Abstract
Background
A common functional polymorphism of the brain‐derived neurotrophic factor gene (BDNF Val66Met) was previously associated with diminished episodic memory performance in healthy people. As cognitive function is commonly impaired in patients with systemic lupus erythematosus (SLE), the association of the BDNF Val66Met with neurocognitive function was studied.
Objective
To study the association of the BDNF Val66Met with neurocognitive function in a cohort of patients with SLE.
Methods
Cognitive function was assessed in 59 patients with SLE with no previous or current central nervous system involvement. Cognitive tests were grouped into five domains (memory, attention/executive function, visuospatial skills, motor function and psychomotor speed) and used to obtain domain Z scores, reflecting the difference between averaged scores of performance on individual tests and published norms in each domain. Genotyping was carried out using a 5′‐nuclease assay with 99.9% accuracy. Unpaired t test was used to assess the relationship between genotypes and cognitive function, whereas the effect of possible confounders was assessed in a multivariate analysis.
Results
Patients carrying the Met66 allele scored significantly higher on psychomotor, attention/executive and motor function tests, resulting in significantly higher domain Z scores for the psychomotor (p = 0.005) and motor (p = 0.002) domains.
Conclusions
The BDNF Met66 allele was associated with better cognitive functioning in the psychomotor and motor domains, even after controlling for differences in ethnicity, sex, depression status and prednisone treatment. These data suggest that the BDNF Met66 allele confers protection against the decline of motor and psychomotor cognitive functions in patients with longstanding SLE.
Systemic lupus erythematosus (SLE) is a chronic systemic autoimmune disease that is characterised by abnormal autoantibody production and involvement of major organs including the central nervous system (CNS). Cognitive dysfunction is reported to occur in 20–80% of patients.1,2,3 CNS involvement in lupus is heterogeneous and may be due to multiple factors, such as vascular damage,4,5 demyelinating processes, effects of drugs,6,7 and neuronal degeneration caused by autoantibodies and cytokines.1,8,9 Despite the previous implications of multiple agents in its pathogenesis, cognitive dysfunction in lupus often has no identifiable connection to any of the exogenous or endogenous factors known to lead to a neuronal insult.
Clinical studies describing cognitive dysfunction in patients with SLE pointed to the impairment in memory,10 visuospatial skills11 and learning. Recently, we found cognitive dysfunction in 46% of patients in a lupus cohort, with changes in memory and visuospatial skills being most pronounced.12 Cognitive dysfunction in SLE has no uniform pattern of appearance or progression and in most cases is not connected with other manifestations of lupus. We hypothesised that genetic polymorphisms may influence the development or progression of cognitive dysfunction in SLE.
Brain‐derived neurotrophic factor (BDNF) is abundantly expressed in the mammalian CNS, with the highest level of expression occurring in the hippocampus.13,14 Along with other members of the neurotrophin family, including nerve growth factor, neurotrophin‐3 (NT‐3), neurotrophin 4/5 (NT‐4/5) and neurotrophin‐6 (NT‐6), BDNF is of crucial importance in neuronal differentiation and survival during embryonic development, as well as in maintenance of viability of neurones in adulthood both in the CNS and in the peripheral nervous system.15,16,17 BDNF is associated with the modulation of activity‐dependent synaptic plasticity, learning and memory processing both in vitro and in animal studies.18 Memory consolidation is thought to be related to short‐term changes in the electrical properties (early‐phase long‐term potentiation), and long‐term structural changes of the synapses (late‐phase long‐term potentiation), and BDNF seems to have a critical role in both processes.19,20,21
The human BDNF gene maps to 11p13 and is composed of six 5′ exons that are differentially spliced to a single 3′ terminal exon that encodes the entire sequence of mature BDNF. A common functional polymorphism (G196A transition) resulting in a Val66Met non‐conservative missense substitution in the proregion has been identified in BDNF. The frequency of the variant Met66 allele exhibits a high degree of population variation, with the frequency of the Met66 allele being 0.18–0.22 in Caucasians, 0.41 in Japanese and 0.05 in African‐Americans in various studies.22 These frequency data are consistent with those provided by the International HapMap Project. Several lines of evidence have shown functional differences between the Val66 and the variant allele.
