (PCR) product were deactivated using shrimp alkaline phosphatase. A primer extension reaction was then carried out using the mass extend primer and the appropriate termination mix. The primer extension products were then cleaned and spotted onto a SpectroChip. The chip was scanned using a mass spectrometry workstation (Bruker), and the resulting spectra were analysed using the Sequenom SpectroTYPER‐RT software.
With respect to the meta‐analysis, the literature was searched up to and including October 2004. All articles pertaining to TNFα polymorphisms in psoriatic arthritis were searched in Medline and Embase. In addition, the reference lists of the articles identified were also a searched. We then selected studies that examined white populations and provided enough data for the numbers of minor alleles and the total number of alleles to be determined. The data was extracted by PR and then cross checked by CB.
Statistical analysis
Logistic regression was used to study the relation between typing information and case/control status. The results are summarised in terms of odds ratios and significance tests. In addition an exploratory haplotype analysis based on EM imputation15 was conducted.
Pairwise linkage disequilibrium was assessed for all combinations of TNFα variants using χ2 tests. Strong associations were noted between −1031 and the other variants. The most notable association was between −863 and −1031 (p<0.001), which was present in separate populations, both Newfoundland (p<0.001) and Toronto (p<0.001). For this reason −1031 was dropped from the haplotype analysis, which was based on the −238, −308, −857, and −863 variants.
Meta‐analyses were based on random effects analyses.
Results
We examined 237 psoriatic arthritis patients and 103 controls from Newfoundland and 203 patients and 101 controls from Toronto. The mean (SD) age of the psoriatic arthritis group in Newfoundland was 50.1 (11.3) years, and in Toronto, 50.5 (13.2) years. Forty nine per cent of patients were female in the Newfoundland cohort and 41% were female in the Toronto cohort. With respect to the subtype of psoriatic arthritis, overall for both populations 61% had the polyarticular pattern, 27% the oligoarticular pattern, 5.2% the isolated spondyloarthropathy, and 7% were the remaining patterns. All probands had psoriasis vulgaris.
For the genotypes in the controls, there was no evidence of a departure from Hardy–Weinberg equilibrium (HWE) in the Newfoundland control population (p values were 0.13, 0.31, 0.29, 0.43, and 0.15 for 238(A), 308(A), 857(T), 863(A), and 1031(C), respectively). For the Toronto control population the comparable p values were 0.16, 0.04, 0.08, 0.24, and 0.23. While there is some weak evidence of departure from HWE for the 308(A) and possibly the 857(T) polymorphisms, this evidence would not survive any form of adjustment for multiple testing.
The genotypes and allele frequencies for psoriatic arthritis subjects and controls for TNFα polymorphisms in the Newfoundland and Toronto population are presented in tables 2 and 3, respectively. For completeness these tables present significance levels based on Fisher's exact test for an association between allelic frequency and disease status in the separate populations, but formal comparison of these must be based on tests of heterogeneity reported subsequently.
Table 2 TNFα genotypes and minor allele frequencies of the Newfoundland population.
| Genotype | NF PsA patients | Controls | OR | p Value | |
|---|---|---|---|---|---|
| G/G | 181 (79.4%) | 93 (90.3%) | |||
| TNF −238 | G/A | 45 (19.7%) | 9 (8.7%) | ||
| A/A | 2 (0.9%) | 1 (1.0%) | |||
| Allele F (A) | 10.7% | 5.3% | 2.13 | 0.03 | |
| G/G | 146 (64.9%) | 67 (65.1%) | |||
| TNF −308 | G/A | 75 (33.3%) | 33 (32.0%) | ||
| A/A | 4 (1.8%) | 3 (2.9%) | |||
| Allele F (A) | 18.4% | 18.9% | 0.97 | 0.91 | |
| C/C | 191 (86.8%) | 88 (85.4%) | |||
| TNF −857 | T/C | 28 (12.7%) | 14 (13.6%) | ||
| T/T | 1 (0.5%) | 1 (1.0%) | |||
| Allele F (T) | 6.8% | 7.8% | 0.87 | 0.74 | |
| C/C | 167 (72.3%) | 70 (68.0%) | |||
| TNF −863 | A/C | 60 (26.0%) | 30 (29.1%) | ||
| A/A | 4 (1.7%) | 3 (2.9%) | |||
| Allele F (A) | 14.7% | 17.5% | 0.82 | 0.36 | |
| TNF −1031 | T/T | 128 (58.2%) | 63 (61.2%) | ||
| T/C | 77 (35.0%) | 33 (32.0%) | |||
| C/C | 15 (6.8%) | 7 (6.8%) | |||
| Allele F (C) | 24.3% | 22.8% | 1.09 | 0.69 |
NF, Newfoundland; OR, odds ratio; PsA, psoriatic arthritis; TNF, tumour necrosis factor.
