Abstract
Cell death plays important roles in the development and defense of plants as in other multicellular organisms. Rapid production of reactive oxygen species often is associated with plant defense against pathogens, but their molecular mechanisms are not known. We introduced the constitutively active and the dominant negative forms of the small GTP-binding protein OsRac1, a rice homolog of human Rac, into the wild type and a lesion mimic mutant of rice and analyzed H2O2 production and cell death in transformed cell cultures and plants. The results indicate that Rac is a regulator of reactive oxygen species production as well as cell death in rice.
Cell death is important in the development and defense of multicellular organisms (1). Cell death occurs in normal plant development and during infection by avirulent pathogens (2–5). It plays a key role in plant defense and shares several features with apoptosis in mammalian cells (3, 5, 6). Rapid production of reactive oxygen species (ROS) often is associated with cell death during resistance reactions to pathogens, and a plasma membrane NADPH oxidase similar to the neutrophil enzyme is suggested to be responsible for ROS production (5–7). However, the molecular mechanisms for ROS production and cell death in plants are largely unknown.
In phagocytic cells, activation of NADPH oxidase leads to production of superoxide, which effectively kills invading microorganisms (8, 9). The neutrophil NADPH oxidase is a multicomplex enzyme consisting of two membrane proteins, gp91phox and p22phox, and three cytosolic factors, p47phox, p67phox, and Rac (8, 9), and the plant enzyme is thought to be similar to the neutrophil enzyme (10–13). Genes whose deduced amino acid sequences are similar to those of the Rho/Rac family of the small GTP-binding proteins have been isolated in plants (14–18), and some were shown to play a role in the control of pollen tube growth (19). However, functions of most of these genes are not known. Genes whose amino acid sequences are similar to that of the animal gp91phox also have been isolated from plants (20, 21), but their functions have not been investigated. In this study we analyzed functions of Rac in rice by expressing constitutively active and dominant negative Rac genes in transgenic cell cultures and plants and found that Rac plays an important role in ROS production as well as cell death in rice.
MATERIALS AND METHODS
Biochemical Analysis of Recombinant OsRac1.
OsRac1 cDNA was cloned in an expression vector, pGEX-4T-1 (Pharmacia), and transformed into Escherichia coli. The glutathione S-transferase (GST)-fusion protein was purified by glutathione-Sepharose beads (Pharmacia) and used for assays of GTP-binding and GTPase activities according to published protocol (22). For the assay of the GTP-binding activity, the binding of [35S]GTPγS to GST-OsRac1 was measured. For the assay of the GTPase activity, GST-OsRac1 was loaded with [γ-32P]GTP and hydrolysis of GTP was measured at different time points.
Rice Transformation.
OsRac1-G19V was made by substitution of the glycine corresponding to G12 of human Rac1 by valine by the use of an LA PCR in vitro Mutagenesis kit (Takara Shuzo, Kyoto). OsRac1-T24N similarly was made by substitution of threonine at the 24th position to asparagine. 35S-OsRac1-G19V and 35S-OsRac1-T24N were introduced in a Ti-based vector, pMSH1, and Agrobacterium-mediated transformation of rice callus was performed according to a published protocol (23). Transformed calli were selected by hygromycin resistance, and plants were regenerated from transformed callus cultures. Suspension cell lines were made from transformed calli and maintained in R2S medium (24) for various analyses. The sl mutant used in this study carried the CM265 allele in the Kinmaze background.
Terminal Deoxynucleotidyltransferase-Mediated UTP End Labeling (TUNEL) and Diaminobenzidine (DAB) Staining.
TUNEL staining was performed by the use of a fluorescein-dUTP-based in situ death detection kit (Boehringer Mannheim) as described previously (25). For DAB staining, the concentration of 1 mg/ml (Wako) was used.
Electron Microscopy.
For electron microscopy, cultured cells were fixed in 2.5% glutaraldehyde in cacodylate buffer, pH 7.2, and then treated with OsO4, dehydrated, and embedded in Spurr’s resin. Sections were stained with uranylacetate and lead citrate and examined by Hitachi H7100 electron microscopy.
RESULTS AND DISCUSSION
OsRac1 Encodes a GTPase.