Cultured hippocampal neurones expressing the variant Met‐BDNF exhibited impaired intracellular trafficking and localisation to secretory granules.23 Activity‐dependent secretion of BDNF was also reduced, whereas constitutive secretion remained unchanged.23
In human subjects carrying the Met66 allele, lower levels of hippocampal N‐acetyl aspartate (an indirect measure of neural integrity and synaptic abundance) were detected by magnetic resonance imaging spectroscopy.24 Moreover, in healthy volunteers, the Met66 allele was associated with reduced episodic memory performance and abnormal hippocampal activation assessed by functional magnetic resonance imaging.23 Met66 carriers have been found to exhibit diminished hippocampal engagement during both encoding and retrieval processes compared with Val66 homozygotes, providing direct neurophysiological evidence for the effect of BDNF alleles on hippocampal memory processing.25 In morphological studies, reduced grey matter volumes have been detected in the hippocampi and the lateral convexity of the frontal lobes, with peak values in the dorsolateral prefrontal cortex of Met66 carriers compared with Val66 homozygotes in healthy volunteers.26,27
As BDNF seems to have a pivotal role in activity‐dependent neural plasticity underlying learning and memory processing both in vitro and in vivo, and the Val66Met variant affects human cognitive functioning, we studied the association of BDNF Val66Met, a common functional polymorphism, with neurocognitive dysfunction in patients with SLE.
Patients and methods
Fifty nine of the 60 consecutive patients participating in a study investigating cognitive dysfunction in SLE conducted at the National Institute of Arthritis, Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, Maryland, USA, were enrolled in this genetic study. DNA from one patient was not available. DNA was isolated from peripheral blood after obtaining informed consent in accordance with National Institutes of Health approved protocols with human subjects. The full eligibility criteria have been described elsewhere.12 In brief, all the enrolled patients fulfilled the 1997 updated American College of Rheumatology criteria28 for the diagnosis of SLE. Patients were enrolled if they had no history of neurological diseases, including head injury resulting in loss of consciousness, strokes, seizures and toxic exposure; had no history of clinically documented transient ischaemic episodes within 6 months of screening visit; had not been treated with anticonvulsant agents; and if their proficiency in English language did not limit their participation in the neuropsychological testing.
Patients' evaluation included clinical interview, physical examination, routine laboratory tests, psychiatric evaluation for depression with Beck Depression Inventory—second edition, and neurocognitive testing.12
Assessment of cognitive function
Cognitive function was assessed in five domains: memory, attention and executive function, visuospatial, motor, and psychomotor domains. Neurocognitive tests were grouped a priori to assess the domain function as follows:
Memory domain: California Verbal Learning Test, Rey‐Osterrieth Complex Figure Test (recall);
Attention and executive function domain: Stroop Colour‐Word test, Trail Making Test (Part B), Semantic clustering part of the California Verbal Learning Test, Controlled Oral Word Association Test (F‐A‐S);
Visuo‐spatial domain: Rey‐Osterrieth Complex Figure Test (copy);
Motor domain: Grooved Pegboard Test for dominant and non‐dominant hands; and
Psychomotor domain: Wechsler Adult Intelligence Scale‐R/III, Digit Symbol Substitution Test, Stroop Word and Stroop Colour tests.
Grading
Raw scores of performance on each test were compared with the published norms derived from age‐matched and sex‐matched populations, and transformed into Z scores to express the deviation from the normal mean. Mean domain Z scores were determined as the average of the Z scores from the tests comprising each domain.
5′‐exonuclease genotyping
Genomic DNA was extracted from peripheral blood mononuclear cells and diluted to a concentration of 10 ng/μl. The 5′‐exonuclease genotyping assay (TaqMan, Foster City, California, USA) is a rapid and accurate method for high‐throughput genotyping of single nuclear polymorphisms,29 combining polymerase chain reaction amplification and sequence variant detection into a single step. Locus‐specific primers and fluorogenic allele‐specific probes were designed and manufactured by Applied Biosystems (ABI) Assays‐On‐Demand service (Foster City, California, USA). The identification number of the assay used is C_11592758_10.
