Abstract
Objective
To determine whether low mannan‐binding lectin (MBL) and C4A null alleles (C4AQ0) are associated with systemic lupus erythematosus (SLE) in multicase families with SLE.
Methods
Low MBL level was determined by measuring serum levels and by genotyping for mutant structural (B/C/D, designated as 0) and promoter (LX) alleles (by real‐time polymerase chain reaction). C4AQ0 was detected by protein electrophoresis and corroborated with haplotype and genotype analysis. In nine Icelandic families, 24 patients with SLE were compared with 83 first‐degree and 23 second‐degree relatives without SLE. Twenty four unrelated family members and a population group of 330 Icelanders served as controls.
Results
Overall, the frequency of low MBL genotypes (0/0, LX/0 and wild‐type/0) tended to be higher in patients with SLE than in their first‐degree and second‐degree relatives (p = 0.06), but the frequency was similar in the families and in the controls (p = 0.6). The frequency of C4AQ0 was, however, increased in patients and their relatives compared with that in the controls (p = 0.04). The combination of low MBL genotypes and C4AQ0 was found more often in the patients than in their relatives (p = 0.03) and controls (p = 0.02). However, low MBL level was observed only in patients and first‐degree relatives in five of the nine multicase families. In these five families, patients with SLE had low MBL genotypes more often (64%) than their first‐degree (38%) and second‐degree (0%) relatives (p = 0.001), and the patients with SLE also had, accordingly, lower MBL levels than their relatives (p = 0.001).
Conclusions
These findings indicate that low MBL levels can predispose people to SLE and highlight the genetic heterogeneity of this disease.
Systemic lupus erythematosus (SLE) is a chronic inflammatory disease that is probably caused by a complex interaction between heterogeneous genetic and environmental factors.1,2 The relative homogeneity of people belonging to multicase families with SLE in well‐defined populations may therefore help to delineate the effect of certain susceptibility alleles on disease expression.3,4,5
The complement system can mediate non‐inflammatory clearance of immune complexes and apoptotic debris in the liver and spleen, but their accumulation may contribute to autoimmunity and inflammation in SLE.2,6,7,8 Complete deficiency of one of the early components of the classical complement pathway is among the strongest known genetic risk factors for SLE, and partial complement deficiencies, such as C4A null alleles (C4AQ0), have also been associated with SLE.2
Mannan‐binding lectin (MBL, also called mannose‐binding lectin) is a serum protein that binds certain surface sugar residue patterns and it also binds various micro‐organisms,9 apoptotic and necrotic cells,10,11,12 nucleic acids,13 phospholipids,14 agalactosylated immunoglobulin (Ig)G15 and IgA.16 Bound MBL activates the complement system through its serine proteases (mannan‐binding lectin‐associated serine proteases) that cleave the complement components C4 and C2.17 MBL mediates non‐inflammatory clearance of apoptotic debris10,11,12 and MBL‐deficient mice show defective apoptotic cell clearance.18 Defects in MBL might, therefore, like other defective early complement components, predispose people to SLE.
MBL normally circulates as a polymer of up to six subunits linked together at their collagenous tails with the sugar‐binding regions at their heads,17 and it must be polymeric to be optimally functional.19,20 The serum concentration of functional MBL is largely genetically determined20,21 and varies widely (0–10 000 μg/l), with a median level around 1000 μg/l in Caucasians,21 including Icelanders.22 The level of MBL is stable for every person over time,23 although it fluctuates slightly during immunosuppressive treatment in patients with SLE with high MBL levels.24
MBL levels <1000 μg/l are mainly associated with three point mutations in exon 1 (in codons 54 (B), 57 (C) and 52 (D)) of the MBL2 gene on chromosome 10 (10q11.2–21). These mutations interfere with the polymeric assembly of the MBL monomers. Low MBL levels can, however, also be associated with the down‐regulating LX promoter polymorphisms, whereas the H and Y promoter alleles are associated with higher MBL levels.17,20 The mutant structural alleles (B, C, D) are commonly designated as 0, in contrast with the wild‐type allele A. People who are homozygous for the A allele (high MBL genotype) have MBL levels >1000 μg/l. Exceptions to this are few and are mostly associated with the LX promoter polymorphisms. Most people who carry the 0 alleles have MBL levels <1000 μg/l, and those who are homozygous for 0, or are heterozygous for 0 and carry the LX promoter polymorphisms on the other chromosome (LXA/0, compound heterozygous), have very low or undetectable levels of MBL.17,20,21 The 0/0, LXA/0, and HYA/0 or LYA/0 genotypes will hereafter be referred to as low MBL genotypes.