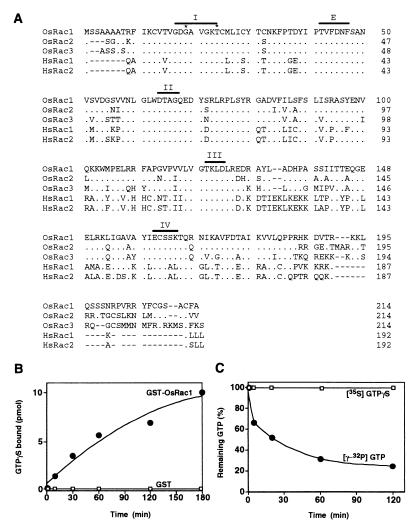
Because Rac plays an important role in the regulation of the NADPH oxidase in phagocytic cells, we sought rice expressed sequence tags that had homology with human Rac and found three such sequences (Fig. 1A; ref. 26). They had deduced amino acid sequences that were ca. 60% identical with those of human Rac proteins. Comparison of their amino acid sequences with those of other Ras-related GTPase proteins indicated high similarity to Rac; thus, we called them OsRac. Particularly, regions I–IV, which are highly conserved among the small GTP-binding protein family (27), and the effector binding site are extremely conserved between humans and rice (Fig. 1A). OsRac1 and OsRac3 are expressed in leaves and roots, but OsRac2 is expressed only in roots (data not shown). The recombinant OsRac1 protein in E. coli was found to have both GTP-binding and GTPase activities, confirming that OsRac1 encodes a GTPase (Fig. 1 B and C), and its binding was specific to GTP and GDP (data not shown). These results indicate that OsRac1 codes for a GTPase similar to those in mammals.
Figure 1.
Deduced amino acid sequences and biochemical analysis of OsRac. (A) Amino acid alignment of predicted OsRac proteins with human Rac proteins (26). Identical residues are indicated by a dot, and gaps are shown by a dash. The conserved regions are indicated by bars above the sequence; regions I and II are the GTPase region, regions III and IV are the GTP/GDP-binding region, and E denotes the effector region. The glycine and threonine residues marked by asterisks in the region I were changed to make a constitutively active and a dominant negative form of OsRac genes, respectively. (B) GTP-binding activity of the recombinant OsRac1 protein. For the assay, the binding of [35S]GTPγS to GST-OsRac1 was measured at different time points. (C) GTPase activity of the recombinant OsRac1 protein. For the assay, GST-OsRac1 was loaded with [γ-32P]GTP and hydrolysis of GTP was measured at different time points.
Constitutively Active OsRac Induces ROS Production in Cultured Rice Cells.
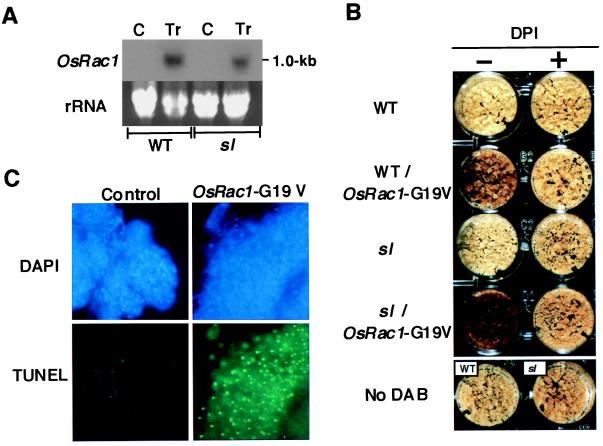
To examine whether OsRac regulates ROS production in rice, we made a constitutively active form of OsRac1 by substituting glycine at position 19 corresponding to G12 of the human Rac (Fig. 1A) with valine and fused with the CaMV35S promoter. The construct was introduced into seed-derived calli of the wild type (cv. Kinmaze) and a lesion-mimic mutant of rice, sl (Sekiguchi lesion), by Agrobacterium-mediated transformation (23), and suspension cell cultures were produced from transformed calli. The sl mutant is a propagation type of the lesion-mimic mutants, and large, orange lesions are induced in the leaf by a number of biotic and abiotic stimuli (28, 29). In the transformed cell lines of the wild type and the sl mutant, OsRac1-G19V was overexpressed (Fig. 2A). Staining with DAB (30) revealed H2O2 production in both transformed wild-type and sl cells. It was, however, slightly higher in the sl mutant than in the wild type (Fig. 2B), suggesting that the sl mutant has biochemical alterations upstream of ROS production. The observed H2O2 production was inhibited by diphenylene iodonium (DPI), an inhibitor of the neutrophil NADPH oxidase, both in the transformed wild-type and sl cells. No H2O2 production was detected in the untransformed control cells. No differences in activities of catalase and peroxidase were detected between the untransformed control cells and the transformed cells, suggesting that the H2O2 production did not result from decreased scavenging activities (data not shown). These findings indicated that OsRac1-G19V induces ROS production in rice cells and suggest that a NADPH oxidase similar to the neutrophil enzyme is involved in Rac-induced H2O2 production.
