Abstract
Human and bovine Streptococcus bovis strains had the same 16S ribosomal DNA restriction fragment length polymorphism and often had the same patterns of starch, mannitol, lactose, and raffinose utilization. PCRs of BOX sequences differed, but numerical analyses indicated that some human strains clustered with bovine strains. However, human and bovine strains had distinctly different sensitivities to lysozyme and 2-deoxyglucose.
Streptococcus bovis is a gram-positive, facultative anaerobe that causes ruminal acidosis in cattle (12) and meningitis, septicemia, and endocarditis in humans (2, 4, 6, 7, 10, 11, 16, 17). There has also been an association between S. bovis and colonic lesions that give rise to cancer (15, 21). However, it is not clear if ruminal S. bovis can colonize the human colon or conversely if human strains can colonize the rumen.
In the 1980s, human S. bovis strains were separated into different biotypes on the basis of differences in substrate utilization (19, 20). Biotype I uses starch and mannitol, but biotype II uses only one of these substrates. Biotype II can be further subdivided. Biotype II/1 uses starch but not mannitol, whereas II/2 strains do not use either mannitol or starch. This classification scheme has been used to diagnose S. bovis infections (5, 8). Ruoff et al. (17) correlated biotype I bacteremia with gastrointestinal lesions, and Clarridge et al. (3) found the II/2 biotype to be prevalent in males suffering from endocarditis, sepsis, and urinary tract infection. Bovine S. bovis seems to be biotype II/1, but only a few strains were examined (3).
Bovine S. bovis strains (Table 1) grew rapidly in an anaerobic medium (described in reference 13) that was supplemented with glucose, and the doubling times were less than 30 min (data not shown). Most human strains (13 of 14) also grew as rapidly on glucose as the bovine strains, but strain 6448 grew poorly (optical density of only 0.4) and did not adapt. Substrate utilization experiments indicated that 13 of 15 human strains were either biotype I or II/1, but two strains were not biotype I or II (positive for mannitol and negative for starch). All of the bovine strains could be classified as biotype II/1, but it should be noted that some human strains were as effective as bovine strains in utilizing starch (Table 2).
TABLE 1.
Origin of bacterial strains
| Strain | Origin | Reference |
|---|---|---|
| 3-01 to 3-10 | Cattle (n = 2) fed a grain-based ration; ruminal pH of 5.6; Ithaca, N.Y.; 1995 | 22a |
| 5-02 to 5-17 | Cattle (n = 2) fed diet that was 50% grain, 50% hay; ruminal pH 6.3; Ithaca, N.Y.; 1995 | 22a |
| 7-02 to 5-24 | Cattle (n = 2) fed hay diet; ruminal pH 6.7; Ithaca, N.Y.; 1995 | 22a |
| JB1 | Cow fed hay diet; ruminal pH 6.8; Davis, Calif.; 1976 | 17a |
| K27FF4 | Cow fed hay; ruminal pH 6.7; Pretoria, South Africa; prior to 1984 | 17a |
| 26, 581AXY2 | Cow fed hay; ruminal pH 6.8; Aberdeen, Scotland; prior to 1984 | 17a |
| ATCC 33317 | Cow feces | 20 |
| ATCC 1535 | Cow fed hay; ruminal pH 6.8; Champaign, Ill.; 1960 | 26 |
| HC5 | Cow fed hay; ruminal pH 6.7; Ithaca, N.Y.; 2000 | 16a |
| FM | Human clinical isolates; meningitis | 10 |
| RG | Human clinical isolate; septic arthritis | 11 |
| V1477, V1388, V1387 | Human clinical isolates; unknown disease; Richmond, Va.; prior to 1993 | 23 |
| 6, 1314, 1499, 2703, 6448, 9410 | Human clinical isolates; colon; Boston, Mass.; prior to 1989 | 17 |
| ATCC 43143, ATCC 43144 | Human blood | 23 |
| ATCC 49133, ATCC 49147 | Human clinical isolates; unknown origin | 23 |
TABLE 2.