The low MBL genotypes are relatively common worldwide, and the B allele is the most common mutant structural allele in Caucasians.9,21,25 Low MBL, detected either as low MBL levels or low MBL genotypes, has been associated with increased susceptibility to infections, especially in immunocompromised patients,9 including those with SLE.24,26 Low MBL is also associated with increased severity of rheumatoid arthritis and SLE,9 and with increased frequency of myocardial infarction in certain groups at risk,23 including patients with SLE.27 Most,24,25,26,28,29,30,31,32,33,34,35,36,37 but not all,38,39,40 previous studies on MBL and SLE have shown that low MBL is associated with a slightly increased risk for SLE, and low MBL and C4AQ0 may have an additive effect.28,30,40
This is, to our knowledge, the first study on multicase families with SLE that deals with the question whether low MBL is associated with SLE. In these families, the frequency of C4AQ0 is increased compared with controls from the general population.3,41 Low MBL was detected in five of the nine families and was significantly associated with SLE in these families. Overall, the combination of low MBL and C4AQ0 was associated with SLE.
Patients and methods
The study was carried out in accordance with the Helsinki Declaration, and was approved by the ethics committee of the University Hospital in Iceland and the Icelandic Computer Database Committee. All participants gave informed consent.
Study group
We examined nine multicase families with SLE from Iceland, which have previously been described in detail.3,42 The total number of patients with SLE studied was 24 (21 women and 3 men), with a mean age of 50 years. All patients fulfilled four or more classification criteria of the American College of Rheumatology for diagnosis of SLE.43 From these nine families, 83 first‐degree and 23 second‐degree relatives without SLE were recruited (56 women and 50 men, mean age 42 years). In addition, 24 unrelated family members (spouses and in‐laws) were included as ethnically, age and to some extent, environmentally matched controls (5 women and 19 men, mean age 52 years). Family members aged >14 years were evaluated through interviews and physical examination by two rheumatologists (GG and KS), and medical records were reviewed following a predefined protocol. Controls for distribution of MBL levels (no DNA samples available for MBL genotyping) were 330 Icelanders aged >18 years, including 130 people who were selected at random from the population and 200 blood donors. We observed no difference in the distribution of MBL levels between these two subgroups.
Genotyping of MBL
A real‐time polymerase chain reaction was carried out in the LightCycler (Roche Diagnostics, Mannheim, Germany), as previously described by Steffensen et al.44 In brief, the three mutant structural alleles (at codons 54 (B), 57 (C) and 52 (D)) in exon 1 were detected in one reaction, and the promoter polymorphisms at position −550 (H→L) and −221 (Y→X) in two reactions. The polymorphism in the 5′ untranslated sequence at position +4 (P/Q) was also detected, but it was not used in the analysis as it was not associated with variability in MBL levels. Due to linkage disequilibrium, only the following seven haplotypes were observed: HYPA, LYPA, LYQA, LXPA, LYPB, LYQC and HYPD. Those who carried the low MBL genotypes—that is, those who were homozygous or heterozygous for the LYPB and HYPD (none with LYQC) haplotypes (0/0, A/0)—were analysed together.
Measurement of serum MBL
A sandwich ELISA system, adopted from Claus Koch at the Statens Serum Institut (Copenhagen, Denmark), was used as previously described.22 The anti‐human MBL detection antibody (Mab clone 131‐01, Statens Serum Institut) used in this assay primarily detects functional polymeric MBL. The lower detection limit of the assay was 20 μg/l.
Complement C4 typing
C4 allotypes were analysed by high‐voltage agarose electrophoresis of serum samples treated with carboxypeptidase (Sigma, Type I) and neuraminidase (Sigma, Type VIII), followed by immunofixation with monoclonal antibodies (Incstar, Stillwater, Minnesota, USA).45 C4A allotypes were determined by the relative intensities of the bands through visual inspection and comparison with control samples. Zygosity was corroborated by haplotype analysis for family members3 and genotype analysis for C4A gene deletion and point mutations in exons 20 and 29 of the C4A gene.41
Statistical analysis
As data were not normally distributed, non‐parametric tests were used to evaluate differences in serum MBL levels between groups. The Mann–Whitney rank sum test was used to compare serum MBL levels between two groups and the Kruskal–Wallis analysis of variance (ANOVA) on ranks was used for comparison of MBL levels among three groups. The frequency of low MBL genotypes and C4AQ0 was analysed using Fisher's exact test when two independent proportions were compared. The χ2 test was used for independence and linear trend between groups when three or four independent proportions were compared. Data were statistically analysed using the Prism 4 (GraphPad, San Diego, California, USA) software. All tests were two sided and the level of significance was set at p<0.05.
