Abstract
Detection of extended-spectrum β-lactamases (ESBLs) in AmpC-producing Enterobacteriaceae is problematic. A modification of the double-disk test (MDDT) has been developed for successful detection of ESBLs in gram-negative bacilli producing well-characterized β-lactamases as well as 212 clinical isolates of Enterobacter cloacae, Enterobacter aerogenes, Serratia marcescens, and Citrobacter freundii. MDDT accurately differentiated between ESBL producers and derepressed chromosomal AmpC mutants. MDDT provides a cost-effective alternative approach for clinical microbiology laboratories for routine susceptibility testing with simultaneous detection of ESBLs in Enterobacteriaceae.
Enterobacteriaceae producing both AmpC β-lactamases and extended-spectrum β-lactamases (ESBLs) have been increasingly reported worldwide (2-6, 12, 16, 18). Since AmpC-producing organisms can act as hidden reservoirs for ESBLs, it is important for clinical microbiology laboratories to be able to detect ESBL production in these organisms on a routine basis (9). The National Committee for Clinical Laboratory Standards (NCCLS) has published guidelines for the detection of ESBLs in clinical isolates of Escherichia coli and Klebsiella spp., but there are currently none available for other genera (10). Since high-level expression of AmpC β-lactamases may mask recognition of ESBLs (17), a unique modification of the double-disk test (MDDT) using a combination of cefepime (FEP) and piperacillin-tazobactam (TZP) was evaluated to detect ESBLs. This evaluation was carried out with well-characterized strains producing either AmpC β-lactamases and ESBLs or either β-lactamase alone. In addition, the MDDT determined the presence of an ESBL in an E. coli isolate that showed a negative result with the NCCLS disk confirmation test.
The following strains producing known β-lactamases were used for this study (Table 1). A total of 212 clinical isolates were also evaluated by MDDT: 94 were E. cloacae, 32 were E. aerogenes, 25 were S. marcescens, and 61 were C. freundii. The isolates were nonrepetitive (one per patient) and were obtained from clinical specimens from Universitas and Pelonomi Hospitals, Bloemfontein, South Africa, over a 9-month period during 1998 and 1999.
TABLE 1.
Control and clinical strains producing well-characterized β-lactamasesa
| Strain | Organism | β-Lactamasea | MDDT result | Source or referenceb |
|---|---|---|---|---|
| RTEM | E. coli | TEM-1 | Negative | a |
| 1752E | E. coli | TEM-2 | Negative | a |
| J53 | E. coli | SHV-1 | Negative | a |
| CF204 | E. coli | TEM-3 | Positive | a |
| K12 | E. coli | TEM-4 | Positive | a |
| CF604 | E. coli | TEM-5 | Positive | a |
| CF804 | E. coli | TEM-6 | Positive | a |
| C600 | E. coli | TEM-7 | Positive | a |
| C600 | E. coli | TEM-8 | Positive | a |
| 2639E | E. coli | TEM-9 | Positive | a |
| C600 | E. coli | TEM-10 | Positive | a |
| C600 | E. coli | TEM-16 | Positive | a |
| CF1104 | K. pneumoniae | TEM-24 | Positive | a |
| PJPQ101 | E. coli | TEM-26 | Positive | a |
| C600 | E. coli | SHV-2 | Positive | a |
| J53 | E. coli | SHV-3 | Positive | a |
| J53-2 | E. coli | SHV-4 | Positive | a |
| ClaNal | E. coli | SHV-5 | Positive | a |
| DH52 | E. coli | SHV-7 | Positive | a |
| C600 | E. coli | MIR-1 | Negative | a |
| NU2936 | K. pneumoniae | MOX-1 | Negative | a, b |
| F12 | K. pneumoniae | FOX-1 | Negative | a, b |
| P20 | K. pneumoniae | LAT-1 | Negative | a, b |
| C600 | E. coli | BIL-1 | Negative | a, b |
| AR15 | E. coli | CTXM-1 | Positive | a |
| Pit 16 | K. pneumoniae | TEM-63 | Positive | 13 |
| Pit 100 | K. pneumoniae | SHV-2 | Positive | 13 |
| Pit 82 | K. pneumoniae | SHV-5 | Positive | 13 |
| Pit 64 | E. coli | TEM-63 | Positive | 13 |
| Pit 56 | E. coli | SHV-2 | Positive | 13 |
| Pit 85 | P. mirabilis | TEM-63 | Positive | 13 |
| 187 | E. aerogenes | AmpC + SHV-3 | Positive | 12 |
| 184 | E. aerogenes | AmpC + SHV-5 | Positive | 12 |
| 200 | E. aerogenes | AmpC + SHV-4 | Positive | 12 |
| 220 | E. aerogenes | AmpC + SHV-4 | Positive | 12 |
| 86 | E. aerogenes | AmpC + TEM-3 | Positive | 11 |
| 94 | E. cloacae | AmpC + SHV-2 | Positive | 11 |
| 142 | E. cloacae | AmpC + TEM-3 | Positive | 11 |
| 029 | E. cloacae | AmpC (WT) | Negative | 11 |
| 108 | E. cloacae | AmpC (DM) | Negative | 11 |
WT, wild type; DM, derepressed mutant.