The 5‐μl reaction mixture consisted of 2.5 μl of Taqman Universal Master Mix (ABI), 0.125 μl of 20× Assay Mix (ABI; 8 μM detection probe for each allele, 36 μM forward and reverse primer each) and 10 ng of genomic DNA diluted in 2.375 μl of Tris EDTA, pH 8.0 (Quality Biological, Gaithersburg, Maryland, USA). Amplification was performed with an ABI Gene Amp PCR System 9700 using 384‐well plates and the following amplification profile: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 92°C for 15 s and 60°C for 1 min. After amplification, endpoint fluorescence intensity was measured directly in the reaction plates, by means of the 7900 ABI Sequence Detector. Genotypes were determined using Sequence Detection System Software V.2.0 (ABI). Four genotyping signal clusters were identified, representing Val66 and Met66 homozygotes, Val66/Met66 heterozygotes and no‐DNA‐template controls.
Genotyping accuracy was determined by triplicate genotyping of 59 DNA samples. The genotyping error rate was 0%. Genotyping completion rate was 100%. No significant deviation from Hardy–Weinberg equilibrium was found (χ2 = 3.53, p = 0.06; table 1).
Table 1 Allele and genotype distributions of the brain‐derived neurotrophic factor Val66Met polymorphism in the cohort as a whole and in the two subgroups into which the cohort was broken down based on ethnicity.
| All participants (n = 59) | African‐Americans (n = 16) | Others (n = 43; 31 Caucasians, 9 Hispanics, 3 Asians) | |
|---|---|---|---|
| Allele | |||
| Val66 (G) | 102 (0.86) | 31 (0.97) | 71 (0.83) |
| Met66 (A) | 16 (0.14) | 1 (0.03) | 15 (0.17) |
| Genotype | |||
| Val66/Val66 | 46 (0.78) | 15 (0.94) | 31 (0.72) |
| Val66/Met66 | 10 (0.17) | 1 (0.06) | 9 (0.21) |
| Met66/Met66 | 3 (0.05) | 0 (0.00) | 3 (0.07) |
| Hardy–Weinberg theorem | |||
| χ2 | 3.53 | 0.03 | 2.71 |
| p Value | 0.06 | 0.857 | 0.1 |
Statistical analysis
Descriptive statistics including means, medians, standard deviations, proportions and graphical displays were computed for all study variables. Mean test Z scores and domain Z scores were compared in patients with and without the Met66 allele. Student's t test was used for unpaired comparisons between the groups. For the analysis of the association of genotype with mean domain Z scores, the level of significance was p<0.01 after the Bonferroni correction for multiple comparisons. A multivariable linear regression model was built to analyse the relationship between the presence of the Met66 allele and the performance in selected cognitive domains using the mean Z scores of the motor and psychomotor domains, as well as controlling for sex, ethnicity, depression status, disease activity and steroid use. StatView V.5.0.1 software and SAS E‐Guide V.3.0 were used for statistical analyses.
Results
Study participants were primarily women, as expected in SLE; 53% were Caucasians and 27% were African‐Americans. The mean level of education was 14.9 years, indicating a high level of expected cognitive abilities in this cohort. Most patients had previously received high doses of steroids but had inactive disease, and were on small doses or no corticosteroids at the time of study evaluation. Demographic and clinical characteristics of the patient cohort as a whole and the two subgroups into which the cohort was broken down on the basis of genotype are shown in table 2. The distribution of the demographic and clinical, and serological characteristics did not differ markedly between the two groups, except that men were over‐represented among Val66/Val66 homozygotes. To exclude the potential confounding effect of sex, it was included in the multivariable regression analysis along with ethnicity, depression and steroid treatment.
