Abstract
Differential delayed-type hypersensitivity skin testing with tuberculin purified protein derivatives from Mycobacterium bovis and M. avium is the standard for diagnosing bovine tuberculosis. However, improved tests based on defined, specific antigens are urgently needed. In the present study, a combination of bioinformatics, molecular biology, and bovine models of infection were used to screen mycobacterial proteins for their potential as diagnostic reagents which could be used in a whole-blood assay for diagnosis of tuberculosis. Initial screening of 28 proteins selected in silico and expressed as recombinants in Escherichia coli indicated that CFP-10, ESAT-6, TB27.4, TB16.2, TB15.8, and TB10.4 induced strong gamma interferon responses in experimentally infected cattle. A more thorough investigation over time in two groups of animals infected with a high (106 CFU) and a low (104 CFU) dose of M. bovis revealed that, for both groups, the strength of the in vitro response to individual antigens varied greatly over time. However, combining the results for ESAT-6, CFP-10, and TB27.4, possibly supplemented with TB10.4, gave sensitivities at different infection stages close to those obtained with M. bovis purified protein derivative. Importantly, while responsiveness to ESAT-6 and CFP-10 correlated strongly for individual samples, the same was not the case for ESAT-6 and TB27.4 responsiveness. The results suggest that combinations of specific antigens such as these have great potential in development of optimized diagnostic systems for bovine tuberculosis.
The current worldwide status of human carrier and clinical tuberculosis (14) has to be linked to the zoonotic threat posed by Mycobacterium bovis, the agent of bovine tuberculosis (28), particularly to immunosuppressed patients (12), and to the epidemiological knowledge that M. bovis infection in cattle is on the increase in some countries, including the United Kingdom (United Kingdom Department for Environment, Food and Rural Affairs Statistics at http://www.defra.gov.uk/animalh/tb). Bovine tuberculosis is also of major importance in many countries as an economic cost to the agricultural industry, requiring massive annual expenditures for its control and eradication (22).
Conventional statutory diagnosis in cattle relies on intradermal skin testing with the purified protein derivative (PPD) of M. bovis (PPDB), often compared to that of M. avium (PPDA), and lacks both specificity and sensitivity (25). As an alternative, the use of an antigen-specific in vitro gamma interferon (IFN-γ) assay has shown promise to detect various infectious diseases in cattle, including tuberculosis (44, 46). This test is based on the detection of IFN-γ released specifically from whole blood cultures after stimulation with PPDB. However, diagnostic tests based on PPDs are limited by the fact that most of the proteins in these preparations are shared among mycobacterial species and that tuberculins are poorly defined, resulting in batch-to-batch variation. Since cattle are commonly sensitized by exposure to nonpathogenic, environmental mycobacteria, the specificity of the classical skin test and in vitro diagnostic tests can be compromised in some circumstances (1). Therefore, because accurate diagnosis plays an important role in the control of tuberculosis, the development of cost-effective, specific diagnostic assays to allow improved detection of disease is urgently needed.
Bovine T-cell epitopes have been characterized for several mycobacterial proteins (32, 33). However, studies of the diagnostic potential of these antigens have generally indicated that they are not recognized either widely enough among tuberculous cattle or constantly enough within individuals for the development of tests with high sensitivity. More recently, the potential of ESAT-6 and CFP-10 as diagnostic candidates has been demonstrated in cattle (30, 42). Both of these proteins are important T-cell targets for tuberculous cattle but are absent from many environmental mycobacteria known to induce nonspecific skin test responses to PPDB (2, 19, 40). ESAT-6 and CFP-10 are therefore good examples of diagnostic antigens for a specific tuberculosis test. However, to make such a test more sensitive and robust among genetically heterogeneous animals, different stages of disease progress, and different exposures to environmental bacteria, it seems likely these proteins should be supplemented with other M. bovis-specific proteins.
The increasing number of genome sequencing projects has presented the opportunity of using database searches as a fast and convenient tool for selecting genes with diagnostic potential. It is now realistic to screen in silico a large number of genes, even whole bacterial genomes, and thereby narrow down the number of candidates to a manageable size. The genome of M. bovis consists of approximately 4,000 genes (http://www.sanger.ac.uk/Projects/M_bovis) coding for almost the same number of proteins, not taking into account multiple transcriptional and translational start and stop sites and posttranslational modifications for each coding region. While it would be a huge task to test all these proteins in a screening process for diagnostic antigens, current progress in genome sequencing of mycobacterial chromosomes allows comparative searches of the entire M. avium, M. bovis, M. leprae, and M. tuberculosis genomes and the almost complete M. marinum, M. paratuberculosis, M. smegmatis, and M. ulcerans genomes. This information can be used to select diagnostic candidates with theoretical specificity to M. bovis.
In this study, we combined bioinformatics and natural-host animal models of tuberculosis infection to identify mycobacterial proteins that could be included in a new M. bovis-specific diagnostic protein cocktail.
MATERIALS AND METHODS
Animals and infection.
Three groups (A, B, and C) of male Friesian cattle aged 4 to 6 months from a tuberculosis-free background were kept in isolation pens and infected intranasally with M. bovis strain AF2122/97, as described previously (27). Group A (six animals) was infected with 104 CFU, and samples from all six animals were collected 126 and 154 days postinfection and from three animals at 184 days postinfection. Groups B and C (six animals each) received 104 and 106 CFU, respectively. All animals were monitored clinically for 7 months postinfection, and blood samples were collected every 2 weeks. Twenty-two cattle from a source known to give nonspecific skin responsiveness to PPD were used as examples of environmentally sensitized individuals to investigate antigen specificity, and samples were processed within 8 h of collection.
IFN-γ assay.
A commercial bovine IFN-γ microplate enzyme-linked immunosorbent assay (ELISA) (Bovigam, CSL, Australia) was used to measure T-cell responses in whole-blood cultures after a 24-h in vitro antigen stimulation. Heparinized blood (200 μl) was incubated on microplates in duplicate for each antigen at a final concentration of 4 μg/ml. Duplicate control wells to which only phosphate-buffered saline was added were also established. Because the coefficient of variation between duplicate wells has been found to be less than 5% (31), culture supernatants were pooled before testing in the IFN-γ ELISA. The optical density for phosphate-buffered saline controls was less than 0.1. As the cutoff for positive antigen responsiveness, the average optical density for the phosphate-buffered saline control in each experiment plus 5 times the standard deviation was used.
Antigen selection and computer analysis.
Since the M. bovis genome has been completely sequenced but the annotations have not been completed, interesting genes were identified in the M. tuberculosis database (http://genolist.pasteur.fr/TubercuList/). M. tuberculosis sequences of interest were compared to the M. bovis genome with Blast to confirm a 99 to 100% DNA sequence match. Subsequently, only proteins with low M. avium homology were selected by comparing the M. bovis gene sequences against the M. avium genome with TBlastN (protein versus translated DNA). After the initial screening, the interspecies distribution of the best-performing proteins was investigated by searching the available mycobacterial genomes and the GenBank bacterial database.