Physiological and phylogenetic characteristics of human and bovine S. bovis strains
| Strain | Biotype | Substrate utilizationa
|
RFLP | BOX-PCR | |||
|---|---|---|---|---|---|---|---|
| Starch | Raffinose | Lactose | Mannitol | ||||
| Bovine | |||||||
| 3-01 | II/1 | + | + | + | − | A | B10 |
| 3-02 | II/1 | + | + | + | − | A | B10 |
| 3-03 | II/1 | + | + | + | − | A | B9 |
| 3-04 | II/1 | + | + | + | − | A | B9 |
| 3-05 | II/1 | + | + | + | − | A | B10 |
| 3-06 | II/1 | + | + | + | − | A | B10 |
| 3-07 | II/1 | + | + | + | − | A | B11 |
| 3-08 | II/1 | + | + | + | − | A | B10 |
| 3-09 | II/1 | + | + | + | − | A | B10 |
| 3-10 | II/1 | + | + | + | − | A | B5 |
| 5-02 | II/1 | + | + | + | − | A | B7 |
| 5-04 | II/1 | + | − | + | − | A | B9 |
| 5-05 | II/1 | + | + | + | − | A | B13 |
| 5-06 | II/1 | + | + | + | − | A | B4 |
| 5-07 | II/1 | + | + | + | − | A | B9 |
| 5-08 | II/1 | + | + | + | − | A | B9 |
| 5-09 | II/1 | + | + | + | − | A | B9 |
| 5-11 | II/1 | + | + | + | − | A | B13 |
| 5-12 | II/1 | + | − | + | − | A | B9 |
| 5-17 | II/1 | + | + | + | − | A | B9 |
| 7-02 | II/1 | + | + | + | − | A | B9 |
| 7-04 | II/1 | + | + | + | − | A | B9 |
| 7-06 | II/1 | + | + | + | − | A | B9 |
| 7-10 | II/1 | + | + | + | − | A | B9 |
| 7-14 | II/1 | + | + | + | − | A | B9 |
| 7-17 | II/1 | + | + | + | − | A | B10 |
| 7-20 | II/1 | + | + | + | − | A | B9 |
| 7-21 | II/1 | + | + | + | − | A | B9 |
| 7-24 | II/1 | + | + | + | − | A | B9 |
| JB1 | II/1 | + | + | + | − | A | B2 |
| K27FF4 | II/1 | + | + | + | − | A | B1 |
| 26 | II/1 | + | + | + | − | A | B8 |
| 581AXY2 | II/1 | + | + | + | − | A | B8 |
| ATCC 33317 | II/1 | + | + | + | − | A | B11 |
| ATCC 15351 | II/1 | + | + | + | − | A | B3 |
| HC5 | II/1 | + | + | + | − | A | B12 |
| Human | |||||||
| FM | II/1 | W | W | + | − | A | H1 |
| RG | I | + | + | + | + | A | H4 |
| V1387 | I | + | + | + | + | A | H4 |
| V1388 | NAb | − | + | + | + | A | H4 |
| V1477 | I | + | + | + | + | A | H4 |
| 6 | II/1 | W | + | + | − | A | H1 |
| 1314 | I | + | + | + | + | A | H4 |
| 1499 | II/1 | + | W | − | − | A | H1 |
| 2703 | I | + | + | W | + | A | H2 |
| 6448 | II/1 | + | + | + | − | A | H5 |
| 9410 | I | + | + | + | + | A | H4 |
| ATCC 43143 | NA | − | + | + | + | A | H4 |
| ATCC 43144 | II/1 | + | + | + | − | A | B10 |
| ATCC 49133 | II/1 | W | + | + | − | A | H3 |
| ATCC 49147 | I | W | + | + | + | A | H4 |
The ability to utilize starch, raffinose, lactose, and mannitol is shown as follows: +, utilizes; −, cannot utilize; W, weak growth. See Fig. 2 for BOX sequence designations.
NA, not applicable.