Results
MBL levels in relation to MBL genotypes
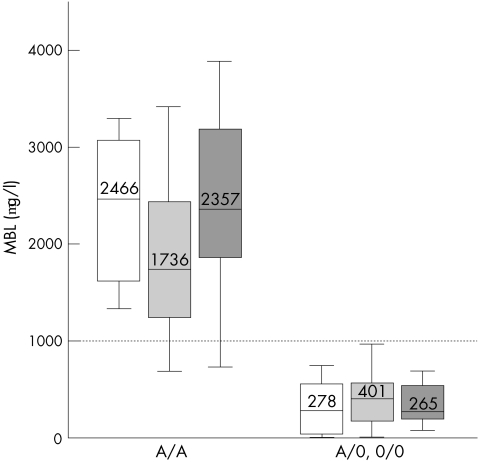
The low MBL genotypes (0/0, LXA/0, LYA/0, HYA/0) were strongly associated with serum MBL levels <1000 μg/l (fig 1) and the distribution of MBL levels in people with and without these low MBL genotypes was similar overall in patients with SLE, in their relatives without SLE and in controls (fig 1). Thus, we found no evidence of MBL consumption. It is also evident from fig 1 that an MBL concentration of 1000 μg/l distinguishes well between people with and those without low MBL genotypes. The few people who carried the high MBL genotype and had MBL levels <1000 μg/l all carried the L promoter polymorphism. Most people with low MBL genotypes carried the B mutant structural allele; a few carried the D allele but no C allele was observed. Only one person who had SLE was homozygous for the 0 alleles (0/0).
Figure 1 Serum levels of mannan‐binding lectin (MBL) for the high MBL (A/A) and low MBL (0/0, LXA/0, HYA/0, LYA/0) genotypes compared in the patients with systemic lupus erythematosus (white), relatives (light grey) and controls (dark grey) using analysis of variance on ranks. Box plots show the median values and interquartiles, and the bars show the 10th centiles of MBL concentrations. Most people carrying high MBL genotypes had MBL levels >1000 μg/l, whereas those carrying low MBL genotypes had MBL levels <1000 μg/l.
MBL and C4AQ0 in all the nine families
Overall, low MBL genotypes were found in 38% of the patients with SLE compared with 26% of first‐degree and 13% of second‐degree SLE relatives without SLE, showing only a marginally significant trend when all the nine families were pooled (p = 0.057; table 1). The frequency of the low MBL genotypes was similar in the families, and in the spouses and in‐laws who served as controls (p = 0.57). The frequency of C4AQ0 is, as previously reported,3 increased in these families compared with controls (p = 0.046), but similar in patients with SLE and in their relatives without SLE (p = 0.46). However, the combination of low MBL genotypes and C4AQ0 was present in 21% of patients with SLE, in 13% of first‐degree relatives and in no second‐degree relative (p = 0.030 for trend) compared with only 4% of controls (p = 0.021) in all the families pooled. A separate analysis of only the 0/0 and LXA/0 genotypes that are associated with very low or undetectable MBL levels showed similar associations with SLE as when they were analysed together with the YA/0 genotypes (data not shown).
Table 1 Frequency of genotypes expressing low levels of mannan‐binding lectin and C4AQ0 in all the nine multicase families with systemic lupus erythematosus.
| Patients with SLE n = 24 (%) | First‐degree relatives n = 83 (%) | Second‐degree relatives n = 23 (%) | p* | p† | Family controls n = 24 (%) | p‡ | p§ | |
|---|---|---|---|---|---|---|---|---|
| Low MBL genotypes | 38 | 26 | 13 | 0.16 | 0.057 | 33 | 0.25 | 0.57 |
| C4AQ0 | 50 | 47 | 39 | 0.73 | 0.46 | 25 | 0.23 | 0.045 |
| Low MBL with C4AQ0 | 21 | 13 | 0 | 0.086 | 0.030 | 4 | 0.079 | 0.021 |
MBL, mannan‐binding lectin, SLE, systemic lupus erythematosus.
*χ2 test for independence between patients with SLE, first‐degree relatives and second‐degree relatives.
†χ2 test for trend between patients with SLE, first‐degree relatives and second‐degree relatives.
‡χ2 test for independence between patients with SLE, relatives and family controls.
§χ2 test for trend between patients with SLE, relatives and family controls.