Table 2 Demographic and clinical characteristics of the cohort as a whole and of the two genotype groups.
| Whole cohort (n = 59) | Met66 carriers (n = 13) | Val66/Val66 homozygotes (n = 46) | |
|---|---|---|---|
| Mean (SD) age (years) | 41 (13) | 38.3 (13.5) | 42.2 (12.8) |
| Sex F:M, n | 46:13 | 12:1 | 34:12 |
| Ethnicity, n (%) | |||
| Caucasian | 31 (53) | 7 (54) | 24 (52) |
| African‐American | 16 (27) | 1 (8) | 15 (33) |
| Asian | 3 (5) | 2 (15) | 1 (2) |
| Hispanic | 9 (15) | 3 (23) | 6 (13) |
| Mean (SD) education (years) | 14.9 (3.0) | 14.3 (3.7) | 15.2 (2.9) |
| Mean (SD) disease duration (years) | 13.8 (10.3) | 13.6 (9.1) | 13.7 (10.6) |
| Anti‐ds DNA Ab pos, n (%) | 32 (54) | 7 (54) | 25 (54) |
| SLEDAI⩽4, n (%) | 46 (78) | 11 (84) | 35 (76) |
| Mean (SD) Beck Depression Inventory Scale | 8 (8) | 5.0 (7.5) | 9.1 (8.3) |
| Mean (SD) current prednisone dose (mg) | 4.5 (6.3) | 3.6 (3.6) | 4.8 (6.9) |
Anti‐ds DNA Abs pos, anti‐double‐stranded DNA antibody positivity; F, female; M, male; SLEDAI, Systemic Lupus Erythematosus Disease Activity Index.
Table 1 gives the allele and genotype distributions as well as χ2 and p values of Hardy–Weinberg equilibria. As the frequency of the Met66 allele is considerably lower in African‐Americans than in Caucasians and Asians, the allele and genotype distributions are also shown in two subgroups into which the cohort was divided based on ethnicity (African‐Americans v others).
Association of the Val66Met polymorphism with cognitive tests and domains
Overall, patients' mean domain Z scores were in the low average to mildly impaired range across all the domains. The lowest scores were seen in the memory (mean Z score (SD): −0.757 (0.943)) and visuospatial domains (−1.256 (2.425)), as opposed to the psychomotor domain, which was least affected (−0.232 (0.628)).
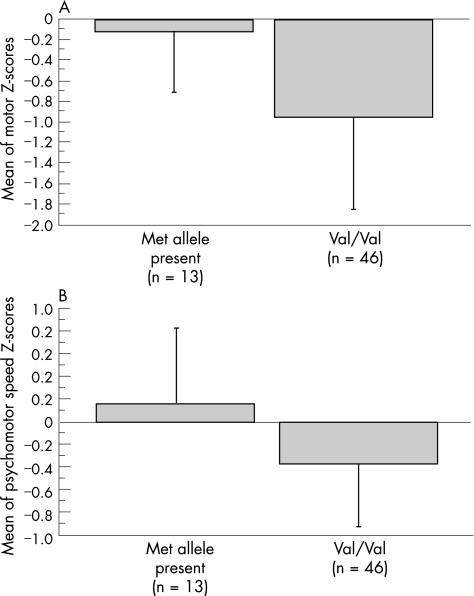
Patients with Val66/Met66 or Met66/Met66 genotypes performed better in the motor and psychomotor domains. Compared with Val66/Val66 homozygotes, both motor and psychomotor domain Z scores were significantly higher in patients carrying the Met66 allele (domain Z score (mean (SD)) for motor domain: −0.103 (0.607) v −0.964 (0.892); p = 0.002 and for psychomotor domain: 0.167 (0.671) v −0.365 (0.56); p = 0.005; fig 1A,B).
Figure 1 Effect of the brain‐derived neurotrophic factor Val66Met polymorphism on performance in the (A) motor (p = 0.002) and (B) psychomotor (p = 0.005) domains. Data are shown as mean domain Z score values. Error bars represent SD.
We observed no significant difference between Met66 carriers and Val66/Val66 homozygotes in the mean Z scores of the memory, attention or executive function and visuospatial domains. Performance on individual tests contributing to the various domains was consistent with our findings at the domain level (data not shown).