Figure 2.
H2O2 production in suspension-cultured rice cells transformed with 35S-OsRac1-G19V. (A) Overexpression of the OsRac1 in transformed wild-type and sl cells. The probe used was specific to OsRac1. (B) H2O2 production by suspension-cultured rice cells transformed with 35S-OsRac1-G19V. Brown color developed after staining with DAB indicates production of H2O2, and light-brown color shows the background. DPI was added to the cell suspension 30 min before H2O2 detection at a concentration of 50 μM. (C) DNA cleavage in sl cells transformed with 35S-OsRac1-G19V.
Constitutively Active OsRac Induces Cell Death in Cultured Rice Cells.
We next analyzed the biochemical and morphological characteristics of the transformed sl cells in culture. Terminal deoxynucleotidyltransferase-mediated UTP end labeling signals indicative of nuclear DNA cleavage were observed in the transformed sl cells but not in untransformed sl cells (Fig. 2C). In the wild-type cells transformed with OsRac1-G19V, cell death was not observed, suggesting that the sl mutation may be required for cell death to occur in the cultured rice cells. Electron microscopy of the transformed sl cells suggested the occurrence of cell shrinkage (Fig. 3B), condensation of the nucleus (Fig. 3C), condensed and fragmented chromatin (Fig. 3D), and blebbing of the plasma membrane (Fig. 3E). No lysis of the mitochondrial membrane was detected (Fig. 3F). None of the morphological changes detected in the transformed sl cells were found in the untransformed sl cells (Fig. 3A). These results suggest that cell death occurs in the OsRac1-G19V-transformed sl cells and that the observed cell death exhibits a set of morphological changes found in apoptosis in mammalian cells (31). To better understand morphological characteristics of Rac-induced cell death in rice cells, a temporal change of their morphology needs to be studied, and such a study is in progress by the use of an inducible promoter system. Our results also suggest a link between ROS production and cell death during plant–pathogen interactions as has been suggested by previous studies (32–35). They also suggest that OsRac1-induced cell death of rice cells occurs through a mechanism similar to that in mammalian cells. However, we were not able to detect laddering or degradation of isolated nuclear DNA in the cultured rice cells undergoing cell death despite repeated experiments (data not shown). The failure to detect any cleavage of isolated nuclear DNA may be due to nonsynchronous cell death occurring in the transformed cells in culture. Alternatively, the type of DNA degradation observed in apoptosis of mammalian cells may not occur in cultured rice cells.
Figure 3.
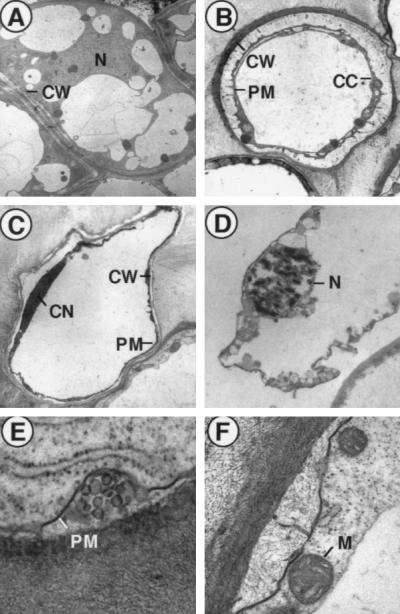
Apoptosis-like cell death in suspension-cultured rice cells transformed with 35S-OsRac1-G19V. (A) Untransformed sl cells. N, nucleus; CW, cell wall. (B) Shrinkage of the plasma membrane (PM) observed in the transformed sl cells. CC, condensed cytoplasm. (C) Condensed nucleus in the transformed sl cells. CN, condensed nucleus. (D) Condensed and fragmented chromatin found in the nucleus of a dead sl cell transformed with 35S-OsRac1-G19V. (E) Blebbing of the plasma membrane found in the transformed sl cells. (F) Intact mitochondrial membrane (M) in a dead, transformed sl cell.