Analysis of five families with low MBL
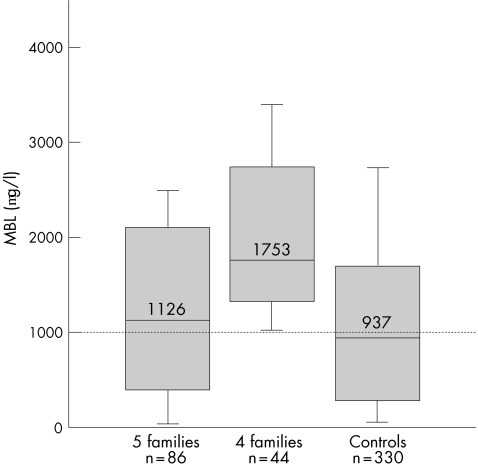
In the course of analysing the data, it became clear that low MBL, detected both as low MBL genotypes and as low serum MBL levels, was detected only in patients and first‐degree relatives in five of the nine families (fig 2). The median MBL concentration in members of the remaining four families was 1753 μg/l (fig 2), which is significantly higher than a median of 937 μg/l in the 330 Icelandic controls (p<0.001), and the distribution of MBL levels in the members of these four families was similar to that observed for the high MBL genotype (fig 1). No member of these four families was homozygous (0/0) or compound heterozygous (LXA/0) for the low MBL alleles, whereas these genotypes were present in all the other five families. However, two second‐degree relatives in one of these four families had low MBL levels (448 and 536 μg/l) and low MBL genotypes (YA/0), but in both cases these were transmitted through in‐laws. C4AQ0 was present in seven of the nine families and one family had neither C4AQ0 nor low MBL.
Figure 2 Serum levels of mannan‐binding lectin (MBL) in the families stratified into five families with and four families without low MBL genotypes compared with 330 adult Icelander controls. Whereas members of the five families with low MBL have MBL levels overall similar to that of the controls (p = 0.2), members of the four families without low MBL genotypes have significantly higher MBL levels (p<0.001) and the distribution is, as expected, similar to that for the high MBL genotype (fig 1). Box plots show the median values and interquartiles, and the bars show the 10th centiles of MBL concentrations.
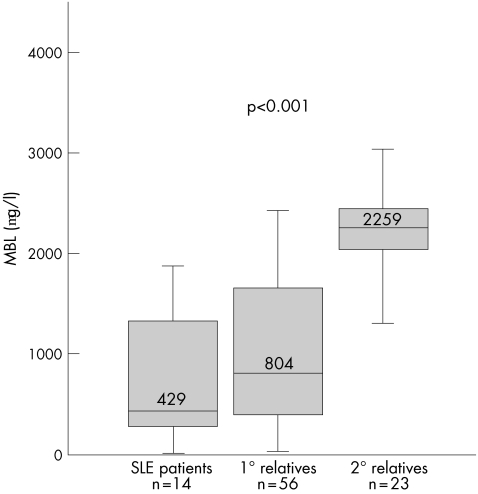
In the five families that had low MBL genotypes, we tested the null hypothesis that the low MBL genotypes were equally transmitted to the members of these families regardless of whether or not they had SLE. However, our findings showed a significant association between SLE and low MBL genotypes in these families (table 2). Thus, 64% of the patients with SLE and 38% of their first‐degree relatives, but none of the second‐degree relatives, had low MBL genotypes (p<0.001 for both independence and trend analysis when all three groups were analysed simultaneously). The combination of a low MBL genotype and C4AQ0 was seen in 36% of the patients and 18% of the first‐degree relatives, but in none of the second‐degree relatives (p = 0.036 for independence and p = 0.011 for trend). Although the distribution of MBL levels in these five families (fig 2) was overall similar to those in the controls (p = 0.2), the patients with SLE had, in accordance with the genetic analysis, much lower MBL levels than their first‐degree and second‐degree relatives (p<0.001, fig 3).
Table 2 Frequency of genotypes expressing low levels of mannan‐binding lectin (MBL) and C4AQ0 in five families after stratification for families with low MBL.
| Patients with SLE, n = 14 (%) | First‐degree relatives, n = 56 (%) | Second‐degree relatives, n = 16 (%) | p* | p* | |
|---|---|---|---|---|---|
| Low MBL genotypes | 64 | 38 | 0 | <0.001 | <0.001 |
| C4AQ0 | 50 | 46 | 44 | 0.94 | 0.73 |
| Low MBL with C4AQ0 | 36 | 18 | 0 | 0.036 | 0.011 |
MBL, mannan‐binding lectin; SLE, systemic lupus erythematosus.
*χ2 test for independence between patients with SLE, first‐degree relatives and second‐degree relatives.
†χ2 test for trend between patients with SLE , first‐degree relatives and second‐degree relatives.
Figure 3 Serum levels of mannan‐binding lectin (MBL) in the five multicase families with systemic lupus erythematosus (SLE) with low MBL. The patients with SLE have lower MBL levels than their first‐degree (1°) relatives, which in turn have lower levels than the second‐degree (2°) relatives. Distribution of MBL levels is compared using analysis of variance on ranks. Box plots show the median values and interquartiles, and the bars show the 10th centiles of MBL concentrations.
This pattern was observed in each of the five families. To test the possibility of a bias due to the different size of the families and variable numbers of relatives, the analysis was repeated with only two first‐degree relatives randomly chosen for each patient and with exclusion of one of two patients who had first‐degree relatives. The findings in this analysis were similar to those for all the members of the five families (data not shown). Whereas all the patients fulfilled ⩾4 classification criteria of the American College of Rheumatology for SLE, some of their relatives fulfilled 1–3 classification criteria and, interestingly, there was an inverse correlation between the number of criteria fulfilled and the MBL levels in the five families who had low MBL genotypes (r = −0.25; p = 0.03).