Although the mean Beck Depression Index Score was lower in patients carrying the Met66 allele (5.0 (7.5) v 9.1 (8.3)), this difference was not significant (p = 0.12). Moreover, no association was found between the presence of Met66 allele and clinically diagnosed depression (data are not shown, p = 0.6), suggesting that the observed difference in cognitive function is not due to the presence of depression.
To further explore the effect of potential confounding factors that may influence the interaction between the BDNF Val66Met polymorphism and mean motor and psychomotor domain Z scores, we carried out a multivariable regression analysis to control for the effects of current and cumulative prednisone treatment, depressive mood, sex, ethnicity and disease activity. As the frequency of the Met66 allele is considerably lower in African‐Americans, we included ethnicity as a binary variable (African‐Americans v others). The effect of genotype on cognitive function in the motor and psychomotor domains remained significant after controlling for depression status, ethnicity, sex and current prednisone treatment (p = 0.02 for motor, p = 0.018 for psychomotor; table 3), as well as for cumulative prednisone dose and disease activity.
Table 3 Multivariable regression analysis of the motor and psychomotor mean domain Z scores.
| Variable | Motor domain | Psychomotor domain |
|---|---|---|
| Presence of Met allele | 0.0206 | 0.0183 |
| Ethnicity (AA v other) | 0.0263 | 0.2344 |
| Beck Depression Scale | 0.4219 | 0.2498 |
| Current prednisone | 0.8464 | 0.4670 |
| Sex | 0.7797 | 0.1394 |
AA, African‐Americans.
p Values of the independent variables are shown.
Discussion
Our results support the hypothesis that in patients with lupus without clinically apparent neurological disease, a polymorphism in the BDNF gene is associated with susceptibility to cognitive dysfunction. Compared with Val66/Val66 homozygotes, the extent of cognitive deterioration in patients carrying one or two copies of the BDNF Met66 allele was smaller in the motor and psychomotor domains, suggesting a protective role for the Met66 allele. The data were analysed using Met66 as dominant because Met66 negatively affects sorting of Val‐Met heterodimers. Thus, Met66 is functionally dominant over Val66 BDNF.30 To our knowledge, this work is the first in which the association of the common functional BDNF Val66Met polymorphism with neurocognitive dysfunction has been explored in patients with lupus.
The role of the Met66 allele as a factor increasing vulnerability to, or conferring protection against, the development of cognitive dysfunction or various psychiatric diseases has not been unequivocally established yet. A substantial amount of evidence suggests that the presence of the Met66 allele represents higher risk for impaired memory function23,25,26,30 as well as comorbid anxiety and depression.31 By contrast, the presence of the Met66 allele, or a haplotype containing the Met66 allele, has been inferred to be protective against different psychiatric disorders.32,33,34,35,36 Although in these genetic association studies functional alterations at the cellular level have not been explored, the apparent protective role of the Met66 allele of BDNF found in other human psychiatric disorders is consistent with our findings.
Hypothetically, there are several mechanisms that could explain how the Met66 allele may be protective in this population. In the brain, BDNF is synthesised as a precursor polypeptide (proBDNF) that is proteolytically cleaved to produce the mature 13.5 kDa form that promotes cell survival through the Trk receptor family.37,38 In contrast with mature BDNF, proBDNF does not bind to TrkB, but rather to the multifunctional neurotrophin receptor, p75. Depending on the cellular context, activation of p75 induces apoptosis39,40 or promotes cell survival.41 Proneurotrophins might account for as much as 40–60% of the total amount of neurotrophins secreted extracellularly in the CNS42; in fact, most of the activity‐dependent BDNF secretion seems to be attributable to the release of the 32‐kDa proBDNF.30,42 Hypothetically, reduced secretion due to impaired intracellular trafficking and release of the proapoptotic proBDNF might confer protection in Met66 carriers, particularly under conditions when a substantial subpopulation of neurones is undergoing apoptotic cell death. Proforms of neurotrophins can also be cleaved extracellularly by the serine protease plasmin and matrix metalloproteinases to form the mature forms.40 Therefore, another possibility is that the rate or extent of extracellular processing of proBDNF to mature BDNF might be influenced by the Val66Met polymorphism altering the proportions of the proapoptotic proBDNF and the antiapoptotic mature BDNF.
