Abstract
Pulsed-field gel electrophoresis of Escherichia coli O157:H7 isolates (n = 228) from 122 healthy animals on 11 farms discriminated 57 types. Most clones were found only on individual farms. Numerous clones were found within each farm, with a prevalent clone normally found in several animals. A variety of clones were found within the different phage types.
Escherichia coli strains that produce Shiga-like toxins may produce two toxins (Shiga toxin 1 [ST1] and/or ST2) with similar biological activities. A subset of Shiga-toxigenic E. coli strains designated as enterohemorrhagic (EHEC) carry sets of virulence genes (e.g., eaeA) that encode factors for attachment to host cells. Recent studies have revealed the existence of two divergent lineages (EHEC 1 and 2) (25). The EHEC 1 lineage consists solely of a few very closely related and geographically disseminated multilocus genotypes each bearing the O157 phenotype (15). Despite this similarity, pulsed-field gel electrophoresis (PFGE) and other molecular techniques have shown a considerable degree of genetic heterogeneity among O157 isolates in several countries, suggesting large evolutionary distances between strains. PFGE is considered the “gold standard” for fingerprinting of O157 strains and forms the basis of a large surveillance database (PulseNet) (24).
Cattle are important reservoirs for this agent. Limited information is available about the genetic diversity of O157:H7 animal strains in the United Kingdom. Several studies have looked at human outbreak isolates, in most cases from Scotland (2, 3, 16, 23). Some studies have described diversity in plasmid profiles in a limited number of animal isolates in comparison with isolates from food and humans (6, 8, 9). To our knowledge, one recent study (5) was the first to investigate the diversity of genotypes among animal isolates. The aim of the present study was to evaluate the genotypic diversity of O157:H7 isolates on a variety of cattle farms.
Bacterial strains.
E. coli O157:H7 (666 isolates) was cultured (7) from bovine fecal samples from 11 epidemiologically unrelated farms dispersed across England and Wales as part of a prevalence and longitudinal study (G. A. Paiba, J. W. Wilesmith, S. J. Evans, S. J. S. Pascoe, R. P. Smith, S. A. Kidd, J. B. M. Ryan, I. M. McLaren, S. A. Chappell, G. A. Willshaw, T. Cheasty, N. P. French, T. J. Jones, H. Buchanan, D. J. Challoner, A. D. Colloff, M. P. Cranwell, I. H. Davies, R. G. Daniel, J. P. Duff, R. A. Hogg, F. D. Kirby, M. F. Millar, J. R. Monies, M. Nicholls, and J. H. Payne, submitted for publication). All the isolates were confirmed as serotype O157 by serum agglutination (21). The presence of the flagellar H7 gene was confirmed by PCR performed in a LightCycler instrument (Roche Diagnostics UK Ltd., Lewes, United Kingdom) in 20-μl reaction mixtures with 1× LightCycler-FastStart DNA Master SYBR Green I (Roche Molecular Biochemicals), primers FLICH7-F and FLICH7-R (14) at 100 ng each, 2.5 mM MgCl2, and 2 μl of bacterial DNA template. The PCR run consisted of 95°C for 10 min (one cycle), followed by 35 cycles of 95°C for 10 s, 65°C for 5 s, and 72°C for 25 s. The nature of the amplicon was determined by melting analysis over a temperature range from 70 to 97°C (0.1°C/s), and the melting temperature (Tm) was 87.5°C. The size of the product (625 bp) was verified by gel electrophoresis. ST production was determined by Vero cell culture assay (10). Phage typing of the isolates was conducted at the Public Health Laboratory Service, Colindale, London, United Kingdom (13).
When the number of isolates available from a farm was lower than 10, all isolates were selected for genetic analysis. For those farms where more than 10 isolates were available, between 24 and 63% of the isolates were fingerprinted. A total of 228 O157:H7 isolates from 122 healthy bovines on 11 farms (farm 1, n = 3 animals; farm 2, n = 23; farm 3, n = 19; farm 4, n = 14; farm 5, n = 5; farm 6, n = 9; farm 7, n = 1; farm 8, n = 12; farm 9, n = 29; farm 10, n = 3; and farm 11, n = 4) were characterized by PFGE. All but 12 of the isolates were ST positive.
PFGE.
Preparation and XbaI digestion of DNA for PFGE and analysis of results were conducted as described previously (5).
Several studies have looked at PFGE analysis of O157 isolates from cattle from Australia (11, 20), the United States (12, 22), and Finland (17). However, most studies have looked at a limited number of strains, strains from a limited number of farms in a particular region, or strains from abattoir surveys. This study intends to present data from 11 farms spread across England and Wales (Paiba et al., submitted). Ten phage types (PTs) (1, 2, 4, 8, 21/28, 31, 32, 34, 43, and 50) were represented in the isolates selected. The most prevalent PT was type 4, present in 70 (30.7%) isolates and on five farms (farms 1, 3, 5, 8, and 9).
