Abstract
Chronic inhalation of low amounts of Cr(VI) promotes pulmonary diseases and cancers through poorly defined mechanisms. SFKs (Src family kinases) in pulmonary airway cells may mediate Cr(VI) signalling for lung injury, although the downstream effectors of Cr(VI)-stimulated SFKs and how they relate to pathogenic gene induction are unknown. Therefore SFK-dependent activation of transcription factors by non-cytotoxic exposure of human bronchial epithelial cells to Cr(VI) was determined. Protein–DNA binding arrays demonstrated that exposing BEAS 2B cells to 5 μM Cr(VI) for 4 and 24 h resulted in increased protein binding to 25 and 43 cis-elements respectively, while binding to 12 and 16 cis-elements decreased. Of note, Cr(VI) increased protein binding to several STAT (signal transducer and activator of transcription) cis-elements. Cr(VI) stimulated acute tyrosine phosphorylation and nuclear translocation of STAT1 over a 4 h period and a prolonged activation of STAT3 that reached a peak between 48 and 72 h. This prolonged activation was observed for both STAT3α and STAT3β. Immunofluorescent confocal microscopy confirmed that Cr(VI) increased nuclear localization of phosphorylated STAT3 for more than 72 h in both primary and BEAS 2B human airway cells. Cr(VI) induced transactivation of both a STAT3-driven luciferase reporter construct and the endogenous inflammatory gene IL-6 (interleukin-6). Inhibition with siRNA (small interfering RNA) targeting the SFK Lck, but not dominant-negative JAK (Janus kinase), prevented Cr(VI)-stimulated phosphorylation of both STAT3 isoforms and induction of IL-6. The results suggest that Cr(VI) activates epithelial cell Lck to signal for prolonged STAT3 activation and transactivation of IL-6, an important immunomodulator of lung disease progression.
Keywords: airway epithelial cell, chromium, interleukin-6, Lck, signal transducer and activator of transcription 3 (STAT3), small interfering RNA (siRNA), Src family kinase
Abbreviations: AM, acetoxymethyl ester; AP-1, activator protein-1; CREB, cAMP-response-element-binding protein; DTT, dithiothreitol; EGFP, enhanced green fluorescent protein; IFN, interferon; GAS, IFN-γ-activated site; IL-6, interleukin-6; ISRE, IFN-stimulated response element; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MTF-1, metal transcription factor-1; NF-κB, nuclear factor κB; NP40, Nonidet P40; pHBE, primary human bronchiolar epithelial; pY, tyrosine phosphorylated; SFK, Src family kinase; SIE, serum-inducible element; siRNA, small interfering RNA; RT, reverse transcriptase; STAT, signal transducer and activator of transcription
INTRODUCTION
Chromium is a ubiquitous metal that exists predominantly in two physiologically relevant valence states: Cr(VI) and Cr(III). As an essential nutrient (reviewed in [1]), Cr(III) may contribute to the efficiency of insulin signalling [2]. In contrast, chronic occupational and environmental exposure to Cr(VI), which occurs predominantly through inhalation, leads to airway disease [3,4] and lung cancer [5,6]. Cr(VI) is the more toxic species, since its structure is similar to sulfate and phosphate [7], allowing it to be easily absorbed and retained by lung cells [8]. Once in the cells, Cr(VI) can be reduced to Cr(III), which is highly reactive with macromolecules, such as proteins and DNA [8–10].
Modification of signalling proteins may play a significant role in the aetiology of Cr(VI)-induced pulmonary diseases. Cr(VI) selectively activates the SFKs (Src family kinases), Lck and Fyn, in pulmonary epithelial cells [11] as well as in lymphocytes [12]. Cr(VI) stimulates recombinant SFK activity in vitro, indicating a direct action on the kinase protein [11]. Downstream of the SFKs are the MAPKs (mitogen-activated protein kinases): JNK (c-Jun N-terminal kinase), ERK (extracellular-signal-regulated kinase) and p38. High levels of Cr(VI) (>10 μM) activate all three MAPKs individually [13–15] or simultaneously [15–17]. Lower concentrations (<10 μM) are more selective in activating JNK in pulmonary epithelial cells [11].
Selective activation of kinase cascades may explain Cr(VI) effects on transcription factors and gene transactivation. While multiple transcription factors have been demonstrated to respond to Cr(VI) exposure, a majority of studies have focused solely on activation of NF-κB (nuclear factor κB) [16–18] or NF-κB in combination with AP-1 (activator protein-1) [19–21]. High concentrations of Cr(VI), as well as other metals, directly disrupt interactions between NF-κB protein and its canonical cis-element in vitro [22]. However, in intact cells, Cr(VI) either has no effect or increases NF-κB binding to DNA [16,18,21]. More importantly, Cr(VI) has been shown to disrupt the transactivational activity of NF-κB and other factors, such as MTF-1 (metal transcription factor-1), by shifting patterns of co-activator interactions [23,24]. Additional transcription factors reported to respond to Cr(VI) by changing DNA binding include Sp-1 [19], p53 [13,24], HIF-1α (hypoxia inducible factor-1α) [14] and MTF-1 [23].