Table 3 TNFα genotype and minor allele frequencies of the Toronto population.
| Genotype | Toronto PsA patients | Controls | OR | p Value | |
|---|---|---|---|---|---|
| G/G | 164 (82.4%) | 90(89.1%) | |||
| TNF −238 | G/A | 35 (17.6%) | 10(9.9%) | ||
| A/A | 0 (0%) | 1 (1.0%) | |||
| Allele F (A) | 8.8% | 5.9% | 1.53 | 0.26 | |
| G/G | 144 (70.9%) | 69 (68.3%) | |||
| TNF −308 | G/A | 53 (26.1%) | 25 (24.8%) | ||
| A/A | 6 (3.0%) | 7 (6.9%) | |||
| Allele F (A) | 16.0% | 19.3% | 0.80 | 0.31 | |
| C/C | 148 (74.4%) | 86 (85.1%) | |||
| TNF −857 | T/C | 43 (21.6%) | 13 (12.9%) | ||
| T/T | 8 (4.0%) | 2 (2.0%) | |||
| Allele F (T) | 14.8% | 8.4% | 1.89 | 0.03 | |
| C/C | 155 (77.5%) | 71 (68.0%) | |||
| TNF −863 | A/C | 41 (20.5%) | 26 (29.1%) | ||
| A/A | 4 (2.0%) | 4 (2.9%) | |||
| Allele F (A) | 12.3% | 16.8% | 0.69 | 0.13 | |
| TNF −1031 | T/T | 126 (63%) | 61 (61.6%) | ||
| T/C | 65 (32.5%) | 32 (32.3%) | |||
| C/C | 9 (4.5%) | 6 (6.1%) | |||
| Allele F (C) | 20.8% | 22.2% | 0.92 | 0.67 |
OR, odds ratio; PsA, psoriatic arthritis; TNF, tumour necrosis factor.
A combined analysis of data from both populations, based on a stratified logistic regression of allelic frequencies, showed a significant association between disease status and the −238(A) variant (p = 0.01). There was no evidence of heterogeneity of the association between the populations (p = 0.49). The observed frequencies in the Newfoundland psoriatic arthritis population and controls for the −238(A) variant were 10.7% v 5.3% respectively, while in the Toronto population they were 8.8% v 5.9%.
For the −857(T) variant, there was no evidence of a relation with disease status based on the combined analysis (p = 0.42). However, there was marginal evidence of heterogeneity between the populations (p = 0.07). Observed allelic frequencies for the −857(T) variant in the Toronto population were14.8% v 8.4% in cases and controls, respectively, whereas no comparable marked difference was found in the Newfoundland population, in which the frequencies were 6.8% v 7.8%.
For the −308(A), −863(A), and −1031(C) variants, there was no evidence of relation with disease status (p values of 0.48, 0.28, and 0.64, respectively) or heterogeneity of effects between populations (p values of 0.53, 0.61, and 0.57, respectively).
Results based on a more conservative genotype analysis16 not influenced by HWE assumptions17 were very similar.
We then conducted a haplotype analysis for SNPs −238, −308, −857, and −863, in the two populations. The most notable result concerns the frequencies of the 1222 haplotype (where (1) indicates the presence of a minor allele, and (2) the presence of a major one) in patients and controls from Newfoundland (p = 0.04). The analysis suggests a greater frequency of this haplotype in cases than in controls. In Toronto, no significance was attached to this haplotype (p = 0.27). In the Toronto population the haplotype that appeared to be associated with the disease expression was 2212 (p = 0.03). This relation was not evident in Newfoundland (p = 0.6). Adjustments for multiplicity are required for interpretation purposes, however. A simple Bonferroni adjustment would generate a significance levels of 0.04×9 = 0.36 and 0.03×7 = 0.21 (as we have nine possible haplotypes in Newfoundland and seven in Toronto). Thus we report these haplotype frequencies as hypothesis‐generating for future studies. Nevertheless, these haplotype analyses do support the univariate SNP analyses, as they indicate that the presence of minor alleles at −238 in Newfoundland and at −857 in Toronto may be of importance for disease expression.
Meta‐analysis
Nine cohorts from eight studies (including our two Canadian cohorts) were identified in the meta‐analysis.7,8,9,10,11,12,13 The leading author, year of publication, population, TNFα variants, and the number of minor and total alleles for each of the variants are listed in table 4.