Complete M. bovis, M. leprae, and M. tuberculosis H37Rv and almost complete M. marinum genome sequences are available at http://www.sanger.ac.uk/projects. Complete M. avium and M. tuberculosis CDC1551 and almost complete M. smegmatis and M. tuberculosis strain 210 genome sequences are available at http://www.tigr.org/tdb, the almost complete M. paratuberculosis genome sequence is available at http://www.cbc.umn.edu/ResearchProjects/AGAC/Mptb/Mptbhome.html, and the almost complete M. ulcerans sequence can be Blast searched at http://genopole.pasteur.fr/Mulc/BuruList.html.
Strains, growth media, and plasmids.
Escherichia coli strain DH5α was used for all cloning steps, and E. coli strain BL21 or BL21 SI was used for protein expression. Luria-Bertani medium with 100 mg of ampicillin per liter was used in plates and cultures for E. coli strains DH5α and BL21, whereas BL21 SI clones were grown in 10 g of yeast extract and 5 g of peptone per liter. The expression vectors pMCT6 (18) and pDest17 (Invitrogen) were used in this study. Both plasmids add an N-terminal His tag to the recombinant protein.
PCR, primer design, and cloning.
The oligonucleotides used for PCR amplification (Table 1) were designed either with restriction sites for cloning of amplified fragments into expression vector pMCT6 or with recombination sites for cloning into pDest17 via pDONR201 (Invitrogen). The gene-specific part of each oligonucleotide was determined with the Primer Express software (PE Applied Biosystems) with standard settings and an annealing temperature of 60°C. All PCRs were conducted in 50 μl with Taq polymerase (Platinum Taq; Invitrogen). Primer concentrations were 0.4 μM, and the DNA source was chromosomal M. tuberculosis H37Rv DNA. The PCRs were initiated by a 3-min 95°C denaturation, followed by 30 to 35 cycles of denaturation (95°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 1 min/kb) and a final extension (72°C, 10 min). Standard protocols were followed for restriction cloning, and the manufacturer's protocol was followed for recombinational cloning (Invitrogen).
TABLE 1.
Primers used for PCR amplification and cloninga
| Rv no. | Forward primer sequence, 5′ → 3′ | Reverse primer sequence, 5′ → 3′ |
|---|---|---|
| 3872 | CTTCCCGGG ATG GAA AAA ATG TCA C | GATGCCATGG TTA GGC GAA GAC GCC GGC |
| 3873 | CTGGGGATCC GC GTG ATC ACC ATG CTG TGG | TGCAAGCTT TCA CCA GTC GTC CTC TTC GTC |
| 3878 | CTT AGA TCT GCT GAA CCG TTG GCC GTC | CCGCTCGAG CTA CAA CGT TGT GGT TGT |
| 1982* | ATC GTG GAC ACA AGC GCC GT | CGC GAC GCC TGG CCA |
| 1988* | GTG TCC GCC CTC GGA C | CCG CCC CTG CCA GT |
| 1573* | ACC ACC ACA CCA GCA CGT | GCG CTC GTC TTC CGC T |
| 1575* | GCG CCG CTG GCC GC | GAT GTG CTG GTG CGC AAC GC |
| 1579* | GTG ACC CCG ATC AAC CGG C | CGA TGG CGA CCC CGC |
| 1580* | GCT GAA ACC CCC GAC CAC | CTG GTC GAC CTC TAT GGT GTC GT |
| 1582* | GCC GAC ATC CCC TAC GGC | ATC ATC GAA ATC ATC GGC CC |
| 1584* | GTG TCG ACC ATC TAC CAT CAT CGC | TCG GGC ACC GCC TGA CC |
| 1585* | AGC CGG CAC CAC AAC ATC GT | GCA GCT ACC ACG CGT TGG GGT |
| 1586* | GTG AGA TAC ACT ACA CCT GTG CGT | TCG CCA ATT CAC CTG CAC |
| 2660* | GTG ATA GCG GGC GTC GA | GTG AAA CTG GTT CAA TCC CAG TA |
| 64* | GAG ACA GGT TCG CCG GGA | ACC GGG CGT TTC GGC |
| 398* | GGA GTC ATT GCC CGC GTT | CCA TGA GCT CGC GGT GGT |
| 450* | GTG AGT ACT AAA TTC GCG AAC GAC T | GAG GCG GTC GCT GCT CAT |
| 559* | AAG GGA ACA AAG CTG GCT GTT | GGT GCC AGC CCG TTG C |
| 569* | AAG GCA AAG GTC GGG GAC | CGT TCC CCT GGC ATG GA |
| 617* | TTA GTG ACG GTG CTG CTC GA | GGT GGT CGT TGG AAT GAG TA |
| 1242* | GTG ATC ATC CCT GAC ATC AAT CT | TTC GCG CAA CGG GTC |
| 1352* | GCC CGC ACG CTT GC | TGA GAT CCG CAT CCA AAG C |
| 2031* | GCC ACC ACC CTT CCC GT | GTT GGT GGA CCG GAT CTG AAT |
| 2253* | TCC GGA CAC CGC AAG AA | AAC GAT CGG TTT GGC CGA |
| 2576* | GGC GTC GGT AAC GCA TCC | CCA GGG ACG AGT CGA GCA |
The M. tuberculosis Rv gene numbering was used (11). Restriction sites used are underlined. An asterisk following an Rv number indicates that forward primers for cloning via recombination into vector pDONR201 all had the recombination sequence 5′-G GGG ACA AGT TTG TAC AAA AAA GCA GGC TTA-3′ upstream from the given sequence and reverse primers all had the recombination sequence 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC CTA-3′ upstream from the given sequence.
Chromosomal DNA was isolated from M. africanum, M. fortuitum, M. gordonae, M. kansasii, and M. szulgai. The internal and flanking oligonucleotide sequences used for detecting the presence or absence of the gene coding for TB27.4 were identical to the oligonucleotides used by Brosch et al. (8). Sequences for detecting the genes coding for TB10.4, TB16.2, and TB15.8 were TB10.4F (5′-T CGC AAA TCA TGT ACA ACT ACC CCG-3′) and TB10.4R (5′-G GGT GCT GGA CAT CGC ATG ATA-3′); TB16.2F (5′-C CAC CTT CGA CAC CCG GTT GAT-3′) and TB16.2R (5′-A AGT CCT TCT GCT CGG TGC GCT-3′); and TB15.8F (5′-A GCG TTT TAG ATA TGA CGT CCG TGC-3′) and TB15.8 (5′-T CTC GCA GTG GAA CTC CGG ATT-3′), respectively.
Recombinant protein expression and purification.
Recombinant proteins CFP-10 and TB10.4 were produced as previously described (40). ESAT-6 was cloned in the Lactococcus lactis expression vector pAMJ752ESAT-6 as an ESAT-6-ESAT-6 dimer and purified directly from the culture supernatant of L. lactis strain PSM631 (Biotechnological Institute, Horsholm, Denmark) transformed with pAMJ752ESAT-6. A growth phase-dependent promoter (P170) upregulated the synthesis of ESAT-6 during the transition to stationary phase, and the signal sequence from the usp45 gene promoted secretion. Recombinant ESAT-6 was purified and concentrated by acid precipitation from the culture supernatant; no further purification was required.