Despite a significant body of literature focused on Cr(VI)-stimulated effects on signalling enzymes and transcription factors, there have been few studies that address the proximal targets of chronic Cr(VI) exposure in the signalling cascades that activate transcription factors involved in pathogenic gene induction. To establish these links in airway epithelial cells, transcription factor arrays were conducted to identify Cr(VI)-dependent patterns of protein–DNA binding. In this process, a novel mechanism for prolonged activation of STAT3 (signal transducer and activator of transcription 3) following Cr(VI) exposure was identified. This translocation of STAT3 in response to Cr(VI) was linked to a non-traditional signalling pathway that requires the SFK, Lck, rather than the traditional phosphorylation of STAT3 by JAK (Janus kinase). The implication of the data linking these changes in cell signalling to increased expression of IL-6 (interleukin-6) is that chronic exposure to Cr(VI) may activate long-term phenotypic changes by promoting prolonged nuclear localization and DNA binding of transcription factors.
MATERIALS AND METHODS
Cell culture and treatment
Human bronchial epithelial cells (BEAS 2B; A.T.C.C., Rockville, MD, U.S.A.) were cultured on a matrix of 0.01 mg/ml fibronectin (Invitrogen, Carlsbad, CA, U.S.A.), 0.029 mg/ml Vitrogen 100 (COHESION, Palo Alto, CA, U.S.A.) and 0.01 mg/ml BSA (Invitrogen) in LHC-9 medium (Invitrogen). The cultures were maintained in serum-free LHC-9 medium at 37 °C and 5% CO2 as described previously [25]. Cultures of pHBE (primary human bronchiolar epithelial) cells isolated from discarded airways were cultured from excess pathological tissue following lung transplantation and organ donation under a protocol approved by the University of Pittsburgh Investigational Review Board. pHBE cells were cultured in modified LHC-9 medium on human placental collagen-coated Corning transwell permeable supports (0.4 μM pore size, 0.33 cm2; Fisher Scientific, Pittsburgh, PA, U.S.A.) at an air/liquid interface as described previously [26,27]. The cells were cultured for 4 weeks to allow for confluence and differentiation to a polarized epithelium. The medium was changed every 2 days and 24 h before experiments. BEAS 2B studies were conducted with 1 day post-confluent cells treated with 5 μM potassium dichromate [Cr(VI); Sigma–Aldrich, St. Louis, MO, U.S.A.] for 1, 4, 24, 48 and 72 h, unless stated otherwise. For the 72 h time point, the medium for both cell types was changed 24 h after adding Cr(VI). The positive control for STAT activation, 0.01 μg/ml human IL-6 (PeproTech, Rocky Hill, NJ, U.S.A.), was added 30 min prior to harvesting.
Cell viability assay
The viability of cells following Cr(VI) treatments was determined by measuring retention of calcein dye (Invitrogen). Briefly, cells were grown in 12-well plates and treated for 72 h, as described above, with 1, 2.5, 5, 10 or 20 μM Cr(VI). Cells were washed twice with PBS and incubated for 15 min at room temperature (20 °C) in PBS containing 1 μM calcein AM (acetoxymethyl ester). Relative fluorescence was measured (λex=495 nm and λem=530 nm) in a fluorescence microplate reader (Invitrogen).
Protein isolation
To terminate experiments, cells were rinsed twice and then scraped in stop buffer (10 mM Tris, pH 7.4, 10 mM EDTA, 5 mM EGTA, 0.1 M NaF, 0.2 M sucrose, 100 μM sodium orthovanadate, 5 mM sodium pyrophosphate and supplemented with protease inhibitors). After pelleting at 400 g for 10 min at 4 °C, the cells were lysed in Dignam's buffer A [10 mM Hepes, pH 7.9, 0.1 mM EDTA, 0.1 mM EGTA, 10 mM KCl and 1% NP40 (Nonidet P40), supplemented with 1 mM DTT (dithiothreitol), 100 μM sodium orthovanadate and protease inhibitors]. An aliquot of the lysate was removed and mixed with 4×SDS electrophoresis buffer for use as total protein lysate. The remainder was incubated for 10 min at 4 °C and then centrifuged at 13000 g for 2 min at 4 °C to pellet the nuclei. The supernatant was collected for cytosolic proteins, and the pellet was rinsed with buffer A and centrifuged again. The nuclear pellet was resuspended in Dignam's buffer C (20 mM Hepes, pH 7.9, 0.42 M NaCl, 1 mM EDTA, 0.1 mM EGTA, 1.0 mM DTT, 100 μM sodium orthovanadate and protease inhibitors), vortex-mixed and then shaken at 4 °C for 15 min. After a 5 min centrifugation at 13000 g and 4 °C, nuclear proteins in the supernatants were recovered and mixed with Dignam's buffer D [20 mM Hepes, pH 7.9, 20% (v/v) glycerol, 0.1 M KCl, 1 mM EDTA, 0.1 mM EGTA, 1% NP40, 1 mM DTT, 100 μM sodium orthovanadate and protease inhibitors]. Protein concentrations in each fraction were determined by the absorbance at 595 nm after addition of Coomassie Blue dye (Pierce Biotechnology, Rockford, IL, U.S.A.) using BSA as a reference standard. The purity of the nuclear and cytosolic fractions was demonstrated by Western-blot analysis for nuclear lamin A/C or cytosolic β-tubulin protein.