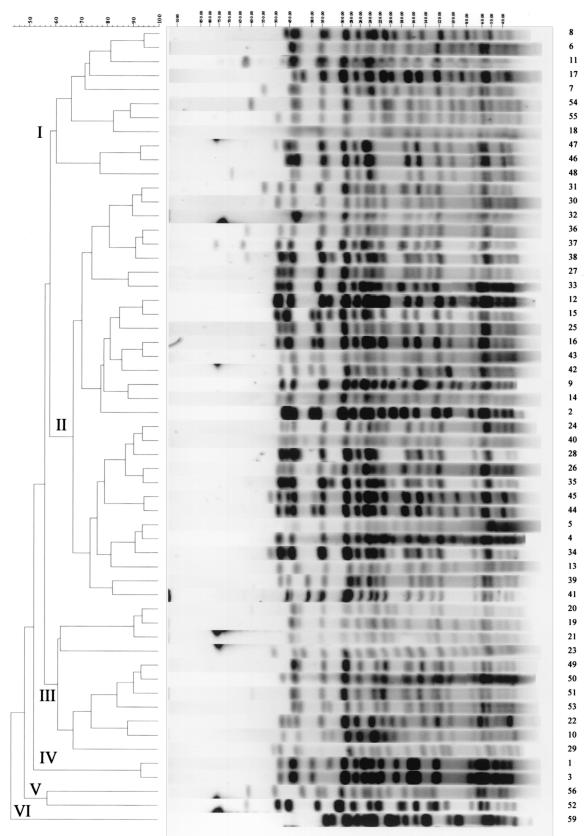
Fifty-seven PFGE types were found among the 228 isolates investigated. The profiles contained between 13 and 18 bands between 50 and 800 kb in length. Bands below 50 kb were ignored in the analysis. Figure 1 shows a representation of the different fingerprint types; clustering analysis shows six distinct clusters (separating at 60% similarity or less).
FIG. 1.
Dendrogram generated with Gelcompar II software showing the relationship of 57 representative PFGE types for 228 bovine E. coli O157 isolates. The analysis of the bands generated was performed using the Jaccard coefficient and the unweighted pair group method with arithmetic averages. The numerals at the top of the figure indicate molecular sizes in kilobase pairs.
We found numerous PFGE profiles within each of the farms in the study. Despite this on-farm diversity, some of the patterns predominated and were found in isolates from several animals on the farm (Table 1). The vast majority of the PFGE types were found only on individual farms, with the exceptions of type X8, found on farms 8 and 11; type X24, found on farms 3, 9, and 11; type X26, found on farms 3 and 9; type X30, found on farms 3 and 8; type X36, found on farms 3 and 10; and finally type X54, found on farms 4, 5, and 6. Table 2 shows a distribution of the different PFGE types found on the different farms. It may be postulated that when a strain appears only in small numbers on a farm this may indicate that there has been a recent addition to the O157:H7 pool on that particular farm.
TABLE 1.
Distribution of prevalent PFGE types on English cattle farms
| Farm no. | Prevalent type (% of isolates on the farm)a | No. of animals (total animals on the farm)b |
|---|---|---|
| 2 | X44 (40.4) | 13 (23) |
| 3 | X24 (48.6) | 11 (19) |
| 4 | X49 (30.7) | 5 (14) |
| 5 | X31 (37.5) | 2 (5) |
| X56 (37.5) | 2 (5) | |
| 6 | X22 (44.5) | 4 (9) |
| 8 | X8 (51.7) | 7 (12) |
| 9 | X24 (48.2) | 16 (29) |
| 11 | X6 (43) | 3 (4) |
Numbers in parentheses indicate the percentage of isolates on the farm belonging to the prevalent PFGE type.
This figure indicates the number of animals from which at least one isolate belonging to the PFGE prevalent type was cultured. The number in parentheses indicates the total number of animals positive for E. coli O157 on the particular farm.
TABLE 2.
Distribution of PFGE types on 11 cattle farms
| Farm no. | No. of isolates tested by PFGE | PFGE type(s) |
|---|---|---|
| 1 | 3 | X14, X29, X43 |
| 2 | 52 | X1, X2, X3, X4, X12, X15, X16, X39, X44, X45 |
| 3 | 35 | X5, X19, X20, X21, X24, X26, X30, X32, X33, X36, X37, X40, X41 |
| 4 | 26 | X46, X47, X48, X49, X50, X51, X52, X53, X54, |
| 5 | 8 | X7, X31, X54, X56 |
| 6 | 9 | X10, X22, X42, X54 |
| 7 | 1 | X13 |
| 8 | 29 | X8, X11, X25, X30, X59 |
| 9 | 54 | X17, X23, X24, X26, X27, X28, X34, X35, X38 |
| 10 | 4 | X24, X36 |
| 11 | 7 | X6, X8, X9, X18 |
Our findings are partially in agreement with results from three Australian dairy farms (11) where it was reported that individual farms harbor populations of predominant strains that are rarely shared between farms. Also, in a study with five dairy farms, interfarm clonal variation for O157:H7 strains was reported (12); interestingly, in that study two farms apparently unrelated and situated 121 km apart had the same PFGE patterns.