Overnight cultures of recombinant E. coli strains were grown to mid-exponential phase and induced with 1 mM isopropylthiogalactopyranoside (IPTG) or 0.3 M NaCl (BL21 SI) for 4 h. For all proteins except TB9.9, TB37.6, and TB27.4, bacteria were harvested, resuspended in 1/50 of B-PER reagent (Pierce) to disrupt the outer membrane, washed twice in B-PER, and resuspended again in 1/50 B-PER reagent. Lysozyme (200 mg per liter) and DNase I (25 mg per liter) were added, and samples were incubated end over end for 1 h at room temperature. Inclusion bodies were harvested by centrifugation and washed twice in a solution containing 20 mM Tris-HCl (pH 8), 0.1 M NaCl, 1 mM EDTA (pH 8), and 0.1% deoxycholic acid. The final pellet was resuspended in wash buffer (4 M guanidine-HCl, 50 mM sodium phosphate [pH 7.2], 300 mM NaCl). Denatured proteins were applied to metal affinity columns (Talon columns; Clontech) and washed with 3 column volumes of wash buffer. Recombinant proteins were eluted by adding 3 column volumes of the wash buffer supplemented with 150 mM imidazole. Flowthrough, wash, and eluates were collected, and selected fractions were pooled after examination on Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. All proteins purified this way were dialyzed against an ammonium-CHAPS (pH 9.5) buffer, and protein concentration was measured (Pierce) with bovine serum albumin as a standard.
Since the purity of recombinants TB9.9, TB37.6, and TB27.4 was not high enough with the above procedure, they were purified by a three- or four-step procedure instead. Purification of TB37.6 has been described elsewhere (L. M. Okkels et al., submitted for publication). Briefly, inclusion bodies were isolated, and recombinant TB37.6 was purified first through a Talon column, then through a Hitrap SP column, and finally through a Hitrap Q column (Amersham Biosciences). Recombinants TB9.9 and TB27.4 were purified in a similar way except that after the metal affinity column, the proteins were applied to a Hitrap Q column in a solution containing 25 mM HEPES (pH 7.0), 10% glycerol, 0.01% Tween 20 (for TB9.9) or in 20 mM bis-Tris (pH 6.0), 1 mM EDTA, 0.1% Tween 20, 10% glycerol, and 3 M urea (for TB27.4). A 0 to 0.5 M linear gradient of NaCl was used for elution of bound proteins. Fractions were analyzed by silver-stained SDS-PAGE and by immunoblots with anti-E. coli antibodies (Dako) (L. M. Okkels, unpublished data). Only fractions containing the recombinant proteins at high purities were pooled and stored in a solution containing 25 mM HEPES (pH 7.5), 0.15 M NaCl, 10% glycerol, and 0.01% Tween 20 at −20°C.
Statistical analysis.
Categorical data were compared by a two-sided Fisher's exact test, and P values below 0.05 were considered statistically significant.
RESULTS
Selection and analysis of antigens.
From the 3,924 open reading frames of M. bovis, 28 genes were selected and tested experimentally (Table 2). First, 148 open reading frames were selected from groups which were either secreted proteins, or from regions of the M. bovis genome known to be deleted in M. bovis BCG. Secreted proteins are known to be important among mycobacterial antigens (2), and therefore a total of 96 proteins were included in this group because they have been identified as secreted either by proteomic studies (21, 36) or by computer analysis with the SignalP and SPScan programs (15). The second group of proteins consisted of proteins known to be deleted in M. bovis BCG strains (4, 16, 24). This group was included to allow vaccination of cattle with BCG while preserving specific diagnosis and contained 52 open reading frames.
TABLE 2.
Characteristics of selected antigens
| Antigen no. | Gene | Rv no.a | Mol wt | Protein | Homology (%) in M. aviumb | Functionc | Commentsd |
|---|---|---|---|---|---|---|---|
| 1 | 3872 | 9,911 | TB9.9 | 28 | Member of PE family | RD1 region | |
| 2 | 3873 | 37,318 | TB37.6 | 32 | Member of PPE family | RD1 region | |
| 3 | lhp | 3874 | 10,780 | CFP-10 | 24 | Unknown | RD1 region |
| 4 | esat-6 | 3875 | 9,889 | ESAT-6 | 29 | Unknown | RD1 region |
| 5 | 3878 | 27,383 | TB27.4 | 24 | Unknown | RD1 region | |
| 6 | 1982 | 14,709 | TB14.7 | 25 | Belongs to UPF0110 family | RD2 region | |
| 7 | 1988 | 19,559 | TB19.6 | 31 | Methyltransferase | RD2 region | |
| 8 | 1573 | 14,564 | TB14.6 | 30 | Probable phage phiRv1 protein | RD3 region | |
| 9 | 1575 | 12,365 | TB12.4 | 34 | Probable phage phiRV1 protein | RD3 region | |
| 10 | 1579 | 11,186 | TB11.2 | 34 | Probable phage phiRv1 protein | RD3 region | |
| 11 | 1580 | 9,734 | TB9.7 | 26 | Probably phage phiRv1 protein | RD3 region | |
| 12 | 1582 | 52,402 | TB52.4 | 14 | Probable phage phiRv1 protein | RD3 region | |
| 13 | 1584 | 7,980 | TB8 | 35 | Unknown | RD3 region | |
| 14 | 1585 | 19,198 | TB19.2 | 27 | Possible phage phiRv1 protein | RD3 region | |
| 15 | 1586 | 51,717 | TB51.7 | 27 | Probable phiRv1 integrase | RD3 region | |
| 16 | 2660 | 7,533 | TB7.6 | 35 | Unknown | RD13 region | |
| 17 | 64 | 107,379 | TB107.4 | 51 | Possible membrane protein | Secreted protein | |
| 18 | 398 | 21,642 | TB21.7 | 58 | Possible exported protein | Secreted protein | |
| 19 | mmpL4 | 450 | 105,223 | TB105.2 | 65 | Transmembrane protein | Secreted protein |
| 20 | 559 | 12,103 | TB12.1 | 54 | Possible N-terminal signal sequence | Secreted protein | |
| 21 | 569 | 9,508 | TB9.52 | 31 | Unknown | Secreted protein | |
| 22 | 617 | 13,921 | TB13.94 | 30 | Unknown | Secreted protein | |
| 23 | 1242 | 15,858 | TB15.87 | 24 | Unknown | Secreted protein | |
| 24 | 1352 | 12,834 | TB12.9 | 43 | Unknown | Secreted protein | |
| 25 | hspX | 2031 | 16,218 | TB16.2 | 28 | Heat shock protein | Secreted protein |
| 26 | 2253 | 17,822 | TB17.8 | 23 | Possible secreted protein | Secreted protein | |
| 27 | 2576 | 15,745 | TB15.8 | 54 | Unknown | Secreted protein | |
| 28 | 288 | 10,374 | TB10.4 | 79 | Unknown | ESAT-6 fam |
The M. tuberculosis gene numbering was used (11).
TBlastN was used with a blosum 62 scoring matrix. To allow homology searches in regions of low compositional complexity, searches were done without any filter.