The association between low MBL genotypes or low serum MBL levels and SLE was observed in the family members regardless of the C4A status, and the same trend was observed in both sexes (data not shown). Only one of the three male patients with SLE belonged to the five families with low MBL, and he carried a low MBL genotype (LXA/0).
Discussion
Most previous studies25,26,28,29,30,31,32,33,34,35,36,37 have shown a rather weak association between low MBL and SLE, but some have reported an additive effect for low MBL and C4AQ0.28,30 However, other studies have failed to confirm such an association,38,39,40 but a recent meta‐analysis, showed a significant overall odds ratio of 1.4 for the mutant structural alleles.46 This is to our knowledge the first study on MBL in multicase families with SLE. When all the nine families were pooled, we found only a borderline association between SLE and low MBL, detected as both low MBL levels and low MBL genotypes, although the combination of low MBL and C4AQ0 showed a major trend association with SLE. Moreover, the frequency of low MBL genotypes was overall similar in the patients with SLE and in the controls. However, when each multicase family was analysed separately, low MBL was associated with SLE in all the five families that showed this defect, whereas low MBL was absent both in the patients with SLE and in their first‐degree relatives in the four remaining families. The frequency of C4AQ0, as previously reported,3,41 was increased overall in the families compared with controls, and the association between low MBL and SLE was independent of C4AQ0. Thus, C4AQ0 may be part of a genetic background predisposing people to SLE, but other genetic components are probably necessary for disease expression. Low MBL genotypes, as observed in five of the nine families, may be one such contributing factor, as well as the PD‐1.3A polymorphism that has previously been associated with SLE in these families.47,48 Our findings may thus provide an example of epistatic genetic effects, highlighting the heterogeneity in predisposition of patients to SLE. Furthermore, the phenotypic effect of low MBL may become apparent only in conjunction with other specific genetic variants or environmental factors.
The strength of this study lies in extended, well‐defined multicase families with SLE, with a high participation rate derived from a relatively homogeneous population of 290 000 inhabitants.49 Furthermore, all family members were evaluated by only two rheumatologists. An additional strength of the study is that the MBL analysis was based on genotyping for all known structural and promoter polymorphisms, corroborated through haplotype analysis,44 and on measurements of serum MBL levels,20,22 and these two methods showed a good agreement (fig 1). Thus, we did not, in accordance with two recent reports,50,51 observe any evidence of MBL consumption in the patients with SLE as has previously been suggested.35 The C4AQ0 allotype was detected by protein electrophoresis and corroborated through haplotype analysis and genotyping for C4A gene deletion and mutations in exons 20 and 29.3,41 Although the study groups are relatively small, the family relations enabled us to evaluate the association between low MBL and SLE susceptibility separately in each of the families. Thus, SLE is clearly associated with transmission of low MBL genotypes in the five families that carried the genetic polymorphism that is associated with low MBL. The two second‐degree relatives in one of the four remaining families are not relevant in this context, as they acquired these genotypes through in‐laws. It should be noted in this connection that a case–control type analysis of the patients participating in this study would probably not have show an association between SLE and low MBL.
The size of the control group of unrelated family members limits the statistical power of the analysis, but these controls had a frequency of low MBL genotypes similar to that observed in another Caucasian population,21 and the distribution of serum MBL levels in our 330 controls was also similar to that in other Caucasians.21 Furthermore, as the unrelated family controls were ethnically and age‐matched members of the families—namely, spouses and in‐laws—they could, to some extent, be regarded as environmentally matched controls.
Various terms have previously been used to describe MBL insufficiency. In this report, a terminology is used that links the genotype to the phenotype—namely, high MBL and low MBL genotypes, which are associated with high and low levels of MBL, respectively. The cut‐off level for insufficient MBL has not been established and probably differs between disease entities.
This study was not designed to elucidate causal mechanisms. The findings are, however, consistent with the notion that high MBL may, similar to an intact classical complement pathway, protect otherwise susceptible people against SLE, presumably by promoting non‐inflammatory clearance of apoptotic debris10,11,12 and immune complexes through the lectin pathway of complement activation.15,16,52 Clearance of apoptotic debris is defective in MBL‐deficient mice, but they did not develop lupus, highlighting that the phenotypic effect of low MBL may become apparent only in conjunction with other specific genetic variants.18
It is concluded that low MBL is overall only marginally associated with SLE, as has been reported in case–control studies, but the combination of low MBL and C4AQ0 is associated with SLE. However, low MBL is clearly associated with SLE within families that show this defect. Our findings in this first family‐based study on MBL in patients with SLE are therefore in accordance with, but extend, previous case–control studies and meta‐analyses of MBL in patients with SLE and highlight the heterogeneous aetiology of SLE.