Constitutively Active OsRac Induces ROS Production and Cell Death in the Leaf of Transgenic Rice.
To test whether H2O2 production and cell death also can be induced at the whole plant level, we introduced 35S-OsRac1-G19V into wild-type and sl plants by Agrobacterium-mediated transformation of rice callus and generated 12 and 14 independent transgenic plants of the wild type and the sl mutant, respectively. DAB staining of leaf sheath cells of transgenic wild-type plants showed H2O2 production, which appeared to be localized at the intercellular space (Fig. 4Ab). In addition, appearance of cytoplasmic granules (Fig. 4Ac), which is the first sign of hypersensitive cell death that occurs after infection of resistant rice cultivars with the avirulent blast fungus (36), also was observed in transgenic rice plants. Neither H2O2 production nor the appearance of cytoplasmic granules was observed in untransformed control plants (Fig. 4Aa). Furthermore, necrotic lesions were found in the leaves of all 12 transgenic wild-type plants at a young stage. During maturation, discrete lesions developed in the leaves (Fig. 4B), and they frequently were observed at the junction of the blade and the sheath of the leaves (Fig. 4Ba) and in the midrib region (Fig. 4Bc). This was observed in 8 of 12 transgenic plants. In transgenic sl mutants, large orange lesions, a characteristic of the sl mutants (28, 29), appeared in leaves of very young plants (Fig. 4Ca), and the lesions progressively spread over the entire plants when they matured (Fig. 4Cb) and then died before the emergence of the spikelet. This was observed in all 14 transgenic sl plants analyzed. In untransformed sl plants, no lesions were detected in plants at the young stage, and spikelets developed normally when they matured. These findings indicated that OsRac1-G19V induces H2O2 production as well as cell death in the leaf of rice plants.
Figure 4.
Cell death and H2O2 production in transgenic rice plants transformed with 35S-OsRac1-G19V. (A) H2O2 production (b) and cytoplasmic granulation (c) in leaf sheath cells of transformed wild-type plants. (a) Untransformed plant. (B) Cell death in wild-type plants transformed with 35S-OsRac1-G19V. Discrete lesions (cell death) were observed at the junction of the blade and the sheath of the leaf (a), part of the leaf blade close to the spikelet (b), and the midrib region (c). (C) Cell death in transformed sl plants. (a) Lesions that showed the size and color characteristics of the Sekiguchi lesion. (b) Lesions developed over the entire plant.
Dominant Negative OsRac1 Blocks H2O2 Production and Cell Death in Transgenic Lesion-Mimic Mutants.
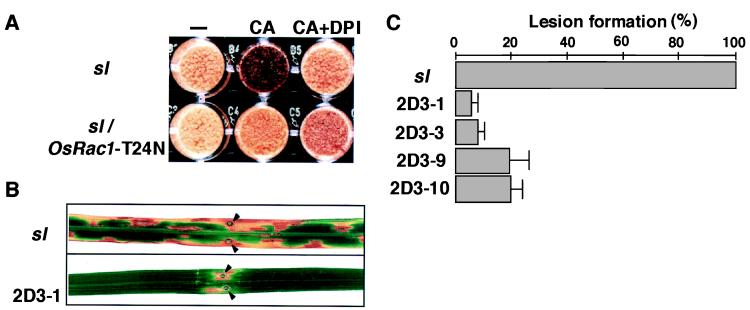
To examine further the role of OsRac1 in ROS production and cell death in rice, we produced a dominant negative form of OsRac1 by changing threonine at the 24th position to asparagine (Fig. 1A), fused with the 35S promoter, and introduced it into the wild type and the sl mutant. In untransformed sl cell cultures H2O2 production was induced by calyculin A (CA), a protein phosphatase inhibitor, and the induction was inhibited by DPI (Fig. 5A), as shown previously with other lesion-mimic mutants of rice (25). In contrast, the CA-induced H2O2 production was inhibited in the sl cells transformed with OsRac1-T24N, indicating that OsRac was required for activation of H2O2 production in the sl cells. To examine whether the dominant negative OsRac1 suppresses lesion formation in sl plants, leaves of untransformed and transformed sl plants were inoculated with the rice blast fungus, M. grisea. It was shown previously that infection of the sl leaf with a blast fungus rapidly induced characteristic lesions distinct from the blast lesion (28, 29). Lesion formation was inhibited strongly in transformed plants (Fig. 4B), and the mean area of lesions developed in leaves of four independent transgenic plants was 10–20% of that found in the untransformed sl mutant (Fig. 5 B and C). Transgenic wild-type plants expressing OsRac1-T24N showed no apparent phenotypic effects (data not shown). These results strongly suggest the role of OsRac in the induction of H2O2 production and cell death in the sl mutant.