From http://www.genolist.pasteur.fr/TubercuList. Proteins in the PE and PPE families share the Pro-Glu (PE) or Pro-Pra-Glu (PPE) motifs at positions 8 and 9 or 8 to 10, respectively.
RD numbering as in reference 4.
All 148 genes from these two groups were used to search the publicly available mycobacterial databases with TBlastX or TBlastN to identify proteins specific to M. bovis. Twenty-six genes without close homologues in the searched genomes of environmental mycobacteria (M. avium, M. paratuberculosis, M. marinum, M. smegmatis, M. leprae, and M. ulcerans) were selected for experimental screening, as listed in Table 2. TB10.4 and TB16.2 were included along with these selected proteins either because of data from studies of human tuberculosis showing a high frequency of recognition (40) or because of the possibility of detecting latently infected animals (37).
Expression and purification of recombinant mycobacterial proteins.
A fusion protein strategy was adopted to increase expression (20), facilitate purification, and improve immunoblot detection in protein extracts and purified fractions. In total, 27 proteins with histidine tags were expressed in E. coli. ESAT-6 was expressed in L. lactis without the tag. Since all formed inclusion bodies in E. coli, they were purified with a denaturing approach. Leader sequences were not cleaved off, and therefore all clones expressed full-length fusion proteins in the range of 7.5 kDa to 107 kDa (Table 2). All cultures grew normally after protein induction with no significant decrease in doubling time or final density. The yields in purified proteins were in the range from 1 to 10 mg of purified protein per liter of substrate, with final concentrations of between 0.05 and 0.5 mg/ml. Based on Coomassie blue-stained gels, all protein preparations were substantially free of contaminating proteins.
Screening of 28 M. bovis antigens.
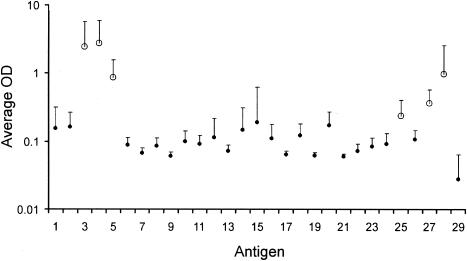
Along with PPD, M. bovis sonicate extract, and pokeweed mitogen as positive controls, the 28 recombinant mycobacterial proteins were screened on animals infected with 104 CFU of M. bovis at 126, 154, and 182 days postinfection. There was marked variation in the recognition of individual antigens (Fig. 1). Based on the amplitude of the IFN-γ responses, the number of responding animals, and the consistency of the response over time, a panel of six antigens was selected: CFP-10, ESAT-6, TB27.4, TB16.2, TB15.8, and TB10.4.
FIG. 1.
Initial screening of 28 recombinant mycobacterial proteins. Average ELISA IFN-γ responses and standard deviation for blood samples isolated from six cattle infected with 104 CFU of M. bovis at 126 and 154 days postinfection and three cattle infected with 104 CFU of M. bovis at 182 days postinfection. Antigen numbers on the x axis correspond to the antigen numbers in Table 2. Antigen 29 is the phosphate-buffered saline control. Open circles mark antigens selected for further study.
It is clear that CFP-10 and ESAT-6 had strong bovine T-cell epitopes, giving rise to a strong response (Fig. 1, antigens 3 and 4). These antigens gave positive responses in 13 and 14, respectively, out of 15 samples tested (data not shown). TB27.4 (antigen 5) is, like ESAT-6 and CFP-10, located in the RD1 region of the chromosome and was strongly and broadly recognized by 13 out of 15 samples (data not shown). TB10.4 (antigen 28) was recognized by 10 samples and was included for further testing. TB15.8 (antigen 27) was recognized by 12 samples and was also included. TB16.2 (antigen 25) was also strongly recognized at 126 and 154 days postinfection, giving positive results in 9 out of 12 samples, and was included for further testing.
Antigen specificity and interspecies distribution.
Twenty-two cattle which were free from tuberculosis but considered sensitized to environmental mycobacteria based on their single intradermal comparative tuberculin test results 2 months earlier (greater delayed-type hypersensitivity responses to M. avium PPD than M. bovis PPD were tested in the IFN-γ assay with both PPDs. The six strongest responders to PPDA were chosen to screen the panel of experimentally selected antigens together with M. bovis sonicate extract, PPDA, and PPDB as controls. All animals had higher IFN-γ responses to PPDA than to PPDB, but none had an IFN-γ response to any of the selected recombinant antigens (Table 3). The TB10.4 antigen, which showed 79% homology to its M. avium ortholog, did not result in an IFN-γ response in these animals, nor did TB16.2, which had orthologues in most of the environmental strains tested (Table 4).
TABLE 3.
Antigen recognition by PPDA-reacting field cattlea
| Antigen | OD in animal no.:
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| PBS | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| PPDA | 0.3 | 1.1 | 2.0 | 0.8 | 1.8 | 1.5 |
| PPDB | 0.1 | 0.2 | 0.3 | 0.1 | 0.4 | 0.2 |
| MBSE | 0.1 | 0.2 | 0.3 | 0.1 | 0.6 | 0.3 |
| PWM | 3.1 | 2.4 | 2.4 | 3.6 | 3.4 | 3.9 |
| TB15.8 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| TB16.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| TB27.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| ESAT-6 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| CFP-10 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| TB10.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
Six field animals showing stronger skin test responses to avian than to bovine PPD were tested in the IFN-γ assay with control and selected antigens. Optical density (OD) values are in bold when above the cutoff value, calculated as the average phosphate-buffered saline (PBS) response + 5 standard deviations. PPDA, M. avium PPD; PPDB, M. bovis PPD; MBSE, M. bovis sonicate extract; PWM, pokeweed mitogen.
TABLE 4.
Interspecies distribution of diagnostic mycobacterial antigens
| Strain | Identitya
|
|||||
|---|---|---|---|---|---|---|
| ESAT-6 | CFP-10 | TB10.4 | TB16.2 | TB27.4 | TB15.8 | |
| M. tuberculosis 210b | + | + | + | + | + | + |
| M. tuberculosis CDC1551 (Oshkosh)b | + | + | + | + | + | + |
| M. tuberculosis clinical isolatesc | + | + | + | ND | + | ND |
| M. tuberculosis H37Rvb | + | + | + | + | + | + |
| M. bovis, 8 strainsbd | + | + | + | + | + | + |
| M. bovis BCG Pasteurd | − | − | + | + | − | + |
| M. bovis BCG Moreaud | − | − | + | + | − | + |
| M. bovis BCG Denmarkd | − | − | + | + | − | + |
| M. africanum, 14 isolatesc | + | + | + | +e | + | +e |
| M. avium sp. aviumb | − | − | (+) | − | − | (−) |
| M. fortuitum | −f | −f | −f | +g | −e | −e |
| M. gordonae | −f | −f | −f | +g | −e | +e |
| M. kansasii | + | + | +e | +e | +e | +e |
| M. marinumb | + | + | + | + | (+) | (+) |
| M. szulgai | +f | +f | +f | +g | +e | −e |
| M. microtih | − | − | ND | ND | + | ND |
| M. ulceransb | − | − | + | + | − | (+) |
| M. smegmatisb | (+) | (+) | (+) | (−) | − | − |
| M. lepraeb | − | (−) | (+) | − | (+) | − |
| M. terrae | −f | −f | −f | ND | ND | ND |
+, 80 to 100% amino acid identity; (+), 60 to 79% identity; (−), 40 to 59% identity; −, below 40% identity. ND, not determined.