Of note, the Met66 allele has consistently been associated with diminished hippocampal activation and volume in all studies dealing with the function or morphology of the hippocampal formation.23,25,26,27 The apparent protective role found in other studies (including the current one) might partly be explained by the fact that in these studies the functions of other brain regions were studied. For instance, in our study the Met66 allele was associated with better performance on tests primarily assessing the function of the frontal lobe as opposed to that of the hippocampus.
Nevertheless, there are several limitations of the study. Firstly, the frequency of the Met66 allele is considerably lower in African‐Americans than in Caucasians and Asians. The small sample size of the cohort did not allow subgroup analyses to be carried out in each ethnic group, but the effect of the Met66 allele on cognitive functioning remained significant in a multivariable regression analysis after controlling for depression, dose of prednisone and ethnicity. Whereas ethnicity had a marked effect on motor but not on psychomotor mean domain Z scores (table 3), genotype remained significant in both analyses, indicating that the effect of the Met66 allele on performance in the psychomotor and motor domains cannot solely be explained by the ethnic heterogeneity of our cohort. Additionally, the genotyping was carried out in a cohort of convenience. The participants were primarily selected for another study12; they had a relatively high level of education, were seen at a tertiary centre and were motivated to participate in this study; thus, our results may not be representative of patients with lupus in other settings. Finally, we cannot rule out the possibility that the association found might be related to another locus, which is in linkage disequilibrium with the Val66Met polymorphism.
In conclusion, whereas the mechanisms through which the Met66 allele might confer protections are hypothetical, this study provides the first evidence for a protective effect against neurocognitive dysfunction of Met66 allele in a cohort of patients with lupus. The study awaits replication and if the association is confirmed in a larger cohort, the polymorphism could serve as a useful biomarker to identify patients with lupus at higher risk of developing cognitive dysfunction.
Abbreviations
BDNF - brain‐derived neurotrophic factor
CNS - central nervous system
SLE - systemic lupus erythematosus
Footnotes
Funding: This study was funded by the intramural research programmes of NIAMS and NIAAA, NIH, Bethesda, Maryland, USA.
Competing interests: None.
This study has been approved by an NIH Institutional Review Board (IRB) located in Bethesda, Maryland, USA.
References
- 1.Kozora E, Thompson L L, West S G, Kotzin B L. Analysis of cognitive and psychological deficits in systemic lupus erythematosus patients without overt central nervous system disease. Arthritis Rheum 1996392035–2045. [DOI] [PubMed] [Google Scholar]
- 2.Brey R L, Holliday S L, Saklad A R, Navarrete M G, Hermosillo‐Romo D, Stallworth C L.et al Neuropsychiatric syndromes in lupus: prevalence using standardized definitions. Neurology 2002581214–1220. [DOI] [PubMed] [Google Scholar]
- 3.Carbotte R M, Denburg S D, Denburg J A. Prevalence of cognitive impairment in systemic lupus erythematosus. J Nerv Ment Dis 1986174357–364. [DOI] [PubMed] [Google Scholar]
- 4.Denburg S D, Carbotte R M, Ginsberg J S, Denburg J A. The relationship of antiphospholipid antibodies to cognitive function in patients with systemic lupus erythematosus. J Int Neuropsychol Soc 19973377–386. [PubMed] [Google Scholar]
- 5.Menon S, Jameson‐Shortall E, Newman S P, Hall‐Craggs M R, Chinn R, Isenberg D A. A longitudinal study of anticardiolipin antibody levels and cognitive functioning in systemic lupus erythematosus. Arthritis Rheum 199942735–741. [DOI] [PubMed] [Google Scholar]
