Abstract
We analyzed eight invasive Haemophilus influenzae strains isolated from individual children vaccinated appropriately for their ages. Five of the strains were type b and three were nontypeable strains. Children infected with nontypeable strains had a protective level of anti-H. influenzae type b antibodies, while only one of the children whose cases represented true vaccine failure did.
A small number of children still develop invasive Haemophilus influenzae type b (Hib) disease despite complete vaccination with anti-Hib conjugate vaccines (2-4). Recently, the incidence of Hib disease in United Kingdom children has increased, with the greatest rise occurring in vaccinated children (2, 10). When a vaccinated child develops an invasive infection due to H. influenzae, a key decision from the public health point of view has to be taken as to whether the infection represents a case of vaccine failure or whether it is a new infection caused by a type of H. influenzae other than type b (noncapsulated or other capsular type). Important serotype discrepancies in Hib detection using conventional slide agglutination serotyping have been reported by the Centers for Disease Control and Prevention (6, 15).
Several predisposing clinical risk factors and immunological conditions have been described for cases of vaccine failures (2-4). However, in the largest published series (12), all of these predisposing factors were present in less than half of the cases.
The aims of this study were to evaluate different molecular techniques for distinguishing true from apparent vaccine failures and to study the molecular epidemiology of strains responsible for a series of consecutive cases of H. influenzae invasive infections in fully vaccinated children compared with prevaccination isolate status.
In 1996, vaccination against Hib with conjugate vaccines was introduced in Spain, first in some Spanish Autonomous Communities and then at the national level. At the beginning of the vaccination program, children >1 year old were vaccinated once with the conjugate vaccine. In Spain, Hib vaccine is usually given at 2, 4, 6, and 18 months of age. Our national Haemophilus reference laboratory set up an active program aimed at the phenotypic and genotypic characterization of possible vaccine failures. Local microbiologists and public health epidemiologists were contacted and asked to send isolates to our laboratory. We also requested a serum sample from each patient to determine anti-Hib antibody status and a clinical protocol including the patient's identity code, clinical data, probable risk factors (premature delivery, underlying diseases, immunosuppression, and congenital abnormalities), dates of anti-Hib vaccination, and vaccine type, brand, and batch numbers.
Cerebrospinal fluid (CSF) and/or blood isolates were identified at the reference laboratory according to standard bacteriological methods (5). A PCR procedure was used for capsule genotyping of the six capsular types (types a to f) (8).
Immunoglobulin G antibody to Hib (polyribosylribitol phosphate [PRP]) was quantified by an enzyme-linked immunosorbent assay test as previously described (12). Serum samples were sent to Helen Griffiths, Radcliffe Hospital, University of Oxford, Oxford, United Kingdom. An antibody determination of ≥1.0 μg/ml was considered to be protective (13).
For the purposes of molecular epidemiological comparisons, in addition to the study strains, we selected eight Hib isolates that had caused invasive infection in children prior to widespread vaccination with Hib conjugate vaccines in Spain. Historical controls were the Hib strains 23095, 1494, 2995, 4194, 11194, 4394, 0994, and 3295. All of these strains were isolated between 1994 and 1995 from patients from similar geographical areas and with similar ages and clinical diagnoses to those of members of the study group of vaccine failure cases. The reference Hib strain Eagan was studied in parallel. All bacterial isolates were examined by pulsed-field gel electrophoresis (PFGE) after digestion of bacterial DNA with SmaI (MBI Fermentas, Vilnius, Lithuania). A cluster analysis was performed, and a similarity index was calculated by the unweighted pair-group average method (16).
Genotyping of Haemophilus strains was performed by Southern blot analysis using the pUO38 cap probe (14), a DNA probe hybridizing to the region of the chromosome involved in the synthesis and expression of capsular type b. The same Hib historical controls as for PFGE were included for comparison. The cluster analysis was performed as described previously (16).
True vaccine failure was defined by Heath et al. (12) as the isolation of an invasive Hib strain from CSF or blood at either >2 weeks after a single dose of vaccine had been given to an infant >1 year of age or >1 week after at least two doses had been given to a child of ≤1 year of age.
From 1997 to 2002, eight consecutive strains of H. influenzae (isolated from the CSF and/or blood of individual children previously vaccinated fully for their ages with conjugate vaccines against Hib) were submitted to our reference laboratory. The children's isolates came from distinct geographical areas of the country. Two cases occurred in the same area (Barakaldo, Basque Country, Spain) but were not epidemiologically linked and occurred 8 months apart. Among the vaccine failure cases, no common pattern of vaccine type, brand, or batch number was found.