Previous researchers attempted to use molecular methods as diagnostic tools to differentiate S. bovis strains. Songy et al. (21) isolated DNA sequences that were specific for biotype I strains, but bovine strains were not examined. Whitehead and Cotta (23, 24) developed 16S ribosomal DNA probes that differentiated bovine and human S. bovis strains, but only a small number of strains were employed.
When S. bovis DNA was amplified and digested with HaeIII and HhaI (as described previously [13]), the same six dominant fragments were always observed, and human and bovine strains could not be differentiated (data not shown). The 16S ribosomal DNA genes were not sequenced, but a BLAST search indicated that human (n = 2) and bovine strains (n = 6) were more than 98% identical, and differences between human and bovine strains were in some cases less than the differences between bovine strains.
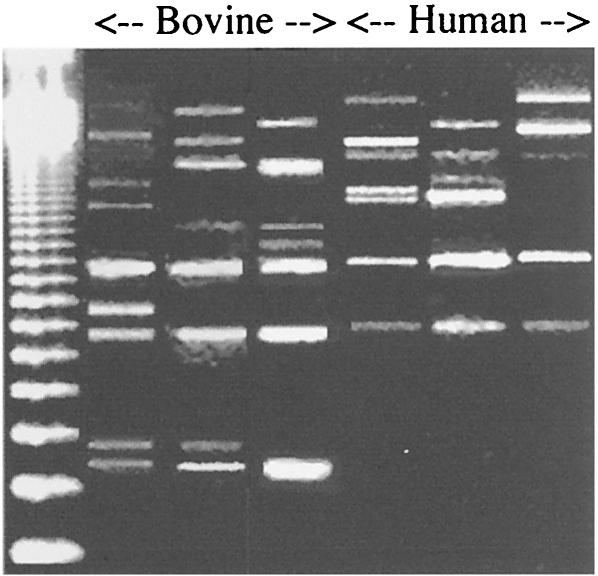
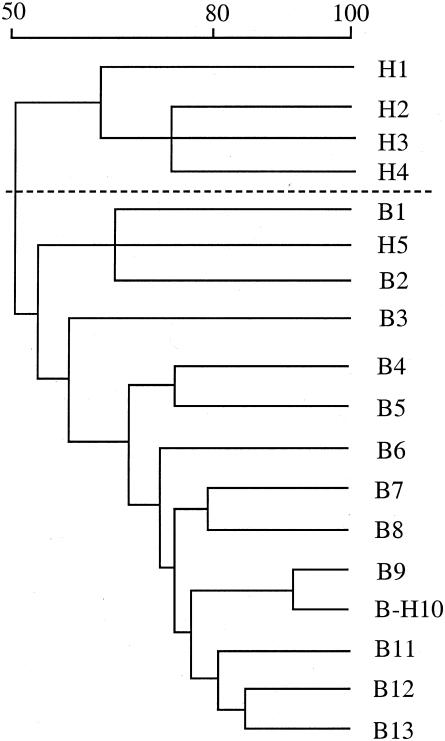
Recent work indicated that bovine strains had different profiles of repetitive DNA (BOX sequences) (13), and BOX-PCR (described in reference 13) indicated that human and bovine strains often could be differentiated (Fig. 1). Unweighted pair group method (UPGAMA) analysis (13) indicated that the dice similarity coefficients differed by as much as 50% (Fig. 2). Most human strains (13 of 15) could be grouped into BOX types that did not include bovine strains, but two human strains clustered more closely with bovine strains than with other human strains.
FIG. 1.
Agarose gel of S. bovis BOX fragments that were obtained by PCR with a BOX A1R primer. Only selected bovine (from left to right, strains JB1, 33317, and 5-11) and human (from left to right, strains 9410, 49133, and FM) are shown, but a complete UPGAMA analysis of all strains is shown in Fig. 2. See Table 2 for other characteristics. A 123-bp ladder is shown in the leftmost lane.
FIG. 2.
Dendrogram of relatedness based on BOX-PCR and weighted average linkages (UPGAMA). BOX type similarity values (expressed as a percentage) are shown over the dendrogram. BOX-type designations for the strains are shown in Table 2.