- 6.Carlomagno S, Migliaresi S, Ambrosone L, Sannino M, Sanges G, Di Iorio G. Cognitive impairment in systemic lupus erythematosus: a follow‐up study. J Neurol 2000247273–279. [DOI] [PubMed] [Google Scholar]
- 7.Ginsburg K S, Wright E A, Larson M G, Fossel A H, Albert M, Schur P H.et al A controlled study of the prevalence of cognitive dysfunction in randomly selected patients with systemic lupus erythematosus. Arthritis Rheum 199235776–782. [DOI] [PubMed] [Google Scholar]
- 8.Denburg J A, Behmann S A. Lymphocyte and neuronal antigens in neuropsychiatric lupus: presence of an elutable, immunoprecipitable lymphocyte/neuronal 52 kd reactivity. Ann Rheum Dis 199453304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denburg J A, Carbotte R M, Denburg S D. Neuronal antibodies and cognitive function in systemic lupus erythematosus. Neurology 198737464–467. [DOI] [PubMed] [Google Scholar]
- 10.Fisk J D, Eastwood B, Sherwood G, Hanly J G. Patterns of cognitive impairment in patients with systemic lupus erythematosus. Br J Rheumatol 199332458–462. [DOI] [PubMed] [Google Scholar]
- 11.Monastero R, Bettini P, Del Zotto E, Cottini E, Tincani A, Balestrieri G.et al Prevalence and pattern of cognitive impairment in systemic lupus erythematosus patients with and without overt neuropsychiatric manifestations. J Neurol Sci 200118433–39. [DOI] [PubMed] [Google Scholar]
- 12.Lapteva L, Nowak M, Yarboro C H, Takada K, Roebuck T, Weickert T.et al Anti‐N‐methyl‐D Aspartate (NMDA) receptor antibodies, cognitive dysfuntion and depression in systemic lupus erythematosus. Arthritis Rheum 2006542505–2514. [DOI] [PubMed] [Google Scholar]
- 13.Ernfors P, Ibanez C F, Ebendal T, Olson L, Persson H. Molecular cloning and neurotrophic activities of a protein with structural similarities to nerve growth factor: developmental and topographical expression in the brain. Proc Natl Acad Sci USA 1990875454–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murer M G, Yan Q, Raisman‐Vozari R. Brain‐derived neurotrophic factor in the control human brain, and in Alzheimer's disease and Parkinson's disease. Prog Neurobiol 20016371–124. [DOI] [PubMed] [Google Scholar]
- 15.Jones K R, Reichardt L F. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci USA 1990878060–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leibrock J, Lottspeich F, Hohn A, Hofer M, Hengerer B, Masiakowski P.et al Molecular cloning and expression of brain‐derived neurotrophic factor. Nature 1989341149–152. [DOI] [PubMed] [Google Scholar]
- 17.Ventimiglia R, Mather P E, Jones B E, Lindsay R M. The neurotrophins BDNF, NT‐3 and NT‐4/5 promote survival and morphological and biochemical differentiation of striatal neurons in vitro. Eur J Neurosci 19957213–222. [DOI] [PubMed] [Google Scholar]
- 18.Yamada K, Mizuno M, Nabeshima T. Role for brain‐derived neurotrophic factor in learning and memory. Life Sci 200270735–744. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, Gottschalk W. Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Prog Brain Res 2000128231–241. [DOI] [PubMed] [Google Scholar]
- 20.Pang P T, Lu B. Regulation of late‐phase LTP and long‐term memory in normal and aging hippocampus: role of secreted proteins tPA and BDNF. Ageing Res Rev 20043407–430. [DOI] [PubMed] [Google Scholar]
- 21.Poo M M. Neurotrophins as synaptic modulators. Nat Rev Neurosci 2001224–32. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu E, Hashimoto K, Iyo M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. Am J Med Genet 2004126B122–123. [DOI] [PubMed] [Google Scholar]
- 23.Egan M F, Kojima M, Callicott J H, Goldberg T E, Kolachana B S, Bertolino A.et al The BDNF val66met polymorphism affects activity‐dependent secretion of BDNF and human memory and hippocampal function. Cell 2003112257–269. [DOI] [PubMed] [Google Scholar]
- 24.Maier M, Ron M A, Barker G J, Tofts P S. Proton magnetic resonance spectroscopy: an in vivo method of estimating hippocampal neuronal depletion in schizophrenia. Psychol Med 1995251201–1209. [DOI] [PubMed] [Google Scholar]