Table 1 summarizes relevant clinical and laboratory data of the study patients. Five patients had meningitis. From the results of PCR capsule genotyping, five cases were shown to be true vaccine failures caused by Hib while the other three were shown to be caused by nonencapsulated strains. All of the children infected by nonencapsulated isolates had antibody levels against PRP of >1.0 μg/ml, whereas only one of the children whose cases involved true vaccine failure did (Table 1). The patient infected by Hib strain 2999 was human immunodeficiency virus (HIV) positive, but she had not developed AIDS, her CD4+ T-cell count was normal, and her viral load was undetectable; no immunoglobulin abnormalities were reported in any of the vaccine failure cases.
TABLE 1.
Characteristics of patients vaccinated against Hib who presented with invasive infections
| Strain | Date of isolation | Age of patient | Clinical diagnosis of patient | Source of isolate | Strain type | Anti-PRP (μg/ml) | Risk factor | No. of vaccine doses |
|---|---|---|---|---|---|---|---|---|
| 90797 | 1/31/97 | 3 yr | Bacteremia | Blood | Nontypeable H. influenzae | 6.24 | No | 1 |
| 70097 | 5/16/97 | 4 yr | Meningitis | CSF | Nontypeable H. influenzae | 2.81 | No | 1 |
| 38398 | 4/20/98 | 15 mo | Bacteremia | Blood | Nontypeable H. influenzae | 4.55 | No | 3 |
| 398 | 4/24/98 | 12 mo | Meningitis | CSF | Hib | <0.15 | No | 3 |
| 2999 | 1/08/99 | 4 yr | Pneumonia | Blood | Hib | 2.29 | HIV positive | 1 |
| 67299 | 9/23/99 | 15 mo | Meningitis | CSF | Hib | 0.17 | No | 3 |
| 52901 | 6/08/01 | 12 mo | Meningitis | CSF | Hib | <0.15 | No | 3 |
| 42402 | 7/18/02 | 19 mo | Meningitis | CSF | Hib | <0.15 | No | 4 |
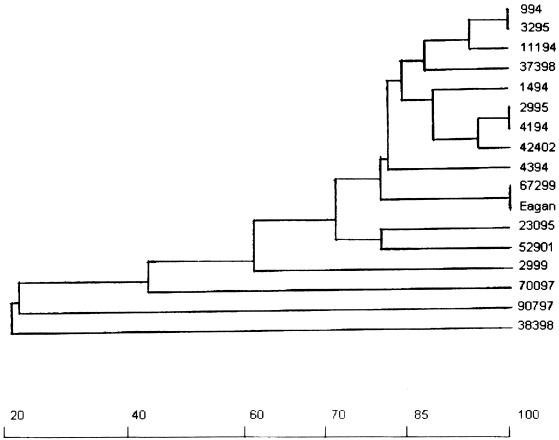
PFGE clearly separated Hib strains from nonencapsulated strains (Fig. 1); most of the Hib strains (including historical controls, the Eagan reference strain, and the Hib vaccine failures) displayed little genetic diversity, as their genetic distances were 0.20 or lower (Fig. 1). The Hib strain 2999 (isolated from a case of true vaccine failure) exhibited an intermediate genetic distance (0.38) between the major Hib clone and the nonencapsulated strains.
FIG. 1.
Dendrogram illustrating the genetic relationship of H. influenzae strains isolated from cases of invasive infections of vaccinated patients (as determined by PFGE) to prevaccination Hib isolates. Strains from vaccinated children: 37398 (b), 2999 (b), 67299 (b), 52901 (b), 42402 (b), 70097 (nontypeable), 90797 (nontypeable), and 38398 (nontypeable). Prevaccination H. influenza type b isolates: 23095, 1494, 2995, 4194, 11194, 4394, 0994, and 3295. The x axis represents percentages of the genetic relationship.