Panomics TranSignal™ protein/DNA arrays
TranSignal™ protein/DNA arrays (Panomics, Redwood, CA, U.S.A.) were used to examine the effects of Cr(VI) on the binding of nuclear proteins to 150 transcription factor cis-elements. Triplicate flasks of BEAS 2B cells were treated with or without Cr(VI) (5 μM) for 4 or 24 h. Pools of nuclear proteins from each treatment group were made to contain 3 μg/μl of protein. Then, 15 μg of each pooled sample was processed according to the manufacturer's directions [28]. Briefly, the nuclear proteins were incubated with biotin-labelled DNA-binding nucleotides to allow protein–DNA complexes to form. The complexes were first separated from the free probe and then DNA was separated from the protein–DNA complexes. The probes were then hybridized to the TranSignal™ array membranes I or II on which complementary oligonucleotides were spotted. The hybridized DNA was detected using streptavidin–horseradish peroxidase conjugates with chemiluminescence substrates and exposure to film. Densitometric analysis of scanned images was performed using Scion Image (Scion, Frederick, MD, U.S.A.). Data are reported as fold change over control after correction for background density.
Western blotting analysis
Changes in protein abundance or phosphorylation were measured by Western blot analysis, as described previously [11,21]. Proteins in cell lysates were separated by SDS/PAGE and transferred to PVDF membranes (Millipore, Bedford, MA, U.S.A.). After blocking, membranes were incubated overnight with primary antibodies at 4 °C. Antibodies recognizing total or phosphorylated forms of STAT1 (pTyr701; pY-STAT1), STAT3 (pTyr705; pY-STAT3), or STAT5 (pTyr694; pY-STAT3) were obtained from Cell Signaling Technology (Danvers, MA, U.S.A.). Other antibodies included STAT5 (BD Biosciences, San Diego, CA, U.S.A.), lamin A/C (Cell Signaling Technology), β-tubulin (CEDARLANE Laboratories, Burlington, NC, U.S.A.), Lck (Upstate Biotechnology, Lake Placid, NY, U.S.A.), JAK2 (Upstate Biotechnology) and β-actin (Sigma, St. Louis, MO, U.S.A.). Antibody binding was detected with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ, U.S.A.), followed by enhanced chemiluminescence (PerkinElmer, Boston, MA, U.S.A.) and exposure to film.
Immunohistofluorescence
Subcellular localization of STAT3 in response to Cr(VI) was imaged by immunofluorescence labelling and confocal microscopy. Cells were grown on coverslips (BEAS 2B cells) or in transwells (pHBE cells) and incubated with or without Cr(VI) for 72 h and then washed three times with cold PBS. Both the BEAS 2B and pHBE cells were fixed for 10 min in PBS containing 2% (w/v) paraformaldehyde and 0.2% glutaraldehyde. pHBE cell transwell membranes were then sliced out of the well holder and placed on glass coverslips. After permeabilizing with −20 °C methanol for 5 min, the cells were washed with TBS and permeabilized further with 0.2% Triton X-100 and 4% (w/v) BSA in PBS. The cells were washed and background fluorescence was quenched by incubating the cells with 0.1% NaBH4 in TBS for 5 min. The cells were washed in TBS and blocked for 1 h at room temperature in PBS containing 20% (v/v) goat serum, 1% BSA and 0.02% NaN3. The cells were then incubated overnight at 4 °C in a humidity chamber with primary antibodies diluted in TBS containing 1% BSA and 0.2% Triton X-100. After washing in TBS, the cells were incubated for 3 h with donkey anti-rabbit conjugated with FITC (Molecular Probes, Eugene, OR, U.S.A.) diluted in TBS containing 1% BSA. The cells were washed twice with TBS followed by a wash in TBS containing the nucleic acid stain TO-PRO3 (Molecular Probes). The coverslips were mounted on to slips with Fluoromount-G (SouthernBiotech, Birmingham, AL, U.S.A.) and confocal images were captured with an Olympus Fluoview BX61 fluorescence microscope in the University of Pittsburgh Center for Biological Imaging. Optical sections through the z-axis enabled separation and visualization of the nuclear and cytosolic compartments. The nuclear slice was classified as the equatorial slice in which the nucleus appeared the largest in most of the cells on the slip.