Acknowledgements
We thank Dr Rudi Steffensen, Aalborg, Denmark, for her guidance with the MBL genotyping, deCODE Genetics for lending the LightCycler Instrument, Kristin Johannesdottir for her technical assistance and Professor Magnus Johannsson for statistical advice.
Abbreviations
ANOVA - analysis of variance
MBL - mannan‐binding lectin
SLE - systemic lupus erythematosus
Footnotes
Funding: This study was supported by grants from The Icelandic Research Fund for Graduate Students, The Science Fund of Landspitali University Hospital, and the Research Fund, University of Iceland.
Competing interests: None.
References
- 1.Illei G G, Tackey E, Lapteva L, Lipsky P E. Biomarkers in systemic lupus erythematosus. I. General overview of biomarkers and their applicability. Arthritis Rheum 2004501709–1720. [DOI] [PubMed] [Google Scholar]
- 2.Pickering M C, Botto M, Taylor P R, Lachmann P J, Walport M J. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol 200076227–324. [DOI] [PubMed] [Google Scholar]
- 3.Kristjansdottir H, Bjarnadottir K, Hjalmarsdottir I B, Grondal G, Arnason A, Steinsson K. A study of C4AQ0 and MHC haplotypes in Icelandic multicase families with systemic lupus erythematosus. J Rheumatol 2000272590–2596. [PubMed] [Google Scholar]
- 4.Johanneson B, Steinsson K, Lindqvist A K, Kristjansdottir H, Grondal G, Sandino S.et al A comparison of genome‐scans performed in multicase families with systemic lupus erythematosus from different population groups. J Autoimmun 199913137–141. [DOI] [PubMed] [Google Scholar]
- 5.Michel M, Johanet C, Meyer O, Frances C, Wittke F, Michel C.et al Familial lupus erythematosus. Clinical and immunologic features of 125 multiplex families. Medicine (Baltimore) 200180153–158. [DOI] [PubMed] [Google Scholar]
- 6.Cline A M, Radic M Z. Apoptosis, subcellular particles, and autoimmunity. Clin Immunol 2004112175–182. [DOI] [PubMed] [Google Scholar]
- 7.Schifferli J A, Ng Y C, Peters D K. The role of complement and its receptor in the elimination of immune complexes. N Engl J Med 1986315488–495. [DOI] [PubMed] [Google Scholar]
- 8.Davies K A, Erlendsson K, Beynon H L, Peters A M, Steinsson K, Valdimarsson H.et al Splenic uptake of immune complexes in man is complement‐dependent. J Immunol 19931513866–3873. [PubMed] [Google Scholar]
- 9.Kilpatrick D C. Mannan‐binding lectin: clinical significance and applications. Biochim Biophys Acta 20021572401–413. [DOI] [PubMed] [Google Scholar]
- 10.Nauta A J, Castellano G, Xu W, Woltman A M, Borrias M C, Daha M R.et al Opsonization with C1q and mannose‐binding lectin targets apoptotic cells to dendritic cells. J Immunol 20041733044–3050. [DOI] [PubMed] [Google Scholar]
- 11.Nauta A J, Raaschou‐Jensen N, Roos A, Daha M R, Madsen H O, Borrias‐Essers M C.et al Mannose‐binding lectin engagement with late apoptotic and necrotic cells. Eur J Immunol 2003332853–2863. [DOI] [PubMed] [Google Scholar]
- 12.Ogden C A, de Cathelineau A, Hoffmann P R, Bratton D, Ghebrehiwet B, Fadok V A.et al C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med 2001194781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palaniyar N, Nadesalingam J, Clark H, Shih M J, Dodds A W, Reid K B. Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose‐binding lectin. J Biol Chem 200427932728–32736. [DOI] [PubMed] [Google Scholar]
- 14.Kilpatrick D C. Phospholipid‐binding activity of human mannan‐binding lectin. Immunol Lett 199861191–195. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra R, Wormald M R, Rudd P M, Fischer P B, Dwek R A, Sim R B. Glycosylation changes of IgG associated with rheumatoid arthritis can activate complement via the mannose‐binding protein. Nat Med 19951237–243. [DOI] [PubMed] [Google Scholar]
- 16.Roos A, Bouwman L H, van Gijlswijk‐Janssen D J, Faber‐Krol M C, Stahl G L, Daha M R. Human IgA activates the complement system via the mannan‐binding lectin pathway. J Immunol 20011672861–2868. [DOI] [PubMed] [Google Scholar]
- 17.Petersen S V, Thiel S, Jensenius J C. The mannan‐binding lectin pathway of complement activation: biology and disease association. Mol Immunol 200138133–149. [DOI] [PubMed] [Google Scholar]