The ability of bacteria to colonize the gastrointestinal tract has been correlated with cell surface properties (22). S. bovis is a Lancefield group D streptococcus, and both human and bovine strains have this serotype (23). Human S. bovis strains use lipoteichoic acids (LTAs) to facilitate their attachment to human gut epithelium (9, 22, 25), but the role of these acids in lysozyme resistance had not been clearly defined. Lysozyme is an antimicrobial enzyme that is found in mammalian secretions, insects, plants, and bacteria.
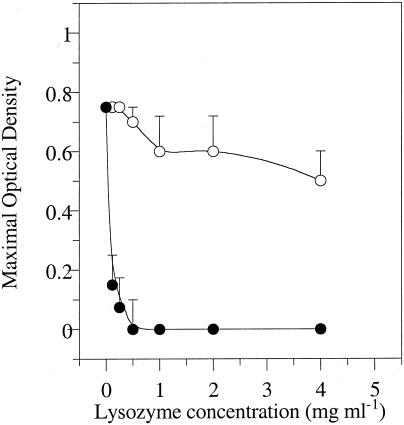
Our results indicated that bovine S. bovis strains were inherently more susceptible to lysozyme than human strains, and most bovine strains were inhibited by as little as 0.13 mg of lysozyme ml−1 (Fig. 3). Even the most resistant bovine strains could not grow if the lysozyme concentration was greater than 0.5 mg ml−1. The bovine strains could be forced to adapt to tolerate higher concentrations of lysozyme by transferring them with sublethal doses of lysozyme (0.06 mg ml−1), but they were initially four- to eightfold more sensitive than human strains.
FIG. 3.
Effect of lysozyme on growth (optical density) of bovine (•) and human (○) S. bovis strains. The inoculum size was 5% (vol/vol), and the incubation period was 24 h. There were 30 bovine strains and 15 human strains. Means ± standard deviations (error bars) are shown.
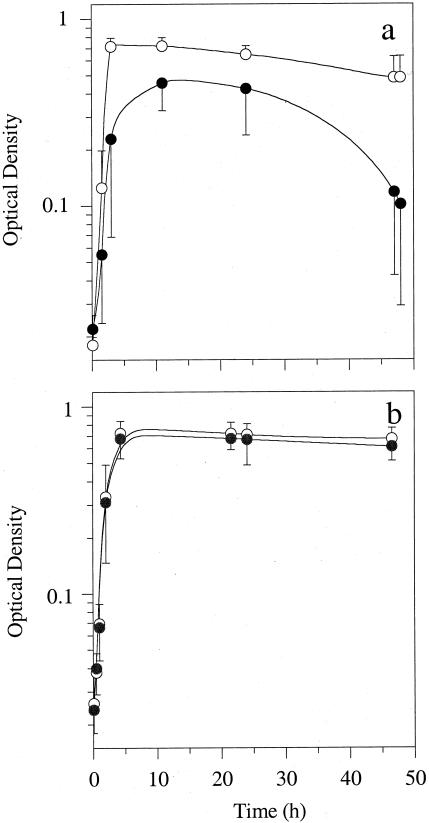
The LTAs of bovine S. bovis strains are an autolytic regulator (1, 18, 25). If 2-deoxyglucose (2DG) is added to the growth medium, the kojibiose moiety of the LTA is not synthesized, the autolysins cannot be inactivated, and the cells lyse (1, 14). Our experiments indicated that all bovine strains eventually lysed if 2DG (2 mg ml−1) was added to the growth medium (Fig. 4a), but none of the human strains were affected (Fig. 4b). Because the bovine strains did not adapt, 2DG sensitivity appeared to be a useful tool for separating human and bovine strains.
FIG. 4.
Effects of 2DG on the growth and lysis of bovine (a) and human (b) S. bovis strains. Cultures were grown in anaerobic medium supplemented with glucose (2 mg ml−1) (○) or with glucose and 2DG (2 mg ml−1 each) (•). There were 30 bovine strains and 14 human strains (strain 6448 was excluded as described in the text). Means ± standard deviations (error bars) are shown.