a, provided by Creighton University, Omaha, Nebr.; b, plasmid-mediated AmpC β-lactamases.
The susceptibility of the isolates was determined by the standard disk diffusion method as described in the NCCLS guidelines (10). Disks for the agar diffusion procedure were obtained from Becton Dickinson Microbiology Systems (Johannesburg, South Africa).
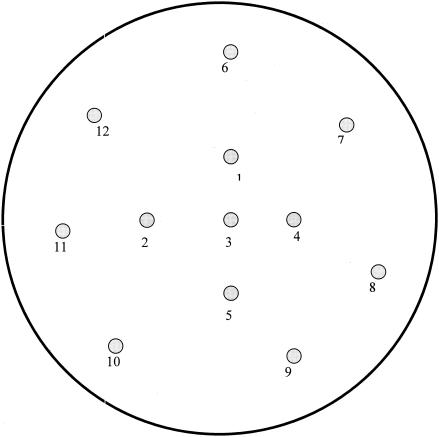
MDDT was performed on both control and clinical strains. Modifications of the original double-disk test were as follows (8). Ceftriaxone (30-μg disk) was replaced with FEP (30-μg disk), the cefotaxime (CTX) (30-μg) disk was placed 20 mm from the amoxicillin (20 μg)-clavulanate (10 μg) (AMC) disk, the aztreonam (ATM) (30 μg) disk was placed at 25 mm, the ceftazidime (CAZ) (30 μg) disk was placed at 30 mm, and the FEP (30 μg) disk was placed at 30 mm (Fig. 1). This modification was incorporated into a gram-negative template, and a piperacillin (100 μg)-tazobactam (10 μg) (TZP) disk was placed 25 mm from FEP (Fig. 1).
FIG. 1.
MDDT and the gram-negative template. The antibiotic disk numbers represent the following antibiotics: 1, ATM; 2, FEP; 3, AMC; 4, CTX; 5, CAZ; 6, imipenem; 7, cefoxitin; 8, cefuroxime; 9, gentamicin; 10, amikacin; 11, TZP; and 12, ciprofloxacin.
Sonic extracts containing β-lactamases were assessed for isoelectric points and substrate and inhibitor profiles in polyacrylamide gels as previously described (13). PCR amplification was performed to determine the presence of blaTEM or blaSHV, and the SHV amplicon was sequenced as previously described (13).
The MDDT detected the presence of ESBLs in all of the well-characterized strains (Table 1). The clinical strains were divided into three groups according to interpretation with MDDT and susceptibilities to CTX and CAZ (Table 2): group 1 (wild type), ESBL negative, sensitive to CTX and CAZ; group 2 (derepressed mutants), ESBL negative, resistant to CTX and CAZ; and group 3 (ESBL producers) ESBL positive, sensitive, intermediate, or resistant to CTX and CAZ.
TABLE 2.