Table 4 Summary of TNFα associations studies in psoriatic arthritis.
| Author (reference) | Year | Population | −238(A) | −308(A) | −857(T) | −863(A) | −1031( C) | 488(A) | |
|---|---|---|---|---|---|---|---|---|---|
| Rahman* | 2005 | NF | Cases | 49/456 | 83/450 | 30/440 | 68/462 | 107/440 | |
| (10.7%) | (18.4%) | (6.8%) | (14.7%) | (24.3%) | |||||
| Controls | 11/206 | 39/206 | 16/206 | 36/206 | 47/206 | ||||
| (5.3%) | (18.9%) | (7.8%) | (17.5%) | (22.8%) | |||||
| Rahman* | 2005 | Toronto | Cases | 35/398 | 65/406 | 59/398 | 49/400 | 83/400 | |
| (8.8%) | (16.0%) | (14.8%) | (12.2%) | (20.8%) | |||||
| Controls | 12/202 | 39/202 | 17/202 | 34/202 | 44/198 | ||||
| (5.9%) | (19.3%) | (8.4%) | (16.8%) | (22.2%) | |||||
| Balding10 | 2003 | Irish | Cases | 70/298 | |||||
| (23.5%) | |||||||||
| Controls | 172/780 | ||||||||
| (22.1%) | |||||||||
| Hohler7 | 2002 | German_1 | Cases | 30/174 | 9/174 | ||||
| (17.2%) | (5.2%) | ||||||||
| Controls | 7/198 | 34/198 | |||||||
| (3.5%) | (17.2%) | ||||||||
| Gonzalez12 | 2002 | Spanish | Cases | 15/162 | 25/162 | ||||
| (9.3%) | (15.4%) | ||||||||
| Controls | 14/220 | 24/220 | |||||||
| (6.4%) | (10.9%) | ||||||||
| Al‐Heresh9 | 2002 | Irish | Cases | 23/248 | 52/248 | 17/248 | |||
| (9.3%) | (21.0%) | (6.9%) | |||||||
| Controls | 14/202 | 33/202 | 14/202 | ||||||
| (6.9%) | (16.3%) | (6.9%) | |||||||
| Gonzalez11 | 2001 | Jewish | Cases | 7/104 | 8/104 | ||||
| (6.7%) | (7.7%) | ||||||||
| Controls | 3/146 | 15/146 | |||||||
| (2.0%) | (10.3%) | ||||||||
| Hamamoto13 | 2000 | Japanese | Cases | 0/40 | 0/40 | 7/40 | 6/40 | 4/40 | |
| (0%) | (0) | (17.5) | (15.0) | (10.0) | |||||
| Controls | 4/174 | 5/174 | 27/174 | 27/174 | 22/174 | ||||
| (2.3%) | (2.9%) | (15.5) | (15.5) | (12.6) | |||||
| Hohler8 | 1997 | German_2 | Cases | 20/124 | 19/124 | ||||
| (16.1%) | (15.3%) | ||||||||
| Controls | 7/198 | 32/198 | |||||||
| (3.5%) | (16.1%) |
*Current study.
NF, Newfoundland.
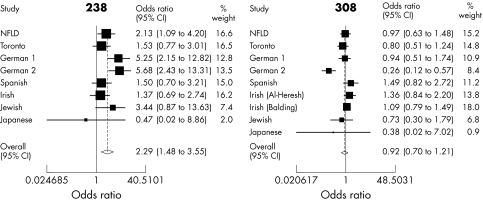
The results of the meta‐analyses are summarised in fig 1 for the TNFα variants −238 and −308, which have been most extensively studied. The figures related to the other variants are not presented as they involve only three populations. Only the −238 variant was noted to have a significant association (odds ratio (OR) = 2.29 (95% confidence interval (CI), 1.48 to 3.55)). If the Japanese study is excluded from the meta‐analysis, the pooled estimates for all the TNF variants were: −238(A), OR = 2.37 (95% CI, 1.52 to 3.69); −308(A), OR = 0.92 (0.69 to 1.23); −857 (T), OR = 1.30 (0.61 to 2.79); −863(A), OR = 0.75 (0.55 to 1.04); and −1031(C), OR = 1.00 (0.75 to 1.33), the latter three being based only on the combined analysis of the Newfoundland and Toronto data. If, however, the Japanese study is included in the meta‐analyses, this did not have a significant impact on the above results, leaving them practically unchanged (see fig 1 for −238(A) and −308(A)). Note that a test for heterogeneity in the −238 meta‐analysis was associated with a marginal significance level of 0.05, probably associated with the higher odds ratios observed in the German studies.7,8 A more significant result was associated with the −308 analysis (p = 0.01) due to the German_2 study's outlying odds ratio.8 This serves as justification for the use of a mixed effects model which adjusts for heterogeneity.