The most prevalent type among all the isolates in our study was X24, which was found in 19.7% of the isolates arising from 28 animals on three different farms (farms 3, 9, and 10). Although we are not aware of any epidemiological links among these farms, those isolates may have been linked in the distant past. This observation highlights the importance of good detailed historical epidemiological data being available for assessment of the results of genetic fingerprinting. In contrast with our findings of a widely distributed clone, data from Germany suggested that there was not a geographically dominant single clone present among isolates from human hemolytic-uremic syndrome patients (19)
In most cases, a particular PFGE profile was found only in isolates from specific PTs (Table 3). However, this was not invariably so. Phage typing did not necessarily correlate with PFGE typing, and PFGE type X8 was found among PT 34 isolates and isolates that reacted but did not conform to type (RDNC), type X24 was found among PT 1 and 4 isolates, and type X42 was found among PT 50 and RDNC isolates. For most PTs, a variety of different PFGE types were found. The genetic diversity of the different PTs remains to be elucidated, as we did not test representative samples from each of them.
TABLE 3.
Relation of PT to PFGE type in E. coli O157 isolates
| PT | No. of isolates | Farm no. | PFGE type(s) |
|---|---|---|---|
| 1 | 2 | 10 | X24 |
| 2 | 55 | 2, 3, 5, 8 | X30, X39, X41, X44, X45, X56 |
| 4 | 70 | 1, 3, 5, 8, 9 | X5, X24, X25, X26, X27, X28, X30, X31, X33, X34, X35, X36, X37, X38, X40, X43 |
| 8 | 3 | 2, 3 | X1, X3, X32 |
| 21/28 | 9 | 1, 2 | X4, X12, X14, X15, X16 |
| 31 | 3 | 11 | X6 |
| 32 | 29 | 2, 3, 4 | X2, X19, X20, X21, X46, X47, X48, X49, X50, X51, X52, X53 |
| 34 | 25 | 1, 5, 6, 7, 8 | X7, X8, X10, X11, X13, X22, X29, X59 |
| 50 | 2 | 6 | X42 |
| RDNC | 5 | 6, 9, 11 | X8, X9, X24, X42 |
| 43 | 14 | 9 | X17, X23 |
Twelve ST-negative O157:H7 isolates were included in the study. Of these, two were of the common PFGE type X24, one each originating from farms 3 and 9, and two were of common type X36 and were found on farm 10. The PFGE types X24 and X36 were also found among ST-positive O157:H7 isolates. Six ST-negative isolates were of type X54, found on farms 4, 5, and 6; this PFGE type was found only in ST-negative isolates. Finally, one isolate from farm 4 was of PFGE type X55 and one isolate from farm 11 was of PFGE type X18; these types were not repeated anywhere else. The question arises whether nontoxigenic O157:H7 strains should be considered a risk to public health. There is some evidence suggesting that toxin genes may be transmitted between strains by phage conversion (4).
PFGE provides a useful tool for outbreak investigations, but only recently have these techniques been used as a surveillance tool. It remains to be demonstrated whether E. coli O157:H7 is genetically stable enough to permit long-term surveillance. Data from Germany suggest a considerable genetic stability among human O157 strains, and molecular fingerprints of outbreak strains have remained identical after several years of storage on agar slants or subcultivation (19). However, this differs from the findings of LeClerc et al. (18) suggesting that some O157 strains may become hypermutable, making them more adaptable to changing environments and increasing the organism's genetic diversity. Also, a Japanese study (1) reported the shift of genetic subtypes on one farm and from one animal. Although there is some evidence that clonal turnover and genetic rearrangement remain rare under environmental situations, it is possible that this is faster in situ than in the laboratory (12).
If a meaningful farm-to-fork surveillance is intended, comprehensive databases must be established. These should include information from environmental, animal, food, and human clinical isolates. Efforts such as PulseNet should be broadened to include veterinary isolates.
Acknowledgments
We thank Tom Cheasty and Geraldine Willshaw (PHLS, Colindale) for phage typing the strains.
This work was supported by DEFRA grant OZ0138.