In recent years, S. bovis has been classified as an “increasingly important pathogen” (21). However, animal models for human S. bovis infections have not been developed, and it is not clear whether bovine strains can pass from cattle to humans to cause infection. Molecular techniques did not readily differentiate bovine S. bovis strains from human strains, but simple growth experiments indicated that they were different. Further work will be needed to see if human and bovine strains should be separated into different species, but it appears that lysozyme and 2DG sensitivities could be useful diagnostic tools.
Acknowledgments
Amina Kurtovic was supported by a Howard Hughes Cornell Undergraduate Research Scholarship.
We thank Terrence Whitehead for providing us many bacterial cultures.
Proprietary or brand names are necessary to report factually on available data; however, the U.S. Department of Agriculture neither guarantees nor warrants the standard of the product, and the use of the name by the U.S. Department of Agriculture implies no approval of the product, and exclusion of others that may be suitable.
REFERENCES
- 1.Bond, D. R., B. M. Tsai, and J. B. Russell. 1999. Physiological characterization of Streptococcus bovis mutants that can resist 2-deoxyglucose-induced lysis. Microbiology 145:2977-2985. [DOI] [PubMed] [Google Scholar]
- 2.Burns, C. A., R. McCaughey, and C. B. Lauter. 1985. The association of Streptococcus bovis fecal carriage and colon neoplasia: possible relationship with polyps and their premalignant potential. Am. J. Gastroenterol. 80:42-46. [PubMed] [Google Scholar]
- 3.Clarridge, J. E., III, S. M. Attorri, Q. Zhang, and J. Bartell. 2001. 16S ribosomal DNA sequence analysis distinguishes biotypes of Streptococcus bovis: Streptococcus bovis biotype II/2 is a separate genospecies and the predominant clinical isolate in adult males. J. Clin. Microbiol. 39:1549-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, L. F., S. A. Dunbar, D. Sirbasku, and J. E. Clarridge III. 1997. Streptococcus bovis infection of the central nervous system: report of two cases and review. Clin. Infect. Dis. 25:819-823. [DOI] [PubMed] [Google Scholar]
- 5.DeVriese, L. A., P. Vandamme, B. Pot, M. Vanrobaeys, K. Kersters, and F. Haesebrouk. 1998. Differentiation between Streptococcus gallolyticus strains of human clinical and veterinary origins and Streptococcus bovis strains from the intestinal tracts of ruminants. J. Clin. Microbiol. 36:3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval, X., V. Papastamopoulos, P. Longuet, C. Benoit, C. Perronne, C. Leport, and J. L. Vilde. 2001. Definite Streptococcus bovis endocarditis: characteristics in 20 patients. Clin. Microbiol. Infect. 7:3-10. [DOI] [PubMed] [Google Scholar]
- 7.Ellmerich, S., M. Scholler, B. Duranton, F. Gosse, M. Galluser, J. P. Klein, and F. Raul. 2000. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis 21:753-756. [DOI] [PubMed] [Google Scholar]
- 8.Facklam, R. R., D. L. Rhoden, and P. B. Smith. 1984. Evaluation of the rapid Strep system for identification of clinical isolates of Streptococcus species. J. Clin. Microbiol. 20:894-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, W. 1988. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29:233-302. [DOI] [PubMed] [Google Scholar]
- 10.Grant, R. J., T. R. Whitehead, and J. E. Orr. 2000. Streptococcus bovis meningitis in an infant. J. Clin. Microbiol. 38:462-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, R. J., W. Y. Shang, and T. R. Whitehead. 1997. Isolated septic arthritis due to Streptococcus bovis. Clin. Infect. Dis. 24:1021. [DOI] [PubMed] [Google Scholar]