Figure 5.
Suppression of H2O2 production and cell death in the sl rice transformed with the dominant negative OsRac1. (A) Inhibition of calyculin A-induced H2O2 production in sl cells transformed with 35S-OsRac1-T24N. Calyculin A (2 μM; Wako) was added to the cell suspensions 30 min before DAB staining. (B) Inhibition of lesion formation in the leaf of the sl plants transformed with 35S-OsRac1-T24N. Inoculation of leaves with the blast fungus was performed on press-injured spots made by a specially designed pressing machine (Fujihara, Osaka, Japan). A piece of agar covered with spores was placed on the injured spots (37). A strain of Magnaporthe grisea, TH67–22 (race 031), which is incompatible with cv. Kinmaze, was used in this experiment. Arrows indicate the sites of inoculation. (C) Quantitative assessment of lesion formation in the leaf of the sl plants transformed with 35S-OsRac1-T24N. Areas of lesions developed were measured with three to five leaves for each transgenic sl plant at 7 days after inoculation.
Concluding Remarks.
Results of our investigations suggest that the small GTP-binding protein Rac regulates the ROS production in rice most likely through a NADPH oxidase, although existence of other routes for ROS production in plants has been suggested (38). Our results also suggest the conservation of the role of Rac to regulate ROS production between plants and animals. They also show that Rac induces cell death in rice cells and that it has biochemical and morphological features similar to apoptosis in mammalian cells. However, the failure to observe laddering or degradation of isolated nuclear DNA in transgenic rice cells might indicate the existence of multiple types of cell death in plants (3, 5). The present findings support a model in which ROS produced by a DPI-inhibitable NADPH oxidase triggers cell death during the defense response of plants (5, 6). Whether the Rac-induced cell death in rice plants has a role in disease resistance remains to be investigated. Analysis of disease resistance in Rac transformants should clarify whether cell death has a causal role in disease resistance in rice. ROS appear to activate cell death in mammalian cells under some circumstances (39). Thus, plants and animals may have in common certain signals for cell death.
Acknowledgments
We thank Drs. S. Kuroda and K. Kaibuchi of Nara Institute of Science and Technology for help in the biochemical assays and valuable suggestions and Ms. M. Nobuhara for excellent technical help. This work was supported in part by a grant (JSPS-RFTF96R16001) from the Research for the Future program by The Japan Society for the Promotion of Science.