- 25.Hariri A R, Goldberg T E, Mattay V S, Kolachana B S, Callicott J H, Egan M F.et al Brain‐derived neurotrophic factor val66met polymorphism affects human memory‐related hippocampal activity and predicts memory performance. J Neurosci 2003236690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pezawas L, Verchinski B A, Mattay V S, Callicott J H, Kolachana B S, Straub R E.et al The brain‐derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci 20042410099–10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szeszko P R, Lipsky R, Mentschel C, Robinson D, Gunduz‐Bruce H, Sevy S.et al Brain‐derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol Psychiatry 200510631–636. [DOI] [PubMed] [Google Scholar]
- 28.Hochberg M C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997401725. [DOI] [PubMed] [Google Scholar]
- 29.Shi M M, Myrand S P, Bleavins M R, de la Iglesia F A. High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol 199952295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z Y, Patel P D, Sant G, Meng C X, Teng K K, Hempstead B L.et al Variant brain‐derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity‐dependent secretion of wild‐type BDNF in neurosecretory cells and cortical neurons. J Neurosci 2004244401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Xu K, Hoberman J, Tian F, Marko A J, Waheed J F.et al BDNF variation and mood disorders: a novel functional promoter polymorphism and Val66Met are associated with anxiety but have opposing effects. Neuropsychopharmacology 2005301353–1361. [DOI] [PubMed] [Google Scholar]
- 32.Hall D, Dhilla A, Charalambous A, Gogos J A, Karayiorgou M. Sequence variants of the brain‐derived neurotrophic factor (BDNF) gene are strongly associated with obsessive‐compulsive disorder. Am J Hum Genet 200373370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sklar P, Gabriel S B, McInnis M G, Bennett P, Lim Y M, Tsan G.et al Family‐based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain‐derived neutrophic factor. Mol Psychiatry 20027579–593. [DOI] [PubMed] [Google Scholar]
- 34.Neves‐Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy J L. The brain‐derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family‐based association study. Am J Hum Genet 200271651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geller B, Badner J A, Tillman R, Christian S L, Bolhofner K, Cook E H., Jr Linkage disequilibrium of the brain‐derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am J Psychiatry 20041611698–1700. [DOI] [PubMed] [Google Scholar]
- 36.Sen S, Nesse R M, Stoltenberg S F, Li S, Gleiberman L, Chakravarti A.et al A BDNF coding variant is associated with the NEO personality inventory domain neuroticism, a risk factor for depression. Neuropsychopharmacology 200328397–401. [DOI] [PubMed] [Google Scholar]
- 37.Chao M V. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci 20034299–309. [DOI] [PubMed] [Google Scholar]
- 38.Friedman W J, Greene L A. Neurotrophin signaling via Trks and p75. Exp Cell Res 1999253131–142. [DOI] [PubMed] [Google Scholar]
- 39.Beattie M S, Harrington A W, Lee R, Kim J Y, Boyce S L, Longo F M.et al ProNGF induces p75‐mediated death of oligodendrocytes following spinal cord injury. Neuron 200236375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee R, Kermani P, Teng K K, Hempstead B L. Regulation of cell survival by secreted proneurotrophins. Science 20012941945–1948. [DOI] [PubMed] [Google Scholar]
- 41.Bui N T, Konig H G, Culmsee C, Bauerbach E, Poppe M, Krieglstein J.et al p75 neurotrophin receptor is required for constitutive and NGF‐induced survival signalling in PC12 cells and rat hippocampal neurones. J Neurochem 200281594–605. [DOI] [PubMed] [Google Scholar]
- 42.Lu B. Pro‐region of neurotrophins: role in synaptic modulation. Neuron 200339735–738. [DOI] [PubMed] [Google Scholar]