BLAST searches against all available mycobacterial databases.
PCR analysis (data from reference 8).
DNA arrays and bacterial artificial chromosome array libraries (data from references 4 and 16).
PCR data from this work.
Southern blots (data obtained from references 2 and 40).
BLAST searches against the Genbank bacteria database.
Bacterial artificial chromosome array libraries (7).
To complement genome searches and determine the distribution of the selected genes within relevant mycobacterial species, gene-specific and region deletion-specific PCRs were performed on genomic DNA from various mycobacterial strains (Table 4). The strains were selected based on how frequently they were isolated from nontuberculous skin test-reactive cattle (30) as well as with the aim to cover broadly the phylogenetic tree of mycobacteria. ESAT-6, CFP-10, and TB10.4 have been investigated previously by Southern blotting (2, 40). The gene coding for TB16.2 is present in most of the included strains, representing a similar theoretical concern as for the TB10.4 homologue in M. avium. As expected, the RD1 gene encoding TB27.4 was not found in any of the three BCG strains, whereas the TB16.2 gene was detected in all three. TB27.4 was also absent from the M. avium and M. terrae genomes, two mycobacterial species that are important from a diagnostic viewpoint.
Investigation of antigen reactivity with disease progression.
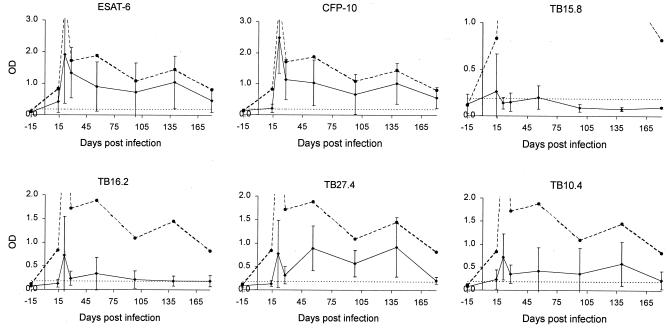
The selected panel of six mycobacterial proteins was retested for 6 to 7 months postinfection on two groups of six animals infected with 104 or 106 CFU of M. bovis, respectively. The result for each antigen is shown in Fig. 2 for the group receiving the higher infection dose. For the group receiving the lower infection dose, the results were similar (data not shown). In neither group was there a significant response to any of the proteins or PPDs before 14 days postinfection. By 21 days postinfection, however, there was a strong response to several antigens as well as to PPD. Considering the recognition pattern over time (Fig. 2), the response to PPDB remained very strong after it developed and detected all six animals at all time points measured. The overall shape of the PPDB curves for the individual animals was similar, with a strong peak early in the infection followed by a gradual decrease over time.
FIG. 2.
Time kinetic of antigen recognition. A group of six animals were infected with 106 CFU of M. bovis, and antigen recognition in blood samples from these animals was followed continuously. IFN-γ levels were measured after stimulation with the appropriate antigen. Results at −15, 14, 21, 28, 56, 98, 140, and 180 days postinfection are shown. The PPDB results are indicated with a stippled line in each figure. Two of the optical density (OD) values for PPDB at 21 days postinfection were above the maximum detection level. The cutoff value (0.188) is indicated by a horizontal dotted line and was calculated as the average of the phosphate-buffered saline group plus 5 standard deviations.
Importantly, however, for individual recombinant antigens, there was not the same pattern of recognition when comparing single animals. Most of the curves had an early peak at 21 days postinfection, but there was a large fluctuation in the response over time, as illustrated by the large standard deviations. CFP-10 and ESAT-6 were both recognized at most of the time points by most of the animals, each detecting 87% of the samples as being infected. The recognition patterns of ESAT-6 and CFP-10 were strikingly similar (Fig. 2), but the response to CFP-10 was generally lower than that to ESAT-6, as illustrated by a 0.8 slope for the best-fit linear regression in Fig. 3.
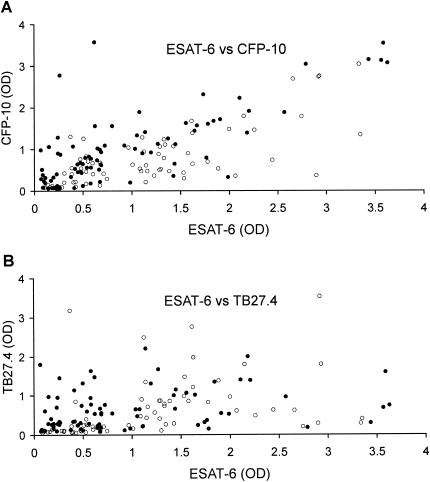
FIG. 3.
Complementation of individual antigens. After the recognition response has developed (14 days postinfection for the group receiving 106 CFU of M. bovis and 21 days postinfection for the group receiving 104 CFU), 144 samples were collected. For each of these, the recognition signal for ESAT-6 is plotted against the recognition signal for CFP-10 (a) or TB27.4 (b). Solid circles are data points from the group infected with 106 CFU of M. bovis (78 samples), and open circles are from the group infected with 104 CFU of M. bovis (66 samples).
TB27.4 gave a level of detection similar to that of ESAT-6 and CFP-10 and detected 84% of the samples in all six animals. TB10.4 and TB16.2 also gave significant levels of recognition, and responses higher than the cut off developed in 54% and 49% of the samples, respectively. In this experiment, however, TB15.8 only gave responses in 17% of the samples.
Animals infected with 104 CFU (data not shown) showed the same general pattern of responses, which varied over time, but CFP-10, ESAT-6, and TB27.4 were recognized by 92, 94, and 78% of the samples, respectively. This should be compared to PPDB, which was recognized by 97% of the samples. Interestingly, one PPDB-negative sample responded strongly to ESAT-6, CFP-10, and TB27.4. Similar to the higher infection dose group, 51% and 42% of the samples recognized TB10.4 and TB16.2, respectively, whereas TB15.8 was recognized by only 28% of the samples.
Due to the large fluctuations in recognition over time, Table 5 is included to give a quick overview of how many animals could be identified at three different time points in the infectious process (early, middle, and late). Both ESAT-6 and CFP-10 detected an M. bovis infection very early in the infection, even with the low infection dose. At the middle stage of infection, TB27.4 most effectively supplemented ESAT-6 and CFP-10, and together the antigens detected all six animals in both groups as being infected. At the late stage of infection, only PPDB still detected all six animals in both groups. At that stage, ESAT-6, CFP-10, and TB27.4 detected all six animals in the group infected with the high dose, but in the low-dose group only four of the six animals were identified.
TABLE 5.