ABBREVIATIONS
- ROS
reactive oxygen species
- DPI
diphenylene iodonium
- GST
glutathione S-transferase
- DAB
diaminobenzidine
Footnotes
References
- 1.Vaux D L, Korsmeyer S J. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg J T. Proc Natl Acad Sci USA. 1996;93:12094–12097. doi: 10.1073/pnas.93.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittler R, Lam E. Trends Microbiol. 1996;4:10–15. doi: 10.1016/0966-842x(96)81499-5. [DOI] [PubMed] [Google Scholar]
- 4.Pennell R I, Lamb C. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morel J-B, Dangl J L. Cell Death Differ. 1997;4:671–683. doi: 10.1038/sj.cdd.4400309. [DOI] [PubMed] [Google Scholar]
- 6.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 7.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal A W, Abo A. Trends Biochem Sci. 1993;18:43–47. doi: 10.1016/0968-0004(93)90051-n. [DOI] [PubMed] [Google Scholar]
- 9.Bokoch G M. Curr Opin Cell Biol. 1994;6:212–218. doi: 10.1016/0955-0674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 10.Tenhaken R, Levine A, Brisson L, Dixon R A, Lamb C. Proc Natl Acad Sci USA. 1995;92:4158–4163. doi: 10.1073/pnas.92.10.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desikan R, Hancock J T, Coffey M J, Neill S J. FEBS Lett. 1996;382:213–217. doi: 10.1016/0014-5793(96)00177-9. [DOI] [PubMed] [Google Scholar]
- 12.Dwyer S C, Legendre L, Low P S, Leto T L. Biochim Biophys Acta. 1996;1289:231–237. doi: 10.1016/0304-4165(95)00156-5. [DOI] [PubMed] [Google Scholar]
- 13.Xing T, Higgins V J, Blumwald E. Plant Cell. 1997;9:249–259. doi: 10.1105/tpc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Watson J C. Proc Natl Acad Sci USA. 1993;90:8732–8736. doi: 10.1073/pnas.90.18.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmer D P, Pear J R, Andrawis A, Stalker D M. Mol Gen Genet. 1995;248:43–51. doi: 10.1007/BF02456612. [DOI] [PubMed] [Google Scholar]
- 16.Winge P, Brembu T, Bones A M. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- 17.Kieffer F, Simon-Plas F, Maume B F, Blein J P. FEBS Lett. 1997;403:149–153. doi: 10.1016/s0014-5793(97)00038-0. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Wu G, Ware D, Davis K R, Yang Z. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Yang Z. Plant Cell. 1997;9:1647–1659. doi: 10.1105/tpc.9.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller T, Damude H G, Werner D, Doerner P, Dixon R A, Lamb C. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres M A, Onouchi H, Hamada S, Machida C, Hammond-Kosack K E, Jones J D G. Plant J. 1998;14:365–370. doi: 10.1046/j.1365-313x.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi A, Nakanishi H, Takai Y. Methods Enzymol. 1995;257:57–70. doi: 10.1016/s0076-6879(95)57010-1. [DOI] [PubMed] [Google Scholar]
- 23.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 24.Ohira K, Ojima K, Fujiwara A. Plant Cell Physiol. 1973;14:1113–1121. [Google Scholar]
- 25.Takahashi A, Kawasaki T, Henmi K, Shii K, Kodama O, Satoh H, Shimamoto K. Plant J. 1999;17:535–545. doi: 10.1046/j.1365-313x.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- 26.Didsbury J, Weber R F, Bokoch G M, Evans T, Snyderman R. J Biol Chem. 1989;264:16378–16382. [PubMed] [Google Scholar]
- 27.Takai Y, Kaibuchi K, Kikuchi A, Kawata M. Int Rev Cytol. 1992;133:187–230. doi: 10.1016/s0074-7696(08)61861-6. [DOI] [PubMed] [Google Scholar]
- 28.Kiyosawa S. Bull Nat Inst Agric Sci Jpn Ser D Physiol Genet. 1970;21:61–71. [Google Scholar]
- 29.Marchetti M A, Bollich C N, Uecker F A. Phytopathology. 1983;73:603–606. [Google Scholar]
- 30.Thordal-Christensen H, Zhang Z, Wei Y, Collinge D B. Plant J. 1997;11:1187–1194. [Google Scholar]
- 31.Martin S J, Green D R, Cotter T G. Trends Biochem Sci. 1994;19:26–30. doi: 10.1016/0968-0004(94)90170-8. [DOI] [PubMed] [Google Scholar]
- 32.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 33.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 34.Bestwick C S, Brown I R, Bennett M H, Mansfield J W. Plant Cell. 1997;9:209–221. doi: 10.1105/tpc.9.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delledonne M, Xia Y, Dixon R A, Lamb C. Nature (London) 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 36.Koga H. Can J Bot. 1994;72:1463–1477. [Google Scholar]
- 37.Iwata M, Suzuki Y, Watanabe T, Mase S, Sekizawa Y. Ann Phytopathol Soc Japan. 1980;46:297–306. [Google Scholar]
- 38.Bestwick C S, Bolwell P, Mansfield J W, Nicole M, Wojtaszek P. Trends Plant Sci. 1999;4:88–89. doi: 10.1016/s1360-1385(99)01385-0. [DOI] [PubMed] [Google Scholar]
- 39.Jacobson M J. Trends Biochem Sci. 1996;21:83–86. [PubMed] [Google Scholar]