- 18.Stuart L M, Takahashi K, Shi L, Savill J, Ezekowitz R A. Mannose‐binding lectin‐deficient mice display defective apoptotic cell clearance but no autoimmune phenotype. J Immunol 20051743220–3226. [DOI] [PubMed] [Google Scholar]
- 19.Matsushita M, Ezekowitz R A, Fujita T. The Gly‐54→Asp allelic form of human mannose‐binding protein (MBP) fails to bind MBP‐associated serine protease. Biochem J 1995311(Pt 3)1021–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garred P, Larsen F, Madsen H O, Koch C. Mannose‐binding lectin deficiency‐revisited. Mol Immunol 20034073–84. [DOI] [PubMed] [Google Scholar]
- 21.Steffensen R, Thiel S, Varming K, Jersild C, Jensenius J C. Detection of structural gene mutations and promoter polymorphisms in the mannan‐binding lectin (MBL) gene by polymerase chain reaction with sequence‐specific primers. J Immunol Methods 200024133–42. [DOI] [PubMed] [Google Scholar]
- 22.Saevarsdottir S, Vikingsdottir T, Vikingsson A, Manfredsdottir V, Geirsson A J, Valdimarsson H. Low mannose binding lectin predicts poor prognosis in patients with early rheumatoid arthritis. A prospective study. J Rheumatol 200128728–734. [PubMed] [Google Scholar]
- 23.Saevarsdottir S, Oskarsson O O, Aspelund T, Eiriksdottir G, Vikingsdottir T, Gudnason V.et al Mannan binding lectin as an adjunct to risk assessment for myocardial infarction in individuals with enhanced risk. J Exp Med 2005201117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi R, Tsutsumi A, Ohtani K, Muraki Y, Goto D, Matsumoto I.et al Association of mannose binding lectin (MBL) gene polymorphism and serum MBL concentration with characteristics and progression of systemic lupus erythematosus. Ann Rheum Dis 200564311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garred P, Madsen H O, Halberg P, Petersen J, Kronborg G, Svejgaard A.et al Mannose‐binding lectin polymorphisms and susceptibility to infection in systemic lupus erythematosus. Arthritis Rheum 1999422145–2152. [DOI] [PubMed] [Google Scholar]
- 26.Garred P, Voss A, Madsen H O, Junker P. Association of mannose‐binding lectin gene variation with disease severity and infections in a population‐based cohort of systemic lupus erythematosus patients. Genes Immun 20012442–450. [DOI] [PubMed] [Google Scholar]
- 27.Ohlenschlaeger T, Garred P, Madsen H O, Jacobsen S. Mannose‐binding lectin variant alleles and the risk of arterial thrombosis in systemic lupus erythematosus. N Engl J Med 2004351260–267. [DOI] [PubMed] [Google Scholar]
- 28.Davies E J, Snowden N, Hillarby M C, Carthy D, Grennan D M, Thomson W.et al Mannose‐binding protein gene polymorphism in systemic lupus erythematosus. Arthritis Rheum 199538110–114. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan K E, Wooten C, Goldman D, Petri M. Mannose‐binding protein genetic polymorphisms in black patients with systemic lupus erythematosus. Arthritis Rheum 1996392046–2051. [DOI] [PubMed] [Google Scholar]
- 30.Davies E J, Teh L S, Ordi‐Ros J, Snowden N, Hillarby M C, Hajeer A.et al A dysfunctional allele of the mannose binding protein gene associates with systemic lupus erythematosus in a Spanish population. J Rheumatol 199724485–488. [PubMed] [Google Scholar]
- 31.Lau Y L, Lau C S, Chan S Y, Karlberg J, Turner M W. Mannose‐binding protein in Chinese patients with systemic lupus erythematosus. Arthritis Rheum 199639706–708. [DOI] [PubMed] [Google Scholar]
- 32.Carthy D, Hajeer A, Ollier B, Tarassi K, Papasteriades C, Boki K.et al Mannose‐binding lectin gene polymorphism in Greek systemic lupus erythematosus patients. Br J Rheumatol 1997361238–1239. [DOI] [PubMed] [Google Scholar]
- 33.Davies E J, Tikly M, Wordsworth B P, Ollier W E. Mannose‐binding protein gene polymorphism in South African systemic lupus erythematosus. Br J Rheumatol 199837465–466. [DOI] [PubMed] [Google Scholar]
- 34.Tsutsumi A, Sasaki K, Wakamiya N, Ichikawa K, Atsumi T, Ohtani K.et al Mannose‐binding lectin gene: polymorphisms in Japanese patients with systemic lupus erythematosus, rheumatoid arthritis and Sjogren's syndrome. Genes Immun 2001299–104. [DOI] [PubMed] [Google Scholar]
- 35.Ip W K, Chan S Y, Lau C S, Lau Y L. Association of systemic lupus erythematosus with promoter polymorphisms of the mannose‐binding lectin gene. Arthritis Rheum 1998411663–1668. [PubMed] [Google Scholar]