Transient transfections
BEAS 2B cells were transfected at 70–80% confluence according to the Lipofectamine™ Plus protocol (Invitrogen, Gaithersburg, MD, U.S.A.), as described previously [25]. For inhibitor studies, cells were co-transfected with 0.2 μg of EGFP (enhanced green fluorescence protein) plasmid (Clontech, Palo Alto, CA, U.S.A.), and 1.0 μg of the dominant-negative JAK2 or pRK5 expression plasmids, which were a gift from Dr David Levy (Department of Pathology, New York University School of Medicine, New York, NY, U.S.A.) via Dr Lu-Hai Wang (Department of Microbiology, Mount Sinai School of Medicine, New York, NY, U.S.A.). Cr(VI) exposure was initiated 24 h after beginning the transfection. Fluorescence of the EGFP (λex=490 nm, λem=509 nm and cutoff 495 nm) was measured in a fluorescence microplate reader (Molecular Devices, Sunnyvale, CA, U.S.A.) to normalize for transfection efficiency. For expression studies, cells were co-transfected with EGFP plasmid and either a STAT3-driven pTal-luc (luciferase) construct or an empty pTal-luc plasmid (Clontech). Cr(VI) exposures of 24 and 48 h were initiated 24 h after transfection. Luciferase activity was measured as described previously [25].
siRNA (small interfering RNA) oligonucleotides
An siRNA duplex specific for blocking human Lck was designed using the standard (AA-N19)™ template described by Dharmacon Research (Boulder, CO, U.S.A.). A BLAST search using the algorithm of Altschul et al. [29] of the corresponding cDNA for the 21 base siRNA sequence (sense 5′-AAGGUCCAAUCCGCCGGUU-3′) demonstrated significant homology only to human Lck mRNA. The Lck siRNA or a random 21 base siRNA (Upstate Biotechnology) was transfected into 70–80% confluent BEAS 2B cells using Mirus™ TransIT-TKO siRNA transfection reagent (Mirus Bio, Madison, WI, U.S.A.) in a minimal volume of LHC-9 for 4 h. Additional LHC-9 was added and the cells were incubated for 24 h before starting Cr(VI) exposure. Knockdown of Lck protein was demonstrated for at least 96 h. No significant cytotoxicity was observed when 200 nM of either selective or random siRNA was transfected into the cells.
RNA isolation and real-time RT (reverse transcriptase)–PCR
Total RNA was harvested and assayed for IL-6 and β-actin mRNA levels as described previously [30]. Specific primer pairs for IL-6 (forward 5′-GCCCAGCTATGAACTCCTTCTC-3′; reverse 5′-GACTTGTCATGTCCTGCAGCC-3′) and β-actin (forward 5′-GGGACCTGACCGACTACCTC; reverse 5′-GGGCGATGATCTTGATCTTC-3′) were used to amplify the specific cDNAs. Gene expression was quantified using standard curves for the respective cDNA products and changes in resulting IL-6 cDNA levels were normalized to changes in β-actin to determine the pg of normalized product per ml of reaction.
Statistics
Statistical analysis was performed on data from triplicate cultures from three separate experiments. One-way ANOVA was used to identify significant differences between treatment groups and controls. The degree of significance between groups was compared using Dunnett or Bonferroni post-hoc test. All statistics were performed using GraphPad Prism, version 4.0 (GraphPad Software, San Diego, CA, U.S.A.). Data are presented as means±S.D. or as a percentage of control.
RESULTS
Low doses of Cr(VI) did not affect cell viability
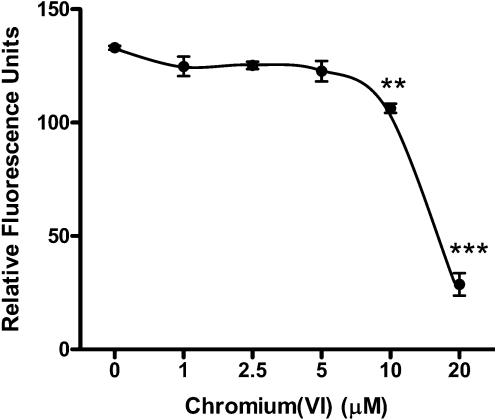
The ability of Cr(VI) to kill cells is both time- and dose-dependent, as well as cell type-specific [31,32]. The cytotoxicity of Cr(VI) in BEAS 2B cells under serum-free culture conditions was determined by evaluating plasma membrane integrity. Calcein AM is a membrane-permeant dye that undergoes intracellular cleavage and is then retained only by live cells. BEAS 2B cells were exposed to Cr(VI) (0–20 μM) for 72 h before testing membrane integrity by calcein retention (Figure 1). Levels of Cr(VI) below 10 μM were not toxic to the cells over this 72 h period. However, a 20 or 78% reduction in dye retention occurred when cells were exposed to 10 or 20 μM of Cr(VI) respectively, indicating significant toxicity. Because of these findings, 5 μM Cr(VI) was chosen as a non-cytotoxic exposure for subsequent studies.
Figure 1. Low doses of Cr(VI) do not affect cell viability.
BEAS 2B cells were exposed to various doses of Cr(VI) ranging from 1 to 20 μM for 72 h. Immediately following this exposure, the cells were incubated with 1 μM calcein AM and washed and retention of dye was measured by fluorescence. The results shown are from two independent experiments performed in triplicate. **P<0.01, ***P<0.001.