Characteristics of β-lactamases produced by different resistant phenotypes
| Resistant phenotypea | (no. of strains) | Susceptibility tob:
|
MDDT result | pI | CTX hydrolysisc | Inhibited byd:
|
Amplification with:
|
Most likely β-lactamasese | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | CLOX | CLAV | TEM | SHV | ||||||
| Group 1 | (126) | S | S | Negative | NDf | ND | ND | ND | ND | ND | AmpC (WT) |
| Group 2 | (57) | R | R | Negative | 8.0-8.9 | Yes | Yes | No | ND | ND | AmpC (DM) |
| Group 3 | (29) | S/I/R | I/R | Positive | 8.0-8.9 | No | Yes | No | AmpC | ||
| 8.2 | Yes | No | Yes | No | Yes | SHV ESBL | |||||
| 7.6 | Yes | No | Yes | No | Yes | SHV ESBL | |||||
| 5.6 | Yes | No | Yes | Yes | No | TEM ESBL | |||||
| Wild typeg | (4) | S | S | Negative | 8.3-8.8 | No | Yes | No | ND | ND | AmpC (WT) |
| Derepressedg | (5) | R | R | Negative | 8.3-8.9 | Yes | Yes | No | ND | ND | AmpC (DM) |
Group 1 consisted of 51 E. cloacae, 18 E. aerogenes, 39 C. freundii, and 18 S. marcescens isolates. Group 2 consisted of 28 E. cloacae, 9 E. aerogenes, 15 C. freundii, and 5 S. marcescens isolates. Group 3 consisted of 15 E. cloacae, 5 E. aerogenes, 7 C. freundii, and 2 S. marcescens isolates.
Disk diffusion. S, susceptible; I, intermediate; R, resistant.
Hydrolysis of 0.75 μg of CTX per ml used in substrate-based IEF overlay technique (13).
Inhibitors used in IEF overlay technique were clavulanic acid (CLAV) and cloxacillin (CLOX) (13).
WT, wild type; DM, derepressed mutant.
ND, not determined.
Reference 12.
Isoelectric focusing (IEF) was performed on the strains in groups 2 and 3. Strains representing the group 2 phenotype produced β-lactamases with pI values ranging from 8.0 to 8.9 and were inhibited by cloxacillin on IEF gels. This correlates with Bush group 1 cephalosporinases (1). The ESBL-positive group produced similar Bush group 1 cephalosporinases (Table 2) as well as additional β-lactamases with pIs of 8.2, 7.6, and 5.6 that were inhibited by clavulanate on IEF gels and showed CTX hydrolysis (0.75 μg/ml) for bands focusing at pIs of 8.2, 7.6, and 5.6. These enzymes have characteristics of Bush group 2e β-lactamases (ESBLs) (14) (Table 2).
TEM- and SHV-specific PCR was performed on DNA obtained from group 3 (Table 2). An 885-bp fragment specific for blaSHV was amplified in organisms producing β-lactamases with pIs of 7.6 and 8.2, and a 971-bp fragment specific for blaTEM was amplified in organisms producing β-lactamases with pI values of 5.6. Taken together, the data obtained from IEF and PCR indicate that these strains produced both AmpC β-lactamases and either SHV or TEM ESBLs. A positive test was present for the organisms in group 3 when the MDDT to ATM, FEP, and/or TZP disks was used. Therefore, the MDDT successfully differentiated between ESBL producers and overexpression of AmpC-derepressed mutants.
An additional E. coli isolate obtained from a urine culture, indicating a negative confirmatory ESBL disk test as recommended by the NCCLS, showed a positive MDDT. This isolate was resistant to cefoxitin and cefpodoxime, intermediate in resistance to CAZ, and sensitive to ceftriaxone. Further evaluation of this strain by IEF revealed the production of two β-lactamases consistent with the presence of Bush group 1 (pI >9.0) and group 2e (pI 7.6). The group 2e enzyme was confirmed as SHV-2 by PCR and sequencing (13, 14).
Several types of TEM and SHV ESBLs have been described in isolates of K. pneumoniae, E. coli, and Proteus mirabilis from South Africa (7, 13). This is the first report of TEM and SHV ESBL production in Enterobacter species, C. freundii, and S. marcescens isolates in South Africa. The characterization of the different enzymes was outside the scope of this study.
In this study, ESBL production among Enterobacter species, C. freundii, and S. marcescens was associated with high levels of resistance to trimethoprim-sulfamethoxazole and gentamicin when compared to derepressed chromosomal AmpC mutants (data not shown). Since ESBL producers express their β-lactamase genes from plasmids, these organisms may also have genes coding for resistance to additional classes of antibiotics (15). This report demonstrates the need to differentiate AmpC overproducers from ESBL-producing strains.