Figure 1 Meta‐analysis of −238 and −308 variants of TNFα.
Discussion
Psoriatic arthritis has a strong heritability, as reflected by a relative risk of 55 among first degree relatives with the disease.18 The TNFα gene has long been considered a major susceptibility gene in immunologically mediated disease, including psoriatic arthritis, for the pathogenesis of which it is of critical importance.2 The associations between the two most widely cited TNFα promoter polymorphisms (−238 and −308 variants) and psoriatic arthritis have been evaluated in several populations.7,8,9,10,11,12 Before our study, a significant association for TNFα and psoriatic arthritis was noted only in German populations.7,8 In this study, we examined five common variants with the TNFα promoter gene, including −238 and −308. We noted a significant association between −238 variant and psoriatic arthritis in the Newfoundland population. The magnitude of our risk (OR = 2) was not as high as reported in the German studies (OR = 5). This may reflect a difference in population diversity, although it is well recognised that early association studies tend to overestimate the magnitude of the association conferred by a genetic polymorphism.19
Our pooled analysis of all TNFα studies in psoriatic arthritis populations also noted a significant association between −238(A) and psoriatic arthritis. The −238(A) variant increased the odds of psoriatic arthritis by twofold in the populations studied. The meta‐analysis failed to support the association with the −308 (A) variant, which was previously reported in one of the German studies.7 The pooled estimates for the other variants showed no significant association, although the estimates for −857, −863, and −1031 were based only on the combined analysis of the two Canadian populations and the small Japanese cohort.
The differing results noted in the Irish9,10 and Spanish11,12 populations may be the result of ethnic admixture causing population stratification, population specific gene–gene or gene–environment interactions, variable linkage disequilibrium between the polymorphisms, and statistical fluctuations.20,21 Another possible explanation is the weak genetic effect of the underlying polymorphisms. In a meta‐analysis of 370 studies addressing 36 genetic associations for various outcomes, it was noted that when a sample size of less than 150 patients was used, there was a sevenfold higher rate of discrepancy between the first and subsequent studies, as compared with studies where the sample size was at least 150.19 Except for our studies in the Canadian cohorts, all other studies reported their finding using fewer than 150 probands. As a result these studies may be underpowered to assess the impact of TNFα polymorphisms in psoriatic arthritis reliably. Furthermore, larger studies may give different results from small studies and thus our inferences here should be interpreted with caution until large scale evidence is generated on these associations.22
As the previous studies were modest in size, we conducted a meta‐analysis to assist in estimating the population‐wide genetic effect of the commonly cited TNFα polymorphisms. Our pooled analysis showed a significant association between −238(A) and psoriatic arthritis. However, we acknowledge that population stratification was not entirely avoided as family based controls were not used in these studies. Furthermore, publication bias may exist in the meta‐analysis, as small negative association studies are often not published. As demonstrated in our study, because meta‐analysis of association studies in complex diseases are helpful in estimating population‐wide genetic effects, further efforts must be made to track all association data for a given polymorphism (positive or negative, small or large, published or not).
Acknowledgement
We thank Dr D Kane and Dr O Fitzgerald for sharing their unpublished data, which was incorporated in the meta‐analyses.
Abbreviations
HWE - Hardy–Weinberg equilibrium
SNP - single nucleotide polymorphism
TNFα - tumour necrosis factor α
References
- 1.Gladman D D, Rahman P. Psoriatic arthritis. In: Ruddy S, Harris ED, Sledge CB, Budd RC, Sergent JS, editors. Kelly's textbook of rheumatology, 6th edition. Philadelphia: WB Saunders Co, 20011071–1079.