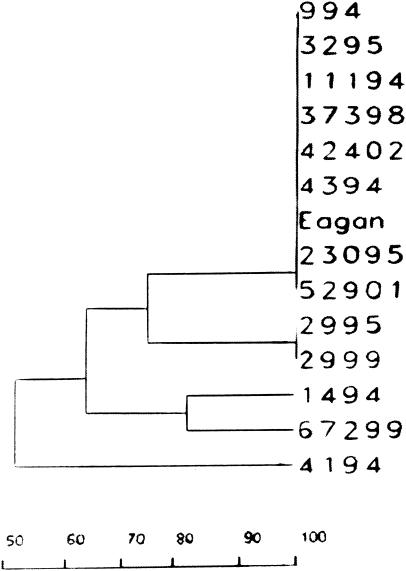
Little genetic variability was found in the cap b genetic structure, as 8 out of 13 of the Hib strains, whether responsible for vaccine failure or not, displayed the same Eagan-like pattern (Fig. 2). Probe hybridization was not observed in the three nonencapsulated strains. Three of five Hib isolates causing vaccine failure had a cap b pattern identical to that of the Eagan strain and five other prevaccination Hib strains (Fig. 2). Hib strain 2999 was distant from the other vaccine failure isolates (genetic distance, 0.75), although its cap pattern was identical to that of another Hib prevaccination control isolated in 1995.
FIG. 2.
Dendrogram illustrating the genetic relationships of Hib strains isolated from cases of invasive infections of vaccinated children (as determined by cap b genotyping) to prevaccination Hib isolates. The Hib strains from vaccinated children were strains 37398, 2999, 67299, 52901, and 42402. The prevaccination Hib isolates were strains 23095, 1494, 2995, 4194, 11194, 4394, 0994, and 3295. The x axis represents percentages of the genetic relationship.
Recently, a resurgence of invasive Hib cases in well-vaccinated children was reported in Alaska (9) and the United Kingdom (10, 11).
In this study, capsulated and nonencapsulated H. influenzae strains were able to be clearly separated by PCR and the two additional molecular epidemiological methods used. Type b strains can lose their capsulated phenotype to become phenotypically nonencapsulated while nevertheless retaining capsulation genes and therefore remaining hybridization positive (14). The fact that these strains were hybridization negative argues strongly that they are not mutants of b-positive strains but, rather, genuine nonencapsulated strains. Vaccine failure was essentially due to the patient's inability to develop an appropriate immune response, as was noted in other reports (3, 12).
Except for one case of a positive carrier of HIV who had not developed AIDS, no underlying predisposing condition was found in the other four cases, in agreement with other authors' observations (13). Other possible reasons for a child with HIV presenting with vaccine failure, such as antibody quality and other immune dysfunctions, might exist. The effectiveness of the Hib conjugate vaccine may be reduced in children with a high prevalence of HIV infection (17). There is a growing concern that some Hib vaccine failures may be the consequence of reduced effectiveness of combined vaccine formulations in which the pertussis component is “acellular” rather than whole-cell pertussis (10).
We used a restrictive definition of vaccine failure. Other Hib-invasive infections may occur in partially vaccinated children for whom insufficient time has elapsed for true vaccine failure to occur; these cases were not included in our study (20). Previously, little genetic diversity had been demonstrated for Hib by cap b genotyping or other epidemiological techniques (1, 19). Van Alphen et al. (21) used phenotypic methods to analyze 16 cases of vaccine failures, of which 7 represented patients who had been fully vaccinated. All isolates belonged to the common prevaccination clones of Hib. In addition, cap genotyping was carried out with a collection of Hib isolates from vaccinated children with no differences in the distribution of cap b genotypes (7). For our patients, the vaccine failure case produced by Hib isolate 2999 may imply support for the hypothesis of a change in the population structure of Hib as a result of vaccine adaptation (20) that occurred as each patient built up a protective anti-PRP antibody response, but the isolate was distantly related to the major clone detected by either PFGE or, to a less accurate extent, cap b genotyping.
Acknowledgments
This work was supported in part by research grants from the Comunidad Autónoma de Madrid (08.2/0007/2001 1) and the Instituto de Salud Carlos III (02/16).
Members of the Collaborative Group for Hib Vaccine Failures are José L. Hernández and Itziar Pochevile, Hospital de Cruces, Barakaldo, Bizkaia, Spain; Natalia Montiel, Hospital Costa del Sol, Marbella, Málaga, Spain; Juan M. Barros, Hospital Clínico Universitario, Santiago de Compostela, Spain; Amós García, DG Salud Pública, Las Palmas de Gran Canaria, Spain; and S. Kroll, Imperial College London, London, United Kingdom.