There are no published guidelines for the detection of ESBLs in organisms other than E. coli and Klebsiella spp. The original double-disk test, Vitek ESBL card, and E-test ESBL strips failed to detect ESBL-producing isolates of C. freundii, E. cloacae, E. aerogenes, S. marcescens, Morganella morganii, and Providentia stuartii (16, 18). The MDDT described in this study detected ESBLs in strains with well-characterized β-lactamases as well as clinical strains of E. cloacae, E. aerogenes, C. freundii, and S. marcescens. Furthermore, the MDDT was able to detect the presence of an ESBL (SHV-2) in an AmpC-producing E. coli isolate that failed the NCCLS ESBL disk confirmation test. This modification has been incorporated into a gram-negative template for routine susceptibility testing with the additional benefits of simultaneous detection of ESBLs. The MDDT provides a cost-effective alternative for ESBL testing in clinical microbiology laboratories, thus negating the need for ESBL confirmation procedures.
Acknowledgments
This work was partly supported by a grant from the Medical Research Council of South Africa.
We thank Ellen Smith Moland and Marilyn Creighton for expert technical assistance.
REFERENCES
- 1.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canton, R., A. Oliver, T. M. Coque, M. del Carmen Varela, J. C. Pérez-Diaz, and F. Baquero. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chanal, C. D., J. P. Sirot, J. P. Romaszko, L. Bret, and J. Sirot. 1996. Survey of extended spectrum β-lactamases among Enterobacteriaceae. J. Antimicrob. Chemother. 38:127-132. [DOI] [PubMed] [Google Scholar]
- 4.Chanawong, A., F. H. M'Zali, J. Heritage, A. Lutitanoud, and P. M. Hawkey. 2001. SHV-12, SHV-5, SHV-29, and VEB-1 extended-spectrum β-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J. Antimicrob. Chemother. 48:839-852. [DOI] [PubMed] [Google Scholar]
- 5.Chanawong, A., F. H. M'Zali, J. Heritage, J.-H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and the French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essack, S. Y., L. M. C. Hall, D. G. Pillay, M. L. McFadyen, and D. M. Livermore. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum β-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Phillipon. 1988. Extended broad-spectrum β-lactamases confirming resistance to newer β-lactamase agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 9.Levison, M. E. 2002. Plasmid-mediated extended-spectrum β-lactamases in organisms other than Klebsiella pneumoniae and Escherichia coli: a hidden resource of transferable resistance genes. Curr. Infect. Dis. Rep. 4:181-183. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial and susceptibility testing: 12th informational supplement (M-100-S12). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Pitout, J. D. D., E. S. Moland, C. C. Sanders, K. S. Thomson, and S. R. Fitzsimmons. 1997. β-Lactamases and detection of β-lactam resistance in Enterobacter spp. Antimicrob. Agents Chemother. 41:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, P. Coudron, and C. C. Sanders. 1998. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob. Agents Chemother. 42:596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, E. S. Moland, and C. C. Sanders. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 42:1350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders, C. C., W. E. Sanders, Jr., and E. S. Moland. 1986. Characterization of β-lactamases in situ on polyacrylamide gels. Antimicrob. Agents Chemother. 30:951-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirot, D. 1995. Extended-spectrum β-lactamases. J. Antimicrob. Chemother. 36(Suppl. A):19-34. [DOI] [PubMed] [Google Scholar]
- 16.Spanu, T., F. Luzzaro, M. Perilli, G. Amicosante, A. Toniolo, G. Fadda, and the Italian ESBL Study Group. 2002. Occurrence of extended-spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implication for resistance to β-lactamases and other antimicrobial drugs. Antimicrob. Agents Chemother. 46:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson, K. S., C. C. Sanders, and E. S. Moland. 1999. Use of microdilution panels with and without β-lactamase inhibitors as a phenotypic test for β-lactamase production among Escherichia coli, Klebsiella spp., Enterobacter spp., Citrobacter freundii, and Serratia marcescens. Antimicrob. Agents Chemother. 43:1393-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzelepi, E., P. Giakkoupi, D. Sofianou, V. Loukova, A. Kemeroglou, and A. Tsakris. 2000. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 38:542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]