Differential effects of Cr(VI) on transcription factor binding
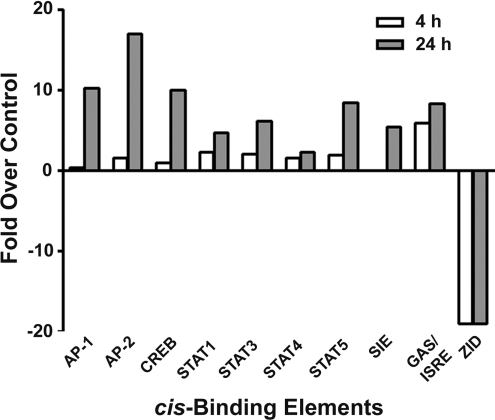
Nuclear extracts from control or Cr(VI)-exposed cells were analysed using Panomics TranSignal™ protein/DNA arrays to identify the simultaneous effects of Cr(VI) on the DNA binding of multiple transcription factors. After a 4 h exposure to Cr(VI), binding to 26 cis-elements increased, while binding to 12 decreased. More changes were seen following a 24 h exposure, with binding to 43 cis-elements increasing and 26 elements decreasing. Binding to the remaining elements was either undetectable or did not change greater than 2-fold. Figure 2 presents densitometric analysis of changes in DNA binding of selected transcription factors in BEAS 2B cells exposed to 5 μM Cr(VI) for either 4 or 24 h relative to control binding. In this subset, binding did not increase at 4 h for most of the transcription factors, with the exception of GAS [IFN-γ (interferon-γ)-activated site]/ISRE (IFN-stimulated response element), the combination of the GAS with the ISRE. However, after 24 h, Cr(VI) increased protein binding to AP-1, AP-2, CREB (cAMP-response-element-binding protein), STAT1, STAT3, STAT4, STAT5 and SIE (sis-inducible element) cis-elements by 2–17-fold over control binding. These changes in AP-1 and CREB are consistent with previously reported effects of both low and high doses of Cr(VI) [19–21]. However, this is the first study to suggest that Cr(VI) activates binding to the STAT1, STAT3 and STAT4 cis-elements, all of which appear to modulate inflammatory processes in the lung defence mechanism [33,34]. Increased protein binding at GAS/ISRE and SIE elements was also consistent with increases in STAT1 and STAT3 binding [35,36]. In comparison, Cr(VI) decreased protein binding to the ZID (zinc finger protein with interaction domain) cis-element after both 4 and 24 h Cr(VI) exposures. It is notable that there were no changes in NF-κB binding (results not shown), which was consistent with our previous studies in pulmonary cells [21].
Figure 2. Cr(VI) affects a variety of transcription factors.
Triplicate flasks of BEAS 2B cells were exposed to 5 μM Cr(VI) for 4 or 24 h, after which nuclear proteins were isolated and quantified. Each treatment group was pooled and samples were analysed for protein binding to DNA cis-elements by the protein/DNA arrays I and II. Densitometric analysis of the blot was performed and results are presented as fold change over control.
Differential activation of STAT proteins in the nucleus
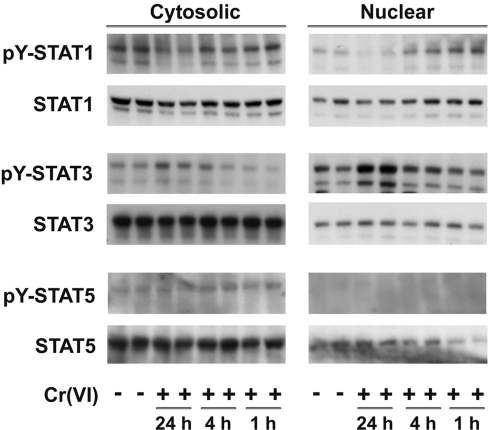
Western blot analysis with antibodies that recognized activated phosphorylated forms of STAT1, STAT3 and STAT5 was used to confirm that Cr(VI) stimulated activation and nuclear translocation of the STAT proteins. There were no changes in the total cellular content of any of the STAT proteins following Cr(VI) exposure (Figures 3 and 4). The data in Figure 3 demonstrate that Cr(VI) did not affect tyrosine-phosphorylation of STAT5 nor its nuclear translocation. In contrast, nuclear levels of both pY-STAT1 and pY-STAT3 increased in response to Cr(VI). However, these increases followed distinct time courses. STAT1 was phosphorylated and translocated to the nucleus within an hour of exposure and nuclear levels of pY-STAT1 remained elevated for at least 4 h. At 24 h, STAT1 had returned to the basal phosphorylation state and was predominantly located in the cytosol. Phosphorylation and translocation of STAT3 lagged behind STAT1 and nuclear levels of pY-STAT3 remained elevated at 24 h (Figure 3). The latent increase in STAT3 was consistent with the lag in SIE protein binding seen in Figure 2 [35].
Figure 3. Cr(VI) induced the phosphorylation of STAT3.