Antigen recognition at selected time pointsa
| Protein | No. of animals recognized (n = 6)
|
|||||
|---|---|---|---|---|---|---|
| Early
|
Middle
|
Late
|
||||
| Low | High | Low | High | Low | High | |
| TB10.4 | 4 | 4 | 0 | 2 | 2 | 2 |
| TB15.8 | 0 | 1 | 2 | 0 | 2 | 0 |
| TB16.2 | 5 | 2 | 2 | 2 | 2 | 2 |
| TB27.4 | 3 | 4 | 5 | 6 | 2 | 3 |
| CFP10 | 5 | 6 | 5 | 5 | 3 | 5 |
| ESAT6 | 5 | 5 | 5 | 5 | 4 | 5 |
| PPDB | 5 | 6 | 6 | 6 | 6 | 6 |
Number of animals recognized by single antigens at three different time points, and two infective doses. Six animals were tested in each group. For the group infected with 104 CFU of M. bovis (low), the number of animals recognized is shown at 20, 91, and 203 days postinfection, corresponding to early, middle, and late, respectively. For the group infected with 106 CFU of M. bovis (high), results are shown for 21, 98, and 180 days postinfection.
Effects of combining individual antigen responses.
An important aspect of investigating new diagnostic antigens is to identify reagents that can complement systems that are already available. ESAT-6 has been recognized as a potential diagnostic antigen in cattle, and lately CFP-10 has also been identified. In the present study, there was a strong correlation between recognition of ESAT-6 and CFP-10, indicated by the fact that most of the data points fell on a diagonal line when the reactivity to ESAT-6 was plotted against the same samples' reactivity to CFP-10 (Fig. 3A). This phenomenon indicates that the level of reactivity to one of these antigens will be mirrored in the other and was especially pronounced for animals receiving the higher-dose inoculum. This could represent a drawback for test sensitivity in field situations for animals with different genetic backgrounds and levels of exposure to infection.
A future diagnostic test should aim to counteract variation among individuals, and one strategy may be to consider diversity within patterns of antigen recognition. A plot of recognition of ESAT-6 versus TB27.4 did not show the same clear association as seen in the comparison with CFP-10 (Fig. 3B). On the contrary, the data were much more scattered and indicated that TB27.4 represents an excellent candidate to supplement ESAT-6 and/or CFP-10 in an antigen cocktail to enhance test sensitivity.
DISCUSSION
The ideal diagnostic tool for bovine tuberculosis would be one that accurately and uniquely detects infection of individuals with M. bovis. The current diagnostic agent, PPDB, although sensitive and inexpensive, has specificity problems, yielding positive results in animals sensitized by environmental, nonpathogenic mycobacteria. Furthermore, the use of PPDB as a skin test reagent is not simple and requires two field visits, first to administer PPDB intradermally and 3 days later to read the reaction. It would therefore also be desirable if future tests were simpler to perform. The present study investigates a noninvasive test format based on the use of whole-blood samples.
It has been recognized that improvements in diagnosis are likely to be based on the use of a combination of specific mycobacterial antigens. Since mycobacterial infections generally induce a strong Th1 response, most work has focused on defining T-cell responses to candidate proteins. ESAT-6 and CFP-10 have previously been identified as antigens for a T-cell-based diagnostic assay for bovine tuberculosis (30, 42). This potential was confirmed in the current study, where levels of recognition of these antigens were remarkably high (Table 5 and Fig. 2). However, to have broad field applicability, the combined sensitivity has to be increased close to the PPDB level while increasing overall specificity. One strategy is to complement ESAT-6 and CFP-10 with other highly specific antigens from M. bovis. In this study, we attempted to identify such proteins.
Genomic data were used to select proteins from two relatively large groups of prospective M. bovis diagnostic antigens: 16 genes from regions deleted in BCG vaccine strains (4) and 11 coding for secreted proteins were selected by homology screening in environmental mycobacteria and tested for T-cell recognition in cattle. TB10.4 does not formally belong to either of these groups but was included in the antigen panel despite the existence of a close orthologue in M. avium because of previous data obtained with human samples (40). Within this antigen panel are genes that are upregulated when mycobacteria are incubated at low oxygen pressure, e.g., TB16.2 (37). These in vitro conditions are considered to mimic the situation within the macrophage during latent infection, thus enhancing the chances for finding proteins that can identify chronically infected animals.
Among the 28 selected genes, 11 encode proteins with no known function or location and 6 encode what seem to be bacteriophage proteins. Since genes without homologues in atypical mycobacteria were selected, it is not surprising to find these categories among the selected genes. All 28 gene products were tested in an in vitro IFN-γ release assay as full-length E. coli recombinant proteins on cattle infected intranasally with 104 CFU of M. bovis, and a panel of six antigens was selected.
The specificity of none of the six antigens had so far been supported by ex vivo experimental data. The three RD1-encoded proteins are prospectively absent from most of the environmental mycobacterial proteomes, whereas TB10.4 and especially TB16.2 have gene homologues in several of the environmental species genomes (Table 4). Because of the common occurrence of these mycobacterial species in soil, the specificity of these two antigens is of concern and has to be addressed before moving into large field trials. The data in the present study, obtained from a very limited number of animals however, gave no indication of specificity problems. A preliminary investigation of antigen specificity was performed with six field cattle with nonspecific responsiveness to PPDA (Table 3). Interestingly, like the other selected antigens, TB10.4 and TB16.2 induced no response in any of these animals. This may be due either to the absence of epitopes common to the M. bovis TB10.4 and its M. avium ortholog (the longest conserved segment is the 12-residue amino terminus), to the lack of expression of the TB10.4 orthologue in M. avium, or to the fact that the field cattle might have been sensitized to a non-M. avium environmental mycobacterium. TB16.2 is upregulated during latency, and a possible explanation for the lack of response in the animals could be that M. avium never establishes a latent infection.
To confirm their immunodominance and to follow the development of recognition patterns, the selected six proteins were followed for 6 to 7 months in two groups of cattle infected with a low and a high dose of M. bovis. The in vitro IFN-γ responses to the tested recombinant proteins varied broadly throughout infection and among animals (Fig. 2). There may be several reasons for this. First, heterogeneity within individual recognition may be ascribed to polymorphisms within major histocompatibility complex class II, TAP, or cytokine receptors (17, 23). Second, because only ESAT-6 and CFP-10 are organized in an operon (5), different levels of in vivo bacterial expression may account for variations in responses to the overall antigen panel. Third, posttranslational glycosylation and acylation in the M. tuberculosis complex have accounted for decreased T-cell reactivity to certain of its E. coli recombinant proteins (35), and the posttranslational modification status of the native antigens within this study is not known.
The RD1 region consists of eight genes, including those for ESAT-6 and CFP-10 (24). These two genes are cotranscribed (5), copurify when expressed in E. coli (13), and colocalize, since both proteins can be found in the culture filtrate from in vitro cultures of M. tuberculosis (41). Lately it has been shown that they form a 1:1 molecular complex (34). This interaction is potentially of direct relevance to the role that the proteins play in tuberculosis, but the function is still unknown. It has been speculated that CFP-10 and ESAT-6 together with TB9.9 and TB37.6 could functionally cluster to form a type IV secretion system (29). The colocalization and complex formation may explain why ESAT-6 and CFP-10 were recognized by the same blood samples (Fig. 3). After tuberculosis infection, these two proteins are likely to be taken up together by antigen-presenting cells and processed together, so that peptides from both proteins are presented to T cells on the same antigen-presenting cells.