REFERENCES
- 1.Allan, I., M. R. Loeb, and E. R. Moxon. 1987. Limited genetic diversity of Haemophilus influenzae (type b). Microb. Pathog. 2:139-145. [DOI] [PubMed] [Google Scholar]
- 2.Booy, R., P. T. Heath, M. P. E. Slack, N. Begg, and E. R. Moxon. 1997. Vaccine failures after primary immunisation with Haemophilus influenzae type-b conjugate vaccine without booster. Lancet 349:1197-1202. [DOI] [PubMed] [Google Scholar]
- 3.Breukels, M. A., L. Spanjaard, L. A. M. Sanders, and G. T. Rijkers. 2001. Immunological characterization of conjugated Haemophilus influenzae type b vaccine failure in infants. Clin. Infect. Dis. 32:1700-1705. [DOI] [PubMed] [Google Scholar]
- 4.Breukels, M. A., E. M. Jol-van der Zijde, M. J. D. van Tol, and G. T. Rijkers. 2002. Concentration and avidity of anti-Haemophilus influenzae type b (Hib) antibodies in serum samples obtained from patients for whom Hib vaccination failed. Clin. Infect. Dis. 34:191-197. [DOI] [PubMed] [Google Scholar]
- 5.Campos, J., and J. A. Sáez-Nieto. 2001. Gram negative infections: Haemophilus and other clinically relevant Gram negative coccobacilli, p. 557-580. In N. Cimolai (ed.), Laboratory diagnosis of bacterial infections. Marcel Dekker, Inc., New York, N.Y.
- 6.Centers for Disease Control and Prevention. 2002. Serotyping discrepancies in Haemophilus influenzae type b disease—United States 1998-1999. Morb. Mortal. Wkly. Rep. 51:706-707. [PubMed] [Google Scholar]
- 7.Falla, T. J., D. W. M. Crook, E. C. Anderson, J. I. Ward, M. Santosham, J. Eskola, and E. R. Moxon. 1995. Characterization of capsular genes in Haemophilus influenzae isolates from H. influenzae type b vaccine recipients. J. Infect. Dis. 171:1075-1076. [DOI] [PubMed] [Google Scholar]
- 8.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galil, K., R. Singleton, O. Levine, et al. 1999. Reemergence of invasive Haemophilus influenzae type b disease in a well-vaccinated population in remote Alaska. J. Infect. Dis. 179:101-106. [DOI] [PubMed] [Google Scholar]
- 10.Garner, D., and V. Weston. 2003. Effectiveness of vaccination for Haemophilus influenzae type b. Lancet 361:395-396. [DOI] [PubMed] [Google Scholar]
- 11.Heath, P. T., and J. McVernon. 2002. The UK Hib vaccine experience. Arch. Dis. Child. 86:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heath, P. T., R. Booy, H. Griffiths, E. Clutterbuck, H. J. Azzopardi, M. P. E. Slack, J. Fogarty, A. C. Moloney, and E. R. Moxon. 2000. Clinical and immunological risk factors associated with Haemophilus influenzae type b conjugate vaccine in childhood. Clin. Infect. Dis. 31:973-980. [DOI] [PubMed] [Google Scholar]
- 13.Kayhty, H., H. Peltola, V. Karanko, and P. H. Makela. 1983. The protective level of serum antibodies to the capsular polysaccharide of Haemophilus influenzae type b. J. Infect. Dis. 147:1100. [DOI] [PubMed] [Google Scholar]
- 14.Kroll, J. S., S. Ely, and E. R. Moxon. 1991. Capsular typing of Haemophilus influenzae with a DNA probe. Mol. Cell. Probes 5:375-379. [DOI] [PubMed] [Google Scholar]
- 15.LaClaire, L. L., M. L. Tondella, D. S. Beall, C. A. Noble, P. L. Ragunathan, N. E. Rosenstein, T. Popovic, and Active Bacterial Core Surveillance Team Members. 2003. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing J. Clin. Microbiol. 41:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, W. H. 1981. Simple method for constructing phylogenetic trees from distance matrices. Proc. Natl. Acad. Sci. USA 78:1085-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madhi, S. A., K. Petersen, M. Khoosal, R. E. Huebner, N. Mbelle, R. Mothupi, H. Saloojee, H. Crewe-Brown, and K. P. Klugman. 2002. Reduced effectiveness on Haemophilus influenzae type b conjugate vaccine in children with a high prevalence of human immunodeficiency virus type 1 infection. Pediatr. Infect. Dis. J. 21:315-321. [DOI] [PubMed] [Google Scholar]
- 18.Mooi, F. R., H. M. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7(Suppl. 3):526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, et al. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 20.Peltola, H. 2000. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13:302-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Alphen, L., A. K. Takala, L. G. Broek, J. Dankert, and J. Eskola. 1992. Changes in the distribution of Haemophilus influenzae type b clones associated with widespread infant vaccination in Finland. J. Infect. Dis. 166:1340-1345. [DOI] [PubMed] [Google Scholar]