Following exposure of BEAS 2B cells to 5 μM Cr(VI) for 1, 4 or 24 h, cytosolic and nuclear protein fractions were isolated and quantified. Then, 15 μg of nuclear protein and 30 μg of cytosolic protein were used for Western blot analysis with antibodies against total and phosphorylated STAT1, STAT3 and STAT5 proteins. The duplicate samples were from separate cell cultures and are representative of duplicates from three independent experiments.
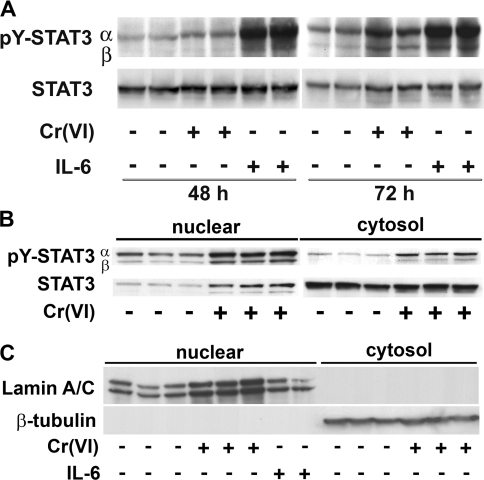
Figure 4. Subchronic exposure to Cr(VI) increases STAT3 phosphorylation.
(A) Total cell lysates were isolated from BEAS 2B cells exposed either to 5 μM Cr(VI) for 48 or 72 h or to 0.01 μg/ml of IL-6 for 30 min prior to the end of the experiment. Antibodies to pY-STAT3 and STAT3 were used in the Western blotting. (B) Cytosolic and nuclear protein fractions from BEAS 2B cells exposed for 72 h to 5 μM Cr(VI). Western-blot analysis was performed on 15 μg of nuclear protein or 30 μg of cytosolic protein. (C) The purity of the nuclear and cytosolic fractions was demonstrated by probing for the presence or absence of the nuclear protein, lamin A/C, or cytosolic β-tubulin. The experiments were performed in duplicate cell cultures and are representative of three independent experiments.
Cr(VI) stimulated prolonged STAT activation
The sustained activation of both STAT3α and STAT3β was examined further by extending the time course of Cr(VI) exposure. The data in Figure 4(A) demonstrated time-dependent increases in Cr(VI)-induced phosphorylation of both STAT3 isoforms in the total protein extract, after both 48 and 72 h of Cr(VI) exposure. In several sets of experiments, including the results shown, the peak increase in Cr(VI)-activated STAT3 occurred between 48 and 72 h. In results not shown, nuclear STAT3 levels remained elevated even after 120 h of exposure. It is also important to note that the Cr(VI) was only added at the initiation of exposure. Adding additional Cr(VI) at 24 h when refreshing the medium had no additional effect on STAT3 activation at 72 h (results not shown).
Cr(VI) stimulated prolonged STAT3 nuclear localization
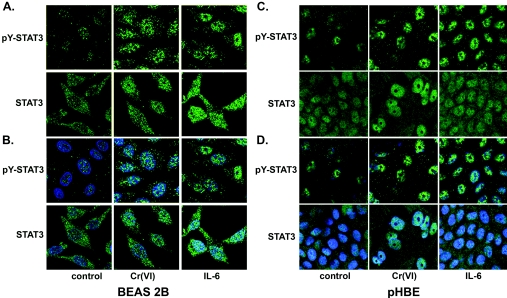
Immunofluorescent confocal microscopy with antibodies that specifically bound pY-STAT3 or total STAT3 was used to confirm that Cr(VI) stimulated phosphorylation and nuclear translocation of STAT3 in both pHBE and BEAS 2B cells. As suggested by the Western-blot analysis in Figure 4(B), both Cr(VI) and IL-6 increase phosphorylation of STAT3 and also increase the amount of total and phosphorylated STAT3 in the nucleus (Figure 5). The responses of pHBE cells cultured in an air/liquid interface and the BEAS 2B cells in conventional cultures to Cr(VI) were essentially the same. The primary STAT3 antibody used for this analysis was not isoform-specific.
Figure 5. Cr(VI) induces phosphorylation of STAT3 in the nucleus.
Immunofluorescence confocal microscopy was used to confirm that Cr(VI) increases nuclear levels of phosphorylated STAT3. BEAS 2B cells grown on coverslips or pHBE cells grown on transwell membranes in air/liquid interface cultures were exposed to 5 μM Cr(VI) (72 h) or to 0.01 μg/ml of IL-6 (30 min). Following fixation and permeabilization, cells were immunostained with antibodies against either pY-STAT3 or total STAT3 (green), as described in the Materials and methods section. (B, D) These Figures show (A) and (C) merged with images of the nuclei stained with TO-PRO3 (blue). These results were from three independent experiments performed in duplicate.