TB37.6, which is a member of the PPE family (proteins sharing a Pro-Pra-Glu N-terminal motif) found just upstream of CFP-10, was included in the initial screening. This allowed investigation of the possibility that the entire RD1 region could be an immunodominant hotspot coding for proteins that would have important roles during tuberculosis pathogenesis. Based on the present results, such a hotspot may exist but would likely not include the entire RD1 region, and CFP-10 could be its upstream boundary. The gene coding for TB27.4 is found downstream from ESAT-6 and was found to be almost as immunodominant as ESAT-6 and CFP-10. The high IFN-γ reactivity to TB27.4 in cattle contrasts with the lower delayed-type hypersensitivity response to TB27.4 in the guinea pig relative to the other RD1-encoded antigens, but such discrepancies between delayed-type hypersensitivity and T-cell assays have been ascribed earlier to distinct subsets of reacting T cells (9). TB27.4 is reportedly a stronger B- than T-cell antigen relative to ESAT-6 and CFP-10 in both human and guinea pig models (9), and this awaits confirmation in cattle. Since TB27.4 is not recognized with the same pattern as ESAT-6 and CFP-10, it is a promising candidate to complement ESAT-6 and CFP-10, although the influence of a homologue in M. terrae requires further investigation.
Our results with antigens from the RD1 region are in general in good agreement with the results found recently by Cockle et al. (10). They also found a high responder frequency to ESAT-6, CFP-10, and TB27.4, but in contrast to this study, they also reported responses to TB9.9 (Rv3872) and TB37.6 (Rv3873). One crucial difference between the two studies is that Cockle et al. used overlapping peptides rather than full-length proteins. This may lead to differences in uptake, processing, and/or presentation of the epitopes. Also, the sampling time postinfection in our screening experiment was in general 4 weeks later than in theirs. The discrepancies illustrate the importance of having a broadly recognizing mix of antigens in order to perform diagnostics in the field.
Twenty-one genes sharing the size and organization of esat-6 have been grouped into a single family, and six of them have been reported to encode strong T-cell antigens in the human, mouse, and guinea pig (39). Three members of the ESAT-6 family known to be immunoreactive in humans (40) were tested in this study: ESAT-6, CFP-10, and TB10.4. TB10.4 has never been evaluated in cattle before and was recognized by 52% of all blood samples collected from 20 to 210 days postinfection, significantly less than ESAT-6 and CFP-10 (90%). TB10.4 has reportedly been detected at similar levels as ESAT-6 (88%) in human tuberculosis patients, but at lower levels in BCG-vaccinated individuals (71%) (40) and latent tuberculosis-infected skin test reactors (58%) (3). The present results from animals that remained asymptomatic are in line with a lower detection level among latent tuberculosis patients. Future studies will show if the presence of a gene homologue in the M. avium genome has implications for test specificity. If not, TB10.4 could be a useful supplement in a diagnostic cocktail.
TB16.2, or HspX (α-crystallin homologue), belongs to the small heat shock protein family. It has been identified as a highly expressed protein under hypoxic conditions mimicking dormancy (38) and upon macrophage infection (26) and is considered as such as a marker for latency (6). Its species specificity and early T-cell immunodominance in the mouse and human are well documented (43, 45), and our novel cattle data are in agreement with these previous reports. Since HspX is expressed primarily during the late stage of infection, it has interesting potential as a diagnostic reagent. However, it was only recognized by 45% of the blood samples, and in the group infected with 104 CFU of M. bovis, the recognition was in general weak. HspX did not provide any additional detection in complement to ESAT-6 and CFP-10 taken together throughout the time points tested.
TB15.8 has no known function but was recognized by 28% of the samples from the group of animals receiving the low infection dose and by 17% of the samples from animals receiving the high dose. None of these samples had any complementing value, which suggested that TB15.8 might not have significant benefit in a future antigenic cocktail.
Based on these results, it seems that a mix of ESAT-6, CFP-10, TB27.4, and TB10.4 could form the basis of a novel diagnostic cocktail for a new generation of tests for bovine tuberculosis. The results of the present study indicate that a combination of these antigens can detect almost the same number of samples as PPDB. Along with the indicated studies to define the practical formulation of a novel antigenic cocktail, larger-scale investigations are needed to define the sensitivity and specificity of such a reagent in the field diagnostic situation. The results of the present study provide an optimistic outlook for the development of improved methods for the detection of bovine tuberculosis and other mycobacterial diseases.
Acknowledgments
C. Aagaard and M. Govaerts contributed equally to this work.
We thank Vivi Andersen, Martyn Girvin, and Kathryn Wattam for excellent technical assistance and Hugh Bassett for provision of samples from environmentally sensitized cattle. Preliminary sequence data were obtained from the Sequencing Group at the Sanger Institute at http://www.sanger.ac.uk/Projects; the Institute for Genomic Research web site at http://www.tigr.org; and from the University of Minnesota M. paratuberculosis genome sequencing web site at http://www.cbc.umn.edu/ResearchProjects/AGAC/Mptb/Mptbhome.html.
This study was partly funded by EU INCO contract ICA4-CT-2000-30023.