- 12.Hungate, R. E., R. W. Dougherty, M. P. Bryant, and R. M. Cello. 1952. Microbiological and physiological changes associated with acute indigestion in sheep. Cornell Vet. 42:423-449. [PubMed] [Google Scholar]
- 13.Jarvis, G. N., A. Kurtovic, A. G. Hay, and J. B. Russell. 2000. The physiological and genetic diversity of bovine Streptococcus bovis strains. FEMS Microb. Ecol. 35:49-56. [DOI] [PubMed] [Google Scholar]
- 14.Kessler, R. E., J. Duke, and I. J. Goldstein. 1984. Interaction of anti-kojibiose antibody with the lipoteichoic acids from Streptococcus faecalis and Streptococcus faecium. Infect. Immun. 46:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, R. S., R. A. Reco, M. T. Catalano, S. C. Edberg, J. I. Casey, and N. H. Steibeigel. 1977. Association of Streptococcus bovis with carcinoma of the colon. N. Engl. J. Med. 297:800-802. [DOI] [PubMed] [Google Scholar]
- 16.Knight, R. G., and D. M. Shalaes. 1985. Physiological characteristics and deoxyribonucleic acid relatedness of human isolates of Streptococcus bovis and Streptococcus bovis (var.). Int. J. Syst. Bacteriol. 35:357-361. [Google Scholar]
- 16a.Mantovani, H. C., D. K. Kam, J. K. Ha, and J. B. Russell. 2001. The antibacterial activity and sensitivity of Streptococcus bovis strains isolated from the rumen of cattle. FEMS Microbiol. Ecol. 37:223-229.
- 17.Ruoff, K. L., S. I. Miller, C. V. Garner, M. J. Ferraro, and S. B. Calderwood. 1989. Bacteremia with Streptococcus bovis and Streptococcus salivarius: clinical correlates of more accurate identification of isolates. J. Clin. Microbiol. 27:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Russell, J. B., and P. H. Robinson. 1984. Compositions and characteristics of strains of Streptococcus bovis. J. Dairy Sci. 67:1525-1531. [DOI] [PubMed]
- 18.Russell, J. B., and J. E. Wells. 1997. The ability of 2-deoxyglucose to promote the lysis of Streptococcus bovis via a mechanism involving cell wall stability. Curr. Microbiol. 35:299-304. [DOI] [PubMed] [Google Scholar]
- 19.Schleifer, K. H., and R. Kilpper-Balz. l987. Molecular and chemotaxonomic approaches to the classification of streptococci, enterococci and lactococci: a review. Syst. Appl. Microbiol. 10:1-19. [Google Scholar]
- 20.Skerman, V. B. D., V. McGowan, and P. H. A. Sneath. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225-420. [Google Scholar]
- 21.Songy, W. B., K. L. Ruoff, R. R. Facklam, M. J. Feraro, and S. Falkow. 2002. Identification of Streptococcus bovis biotype I strains among S. bovis clinical isolates by PCR. J. Clin. Microbiol. 40:2913-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Von Hunolstein, C., L. M. Ricci, and G. Orefici. 1993. Adherence of glucan-positive and glucan-negative strains of Streptococcus bovis to human epithelial cells. J. Med. Microbiol. 39:53-57. [DOI] [PubMed] [Google Scholar]
- 22a.Wells, J. E., D. O. Krause, T. R. Callaway, and J. B. Russell. 1997. A bacteriocin-mediated antagonism by ruminal lactobacilli against Streptococcus bovis. FEMS Microbiol. Ecol. 22:237-243.
- 23.Whitehead, T. R., and M. A. Cotta. 1993. Development of a DNA probe for Streptococcus bovis by using a cloned amylase gene. J. Clin. Microbiol. 31:2387-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehead, T. R., and M. A. Cotta. 2000. Development of molecular methods for identification of Streptococcus bovis from human and ruminal origins. FEMS Microb. Lett. 182:237-240. [DOI] [PubMed] [Google Scholar]
- 25.Wicken, A. J., and K. W. Knox. 1975. Lipoteichoic acids: a new class of bacterial antigen. Science 187:1161-1167. [DOI] [PubMed] [Google Scholar]
- 26.Wolin, M. J. 1960. Some factors affecting the growth of Streptococcus bovis on chemically defined media. J. Dairy Sci. 43:825-830.