- 36.Villarreal J, Crosdale D, Ollier W, Hajeer A, Thomson W, Ordi J.et al Mannose binding lectin and FcgammaRIIa (CD32) polymorphism in Spanish systemic lupus erythematosus patients. Rheumatology (Oxford) 2001401009–1012. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y F, Wang W, Han J Y, Wu X W, Zhang S T, Liu C J.et al Increased frequency of the mannose‐binding lectin LX haplotype in Chinese systemic lupus erythematosus patients. Eur J Immunogenet 200330121–124. [DOI] [PubMed] [Google Scholar]
- 38.Garcia‐Laorden M I, Manzanedo A, Figuerola A, Sanchez‐Garcia F, Rodriguez‐Gallego C. Mannose‐binding lectin polymorphisms in a Canary Islands (Spain) population. Genes Immun 20012292–294. [DOI] [PubMed] [Google Scholar]
- 39.Horiuchi T, Tsukamoto H, Morita C, Sawabe T, Harashima S, Nakashima H.et al Mannose binding lectin (MBL) gene mutation is not a risk factor for systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) in Japanese. Genes Immun 20001464–466. [DOI] [PubMed] [Google Scholar]
- 40.Jonsen A, Bengtsson A A, Sturfelt G, Truedsson L. Analysis of HLA DR, HLA DQ, C4A, FcgammaRIIa, FcgammaRIIIa, MBL, and IL‐1Ra allelic variants in Caucasian systemic lupus erythematosus patients suggests an effect of the combined FcgammaRIIa R/R and IL‐1Ra 2/2 genotypes on disease susceptibility. Arthritis Res Ther 20046R557–R562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristjansdottir H, Steinsson K. A study of the genetic basis of C4A protein deficiency. Detection of C4A gene deletion by long‐range PCR and its associated haplotypes. Scand J Rheumatol 200433417–422. [DOI] [PubMed] [Google Scholar]
- 42.Grondal G, Traustadottir K H, Kristjansdottir H, Lundberg I, Klareskog L, Erlendsson K.et al Increased T‐lymphocyte apoptosis/necrosis and IL‐10 producing cells in patients and their spouses in Icelandic systemic lupus erythematosus multicase families. Lupus 200211435–442. [DOI] [PubMed] [Google Scholar]
- 43.Hochberg M, for the Diagnostic and Therapeutic Criteria Committee of the American College of Rheumatology Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus [letter]. Arthritis Rheum 1997401725. [DOI] [PubMed] [Google Scholar]
- 44.Steffensen R, Hoffmann K, Varming K. Rapid genotyping of MBL2 gene mutations using real‐time PCR with fluorescent hybridisation probes. J Immunol Methods 2003278191–199. [DOI] [PubMed] [Google Scholar]
- 45.Sim E, Cross S J. Phenotyping of human complement component C4, a class‐III HLA antigen. Biochem J 1986239763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y H, Witte T, Momot T, Schmitdt R E, Kaufman K M, Harley J B.et al The mannose‐binding lectin gene polymorphisms and systemic lupus erythematosus. Two case‐control studies and a meta‐analysis. Arthritis Rheum 2005523966–3974. [DOI] [PubMed] [Google Scholar]
- 47.Prokunina L, Castillejo‐Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V.et al A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 200232666–669. [DOI] [PubMed] [Google Scholar]
- 48.Lindqvist A K, Steinsson K, Johanneson B, Kristjansdottir H, Arnasson A, Grondal G.et al A susceptibility locus for human systemic lupus erythematosus (hSLE1) on chromosome 2q. J Autoimmun 200014169–178. [DOI] [PubMed] [Google Scholar]
- 49.Helgason A, Nicholson G, Stefansson K, Donnelly P. A reassessment of genetic diversity in Icelanders: strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann Hum Genet 200367281–297. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi R, Tsutsumi A, Ohtani K, Goto D, Matsumoto I, Ito S.et al Anti‐mannose binding lectin antibodies in sera of Japanese patients with systemic lupus erythematosus. Clin Exp Immunol 2004136585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seelen M A, Trouw L A, van der Hoorn J W, Fallaux‐van den Houten F C, Huizinga T W, Daha M R.et al Autoantibodies against mannose‐binding lectin in systemic lupus erythematosus. Clin Exp Immunol 2003134335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saevarsdottir S, Vikingsdottir T, Valdimarsson H. The potential role of mannan‐binding lectin in the clearance of self‐components including immune complexes. Scand J Immunol 20046023–29. [DOI] [PubMed] [Google Scholar]