REFERENCES
- 1.Akiba, M., T. Sameshima, and M. Nakazawa. 1999. The shift of genetic subtypes of Escherichia coli O157:H7 isolates from cattle. Epidemiol. Infect. 122:343-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, L., A. Stirrat, and F. M. Thomson-Carter. 1998. Genetic heterogeneity of Escherichia coli O157:H7 in Scotland and its utility in strain subtyping. Eur. J. Clin. Microbiol. Infect. Dis. 17:844-848. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, C., L. Metherell, G. Willshaw, A. Maggs, and J. Stanley. 1999. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J. Clin. Microbiol. 37:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asakura, H., S. I. Makino, H. Kobori, M. Watarai, T. Shirahata, T. Ikeda, and K. Takeshi. 2001. Phylogenetic diversity and similarity of active sites of Shiga toxin (Stx) in Shiga toxin-producing Escherichia coli (STEC) isolates from humans and animals. Epidemiol. Infect. 127:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery, S. M., E. Liebana, C. A. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, P. A., A. T. Cerdan Malo, M. Ellin, R. Ashton, and M. A. Harkin. 2001. Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int. J. Food Microbiol. 64:139-150. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, P. A., and C. A. Siddons. 1996. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from cases of bloody diarrhoea, non-bloody diarrhoea and asymptomatic contacts. J. Med. Microbiol. 44:267-271. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 2000. A one year study of Escherichia coli O157 in raw beef and lamb products. Epidemiol. Infect. 124:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, P. A., C. A. Siddons, A. T. Cerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman, P. A., D. J. Wright, and P. Norman. 1989. Verotoxin-producing Escherichia coli infections in Sheffield: cattle as a possible source. Epidemiol. Infect. 102:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobbold, R., and P. Desmarchelier. 2001. Characterisation and clonal relationships of Shiga-toxigenic Escherichia coli (STEC) isolated from Australian dairy cattle. Vet. Microbiol. 79:323-335. [DOI] [PubMed] [Google Scholar]
- 12.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost, J. A., H. R. Smith, G. A. Willshaw, S. M. Scotland, R. J. Gross, and B. Rowe. 1989. Phage-typing of Vero-cytotoxin (VT) producing Escherichia coli O157 isolated in the United Kingdom. Epidemiol. Infect. 103:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon, V. P. J., S. D'Souza, T. Graham, R. K. King, K. Rahn, and S. Read. 1997. Use of flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J. Clin. Microbiol. 35:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause, U., F. M. Thomson-Carter, and T. H. Pennington. 1996. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J. Clin. Microbiol. 34:959-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahti, E., M. Keskimaki, L. Rantala, P. Hyvonen, A. Siitonen, and T. Honkanen-Buzalski. 2001. Occurrence of Escherichia coli O157 in Finnish cattle. Vet. Microbiol. 79:239-251. [DOI] [PubMed] [Google Scholar]
- 18.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 19.Liesegang, A., U. Sachse, R. Prager, H. Claus, H. Steinruck, S. Aleksic, W. Rabsch, W. Voigt, A. Fruth, H. Karch, J. Bockemuhl, and H. Tschape. 2000. Clonal diversity of Shiga toxin-producing Escherichia coli O157:H7/H− in Germany—a ten-year study. Int. J. Med. Microbiol. 290:269-278. [DOI] [PubMed] [Google Scholar]
- 20.Midgley, J., and P. Desmarchelier. 2001. Pre-slaughter handling of cattle and Shiga toxin-producing Escherichia coli (STEC). Lett. Appl. Microbiol. 32:307-311. [DOI] [PubMed] [Google Scholar]
- 21.Paiba, G. A., J. C. Gibbens, S. J. Pascoe, J. W. Wilesmith, S. A. Kidd, C. Byrne, J. B. Ryan, R. P. Smith, M. McLaren, R. J. Futter, A. C. Kay, Y. E. Jones, S. A. Chappell, G. A. Willshaw, and T. Cheasty. 2002. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Vet. Rec. 150:593-598. [DOI] [PubMed] [Google Scholar]
- 22.Paros, M., P. I. Tarr, H. Kim, T. E. Besser, and D. D. Hancock. 1993. A comparison of human and bovine Escherichia coli O157:H7 isolates by toxin genotype, plasmid profile, and bacteriophage lambda-restriction fragment length polymorphism profile. J. Infect. Dis. 168:1300-1303. [DOI] [PubMed] [Google Scholar]
- 23.Smith, D., G. Willshaw, J. Stanley, and C. Arnold. 2000. Genotyping of verocytotoxin-producing Escherichia coli O157: comparison of isolates of a prevalent phage type by fluorescent amplified-fragment length polymorphism and pulsed-field gel electrophoresis analyses. J. Clin. Microbiol. 38:4616-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]