REFERENCES
- 1.Amadori, M., S. Tagliabue, S. Lauzi, G. Finazzi, G. Lombardi, P. Telò, L. Pacciarini, and L. Bonizzi. 2002. Diagnosis of Mycobacterium bovis infection in calves sensitized by mycobacteria of the avium/intracellulare group. J. Vet. Med. B Infect. Dis. Vet. Public Health 49:89-96. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, P., M. E. Munk, J. M. Pollock, and T. M. Doherty. 2000. Specific immune-based diagnosis of tuberculosis. Lancet 356:1099-1104. [DOI] [PubMed] [Google Scholar]
- 3.Arend, S. M., A. C. Engelhard, G. Groot, K. de Boer, P. Andersen, T. H. Ottenhoff, and J. T. van Dissel. 2001. Tuberculin skin testing compared with T-cell responses to Mycobacterium tuberculosis-specific and nonspecific antigens for detection of latent infection in persons with recent tuberculosis contact. Clin. Diagn. Lab. Immunol. 8:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Berthet, F.-X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144:3195-3203. [DOI] [PubMed] [Google Scholar]
- 6.Boon, C., R. Li, R. Qi, and T. Dick. 2001. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J. Bacteriol. 183:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Sanger, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusasca, P. N., R. Colangeli, K. P. Lyashchenko, X. Zhao, M. Vogelstein, J. S. Spencer, D. N. McMurray, and M. L. Gennaro. 2001. Immunological characterization of antigens encoded by the RD1 region of the Mycobacterium tuberculosis genome. Scand. J. Immunol. 54:448-452. [DOI] [PubMed] [Google Scholar]
- 10.Cockle, P. J., S. V. Gordon, A. Lalvani, B. M. Buddle, R. G. Hewinson, and H. M Vordermeier. 2002. Identification of novel Mycobacterium tuberculosis antigens with potential as diagnostic reagents or subunit vaccine candidates by comparative genomics. Infect. Immun. 70:6996-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. Mclean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Daborn, C. J., and J. M. Grange. 1993. HIV/AIDS and its implications for the control of animal tuberculosis. Br. Vet. J. 149:405-417. [DOI] [PubMed] [Google Scholar]
- 13.Dillon, D. C., M. R. Alderson, C. H. Day, T. Bement, A. Campos-Neto, Y. A. Skeiky, T. Vedvick, R. Badaro, S. G. Reed, and R. Houghton. 2000. Molecular and immunological characterization of Mycobacterium tuberculosis CFP-10, an immunodiagnostic antigen missing in Mycobacterium bovis BCG. J. Clin. Microbiol. 38:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolin, P. J., M. C. Raviglione, and A. Kochi. 1994. Global tuberculosis incidence and mortality during 1990-2000. Bull. W.H.O. 72:213-220. [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez, M., S. Johnson, and M. L. Gennaro. 2000. Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect. Immun. 68:2323-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli with bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 17.Grosse, W. M., S. M. Kappes, W. W. Laegreid, J. W. Keele, C. G. Chitko-McKown, and M. P. Heaton. 1999. Single nucleotide polymorphism (SNP) discovery and linkage mapping of bovine cytokine genes. Mamm. Genome 10:1062-1069. [DOI] [PubMed] [Google Scholar]
- 18.Harboe, M., A. S. Malin, H. S. Dockrell, H. G. Wiker, G. Ulvund, A. Holm, M. C. Jorgensen, and P. Andersen. 1998. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect. Immun. 66:717-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harboe, M., T. Oettinger, H. G. Wiker, I. Rosenkrands, and P. Andersen. 1996. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harth, G., B. Y. Lee, and M. A. Horwitz. 1997. High-level heterologous expression and secretion in rapidly growing nonpathogenic mycobacteria of four major Mycobacterium tuberculosis extracellular proteins considered to be leading vaccine candidates and drug targets. Infect. Immun. 65:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 22.Krebs, J., R. Anderson, T. Clutton-Brock, I. Morrison, D. Young, and C. Donnely. 1997. Bovine tuberculosis in cattle and badgers: report to the Rt. Hon. Dr. Jack Cunningham MP. MAFF Publications, London, UK.
- 23.Lewin, H. A., G. C. Russell, and E. J. Glass. 1999. Comparative organization and function of the major histocompatibility complex of domesticated cattle. Immunol. Rev. 167:145-158. [DOI] [PubMed] [Google Scholar]
- 24.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monaghan, M. L., M. L. Doherty, J. D. Collins, J. F. Kazda, and P. J. Quinn. 1994. The tuberculin test. Vet. Microbiol. 40:111-124. [DOI] [PubMed] [Google Scholar]
- 26.Monahan, I. M., J. Betts, D. K. Banerjee, and P. D. Butcher. 2001. Differential expression of mycobacterial proteins following phagocytosis by macrophages. Microbiology 147:459-471. [DOI] [PubMed] [Google Scholar]
- 27.Neill, S. D., J. Hanna, J. J. O'Brien, and R. M. McCracken. 1988. Excretion of Mycobacterium bovis by experimentally infected cattle. Vet. Rec. 123:340-343. [DOI] [PubMed] [Google Scholar]
- 28.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuberc. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 29.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily-and a new Gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 30.Pollock, J. M., and P. Andersen. 1997. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J. Infect. Dis. 175:1251-1254. [DOI] [PubMed] [Google Scholar]
- 31.Pollock, J. M., and P. Andersen. 1997. Predominant recognition of the ESAT-6 protein in the first phase of interferon with Mycobacterium bovis in cattle. Infect. Immun. 65:2587-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock, J. M., A. J. Douglas, D. P. Mackie, and S. D. Neill. 1994. Identification of bovine T-cell epitopes for three Mycobacterium bovis antigens: MPB70, 19, 000 MW and MPB57. Immunology 82:9-15. [PMC free article] [PubMed] [Google Scholar]
- 33.Pollock, J. M., A. J. Douglas, D. P. Mackie, and S. D. Neill. 1995. Peptide mapping of bovine T-cell epitopes for the 38 kDa tuberculosis antigen. Scand. J. Immunol. 41:85-93. [DOI] [PubMed] [Google Scholar]
- 34.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, R. G. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 277:21598-21603. [DOI] [PubMed] [Google Scholar]
- 35.Romain, F., C. Horn, P. Pescher, A. Namane, M. Riviere, G. Puzo, O. Barzu, and G. Marchal. 1999. Deglycosylation of the 45/47-kilodalton antigen complex of Mycobacterium tuberculosis decreases its capacity to elicit in vivo or in vitro cellular immune responses. Infect. Immun. 67:5567-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenkrands, I., A. King, K. Weldingh, M. Moniatte, E. Moertz, and P. Andersen. 2000. Towards the proteome of Mycobacterium tuberculosis. Electrophoresis 21:3740-3756. [DOI] [PubMed] [Google Scholar]
- 37.Rosenkrands, I., R. A. Slayden, J. Crawford, C. Aagaard, C. E. Barry, 3rd, and P. Andersen. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol. 184:3485-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skjøt, L. R. M., E. M. Agger, and P. Andersen. 2001. Antigen discovery and tuberculosis vaccine development in the post-genomic era. Scand. J. Infect. Dis. 33:643-647. [DOI] [PubMed] [Google Scholar]
- 40.Skjøt, R. L., T. Oettinger, I. Rosenkrands, P. Ravn, I. Brock, S. Jacobsen, and P. Andersen. 2000. Comparative evaluation of low-molecular-mass proteins from Mycobacterium tuberculosis identifies members of the ESAT-6 family as immunodominant T-cell antigens. Infect. Immun. 68:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sørensen, A. L., S. Nagai, G. Houen, P. Andersen, and Å. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Pinxteren, L. A., P. Ravn, E. M. Agger, J. Pollock, and P. Andersen. 2000. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and CFP10. Clin. Diagn. Lab. Immunol. 7:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vordermeier, H. M., D. P. Harris, R. Lathigra, E. Roman, C. Moreno, and J. Ivanyi. 1993. Recognition of peptide epitopes of the 16, 000 MW antigen of Mycobacterium tuberculosis by murine T cells. Immunology 80:6-12. [PMC free article] [PubMed] [Google Scholar]
- 44.Weynants, V., J. Godfroid, B. Limbourg, C. Saegerman, and J. J. Letesson. 1995. Specific bovine brucellosis diagnosis based on in vitro antigen-specific gamma interferon production. J. Clin. Microbiol. 33:706-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson, R. J., K. A. Wilkinson, K. A. De Smet, K. Haslov, G. Pasvol, M. Singh, I. Svarcova, and J. Ivanyi. 1998. Human T- and B-cell reactivity to the 16kDa alpha-crystallin protein of Mycobacterium tuberculosis. Scand. J. Immunol. 48:403-409. [DOI] [PubMed] [Google Scholar]
- 46.Wood, P. R., L. A. Corner, and P. Plackett. 1990. Development of a simple, rapid in vitro cellular assay for bovine tuberculosis based on the production of gamma interferon. Res. Vet. Sci. 49:46-49. [PubMed] [Google Scholar]