Cr(VI)-stimulated STAT3 transactivation was consistent with nuclear localization
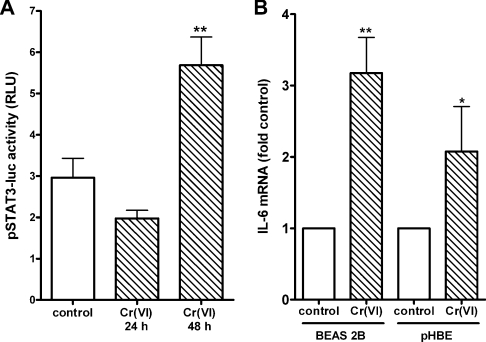
To confirm that nuclear increases in STAT3 were functional, BEAS-2B cells were transfected with a plasmid reporter construct that expresses luciferase in response to STAT3 transactivation. In keeping with the slow progressive increases in STAT3 tyrosine phosphorylation and nuclear translocation, luciferase expression did not change before 24 h and was highly significant by 48 h (Figure 6A). IL-6 mRNA levels in control and Cr(VI)-exposed BEAS 2B and pHBE cells were compared to determine whether Cr(VI) induced an endogenous gene in the airway cells. Three days after adding Cr(VI), IL-6 levels in both cell types were increased by 2–3-fold relative to control (Figure 6B). There was no difference in the degree of responsiveness to Cr(VI) between the cell types. These results demonstrate that Cr(VI) induced pathogenic STAT3 transactivation in both the immortalized cell line and in primary isolates of human airway epithelium.
Figure 6. Cr(VI) induces STAT3-dependent transactivation and IL-6 mRNA levels.
(A) BEAS 2B cells were transfected with pSTAT3-luc. After 24 h, cells were treated with 5μM Cr(VI) for either 24 or 48 h. Luciferase activity in cell lysates was then measured and results are reported as means±S.D. in relative light units (RLU) for three separate experiments. **Significance at P<0.01. (B) BEAS 2B grown under normal culture conditions and pHBE cells cultured in transwells under an air/liquid interface were incubated in the presence or absence of 5 μM Cr(VI) for 72 h. Total RNA was then collected and assayed for IL-6 or β-actin mRNA levels using quantitative real-time RT–PCR. The results are reported as means±S.D. of fold change from respective controls to account for differences in basal levels in IL-6 mRNA between the two cell types and culture conditions. Statistical differences are designated by *P<0.05 and **P<0.01 (n=3 for BEAS 2B cells and n=4 for pHBE cells).
Lck is required for Cr(VI)-stimulated STAT3 phosphorylation and increased IL-6 mRNA levels
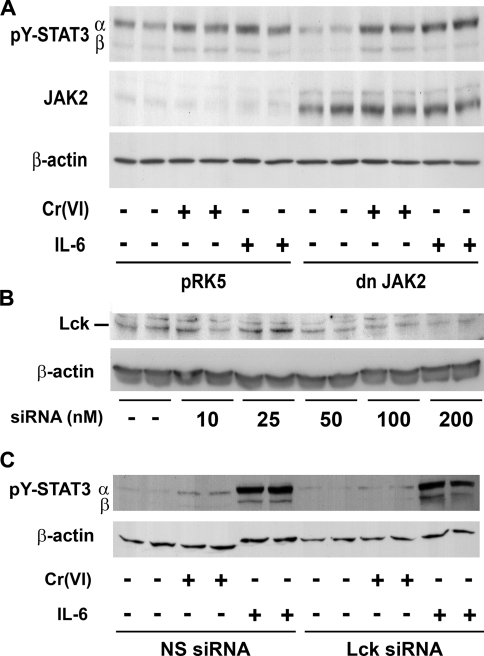
Tyrosine phosphorylation of STAT3 in response to cytokines is catalysed by either JAKs or SFKs. Preliminary experiments demonstrated that only JAK2 is expressed in significant amounts in BEAS 2B cells, relative to JAK1 (results not shown). Transient transfection with dominant-negative JAK2 partially blocked IL-6-stimulated STAT3β phosphorylation, but did not prevent Cr(VI)-stimulated STAT3α or STAT3β phosphorylation (Figure 7A). Cr(VI) stimulates Lck in human lung adenocarcinoma cells (A549) [11], and Lck-has been shown to phosphorylate STAT3 in other cell types [37]. Therefore the role of Lck in mediating the response to Cr(VI) was examined by preventing Lck protein expression with a specific siRNA. As shown in Figure 7(B), treatment with 200 nM of Lck-specific siRNA reduced Lck protein expression. Pretreatment of cells with specific siRNA, but not random siRNA, decreased Cr(VI) phosphorylation of STAT3 (Figure 7C). Neither Lck-specific nor randomized siRNA reduced IL-6-stimulated STAT3 phosphorylation, confirming that Cr(VI) activates STAT3 through a non-classical signalling cascade. In addition to inhibiting Cr(VI)-stimulated STAT3 phosphorylation, knockdown of Lck prevented Cr(VI)-induced IL-6 mRNA, relative to induction in the presence of random siRNA. In cells transfected with 200 nM random siRNA, a 72 h exposure to Cr(VI) increased IL-6 mRNA levels by 3.5±0.6-fold over levels in non-exposed transfected cells (means±S.E.M., difference from control P<0.01, n=6). In contrast, there was no significant Cr(VI)-stimulated increase in IL-6 mRNA in cells transfected with 200 nM Lck-specific siRNA (1.4±0.1-fold stimulation above transfected controls, n=6). Thus Lck appears to be proximal in the Cr(VI)-stimulated signalling cascade and required for STAT3 gene transactivation in response to Cr(VI).