- 2.AM, Cookson WOCM The genetics of psoriasis, psoriatic arthritis, and atopic dermatitis. Hum Mol Genet 200413R43–R55. [DOI] [PubMed] [Google Scholar]
- 3.Kane D, FitzGerald O. Tumor necrosis factor‐alpha in psoriasis and psoriatic arthritis: a clinical, genetic, and histopathologic perspective. Curr Rheumatol Rep 20046292–298. [DOI] [PubMed] [Google Scholar]
- 4.Danning C L, Illei G G, Hitchon C, Greer M R, Boumpas D T, McInnes I B. Macrophage‐derived cytokine and nuclear factor kappaB p65 expression in synovial membrane and skin of patients with psoriatic arthritis. Arthritis Rheum 2000431244–1256. [DOI] [PubMed] [Google Scholar]
- 5.Partsch G, Steiner G, Leeb B F, Dunky A, Broll H, Smolen J S. Highly increased levels of tumor necrosis factor‐alpha and other proinflammatory cytokines in psoriatic arthritis synovial fluid. J Rheumatol 199724518–523. [PubMed] [Google Scholar]
- 6.Mease P. TNFα therapy in psoriatic arthritis and psoriasis. Ann Rheum Dis 200463755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohler T, Grossmann S, Stradmann‐Bellinghausen B, Kaluza W, Reuss E, de Vlam K.et al Differential association of polymorphisms in the TNFα region with psoriatic arthritis but not psoriasis. Ann Rheum Dis 200261213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohler T, Kruger A, Schneider P M, Schopf R E, Knop J, Rittner C.et al A TNF‐alpha promoter polymorphism is associated with juvenile onset psoriasis and psoriatic arthritis. J Invest Dermatol 1997109562–565. [DOI] [PubMed] [Google Scholar]
- 9.Al‐Heresh A M, Proctor J, Jones S M, Dixey J, Cox B, Welsh K.et al Tumour necrosis factor‐alpha polymorphism and the HLA‐Cw*0602 allele in psoriatic arthritis. Rheumatology (Oxford) 200241525–530. [DOI] [PubMed] [Google Scholar]
- 10.Balding J, Kane D, Livingstone W, Mynett‐Johnson L, Bresnihan B, Smith O.et al Cytokine gene polymorphisms: association with psoriatic arthritis susceptibility and severity. Arthritis Rheum 2003481408–1413. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez S, Brautbar C, Martinez‐Borra J, Lopez‐Vazquez A, Segal R, Blanco‐Gelaz M A.et al Polymorphism in MICA rather than HLA‐B/C genes is associated with psoriatic arthritis in the Jewish population. Hum Immunol 200162632–638. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez S, Martinez‐Borra J, Lopez‐Vazquez A, Garcia‐Fernandez S, Torre‐Alonso J C, Lopez‐Larrea C. MICA rather than MICB, TNFα, or HLA‐DRB1 is associated with susceptibility to psoriatic arthritis. J Rheumatol 200229973–978. [PubMed] [Google Scholar]
- 13.Hamamoto Y, Tateno H, Ishida T, Muto M. Lack of association between promoter polymorphism of the tumor necrosis factor‐alpha gene and psoriatic arthritis in Japanese patients. J Invest Dermatol 20001151162–1164. [DOI] [PubMed] [Google Scholar]
- 14.Mossner R, Kingo K, Kleensang A, Kruger U, Konig I R, Silm H.et al Association of TNF −238 and −308 promoter polymorphisms with psoriasis vulgaris and psoriatic arthritis but not with pustulosis palmoplantaris. J Invest Dermatol 2005124282–284. [DOI] [PubMed] [Google Scholar]
- 15.Excoffier L, Slatkin M. Maximum‐likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 199512921–927. [DOI] [PubMed] [Google Scholar]
- 16.Sasieni P D. From genotypes to genes: doubling the sample size. Biometrics 1997531253–1261. [PubMed] [Google Scholar]
- 17.Schaid D J, Jacobsen S J. Biased tests of association: Comparisons of allele frequencies when departing from Hardy‐Weinberg proportions. Am J Epidemiol 1999149706–711. [DOI] [PubMed] [Google Scholar]
- 18.Moll J M, Wright V. Familial occurrence of psoriatic arthritis. Ann Rheum Dis 197332181–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioannidis J P, Ntzani E E, Trikalinos T A, Contopoulos‐Ioannidis D G. Replication validity of genetic association studies. Nat Genet 200129306–309. [DOI] [PubMed] [Google Scholar]
- 20.Hirschhorn J N, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med 20024(2)45–61. [DOI] [PubMed] [Google Scholar]
- 21.Huizinga T W, Pisetsky D S, Kimberly R P. Associations, populations, and the truth: recommendations for genetic association studies in arthritis and rheumatism. Arthritis Rheum 2004502066–2071. [DOI] [PubMed] [Google Scholar]
- 22.Ioannidis J P, Trikalinos T A, Ntzani E E, Contopoulos‐Ionnidis D G. Genetic associations in large versus small studies: an empirical assessment. Lancet 200315361, 567–361, 571. [DOI] [PubMed] [Google Scholar]