Figure 7. Lck is required for full Cr(VI)-stimulated STAT3 phosphorylation.
(A) BEAS 2B cells at 70–80% confluence were transiently transfected with plasmids containing either pRK5 or dominant-negative (dn) JAK2. After 24 h, cells were exposed to Cr(VI) (5 μM) for 72 h or to 0.01 μg/ml of IL-6 for 30 min prior to the end of the experiment. Western-blot analysis was performed on 30 μg of total cell lysate. (B) BEAS 2B cells were transiently transfected with various amounts of siRNA specific to human Lck. Total cell lysates were obtained after 96 h and Lck and β-actin protein was measured by Western-blot analysis. (C) BEAS 2B cells were transiently transfected with 200 nM siRNA specific to human Lck or non-specific, random siRNA (NS). After 72 h, cells were exposed to Cr(VI) (5 μM) for an additional 72 h. IL-6 (0.01 μg/ml) was added for 30 min prior to the end of the experiment. Nuclear proteins were extracted at the end of the experiments and probed for pY-STAT3 and β-actin by Western-blot analysis. The results are representative of three separate experiments.
DISCUSSION
The cellular and molecular effects of Cr(VI) that lead to respiratory diseases are still in question. In addition, few studies of Cr(VI)-stimulated effects on cell signalling have examined longer-term or sustained changes that might lead to pathogenic phenotypic change. The data in the present study indicate that non-cytotoxic exposure of airway epithelial cells to Cr(VI) causes delayed and prolonged activation of STAT3, a transcription factor that plays important roles in lung responses to injury. In addition to this novel observation, the SFK family member Lck was identified as an upstream target of Cr(VI), as well as the mediator of the prolonged STAT3 nuclear translocation and transactivation of IL-6. The significance of the observations are increased by confirmation of Cr(VI)-stimulated STAT3 nuclear translocation (Figure 5) and increased IL-6 mRNA levels (Figure 6) in primary airway epithelial cells. Thus Cr(VI) appears to have a uniform effect on airway epithelial cells, since it stimulates Lck tyrosine kinase activity in alveolar cells [11] and Lck-dependent signalling in epithelial cells from the upper airways (Figure 7 and effect on IL-6 mRNA levels).
Multiple transcription factors have been shown to respond to a range of Cr(VI) exposures. The analysis represented in Figure 2 expands appreciation of the extent and context of Cr(VI)-stimulated transcription factor changes. The protein/DNA binding arrays demonstrated that a number of transcription factors decreased in nuclear abundance, in addition to those that increased. This is consistent with our previous demonstration that 10 μM Cr(VI) altered the transcription of 44 genes in BEAS 2B cells [38]. In keeping with the demonstration of both positive and negative changes in nuclear transcription factor DNA binding, 90% of the altered genes showed reduced expression, while 10% showed positive changes [38]. In contrast, high levels of Cr(VI) (300 μM) increased the expression of 150 genes and reduced the expression of 70 genes in A549 cells [39].
Increased protein binding to STAT cis-elements is an example of how Cr(VI) might differentially affect gene expression. The STAT family of latent transcription factors is aptly named after their dual role in cell signalling [40]. The increased protein binding to the GAS/ISRE, as well as to STAT1, STAT3 and STAT4 cis-elements (Figure 2), may be of particular relevance to injury responses in the lung. Both STAT1 and STAT3 can bind to the GAS/ISRE cis-element [36], suggesting that Cr(VI) may activate multiple STAT proteins to either induce or suppress specific pulmonary genes. Both STAT1 and STAT4 appear to play a role in lung defence mechanisms [33,34]. In addition, activation of STAT1 and STAT3 has been observed in a variety of pulmonary diseases [41–43]. For instance, STAT1 is constitutively activated in bronchial epithelial cells from patients with stable asthma [41]. In animal models, STAT3 is activated in acute lung injury induced by intrapulmonary deposition of IgG immune complexes, lipopolysaccharide administration and acute hypoxia [42,43]. STAT3 is the only STAT gene that is early embryonic lethal in mouse gene knockout studies [44]. STAT3 induction may be protective in the lungs, since conditional STAT3 knockout in respiratory epithelial cells results in increased injury from hyperoxia [45]. The data in the present study indicated that Cr(VI) induced IL-6 expression through activation of STAT3 (Figures 6 and 7). Even though STAT3 transduces IL-6 expression and this cytokine has been implicated in lung disease progression [43,46], this induction may also be an attempt to protect the lung [45]. For instance, the IL-6/STAT3 pathway is an important component of the innate immune response of the lung [46] and has been demonstrated to provide protection in liver injury [47].
Cytokine activation of STAT proteins is the most well-characterized signalling pathway for increasing their nuclear localization and transactivation of genes [48]. The pathway is activated when cytokines bind and stimulate their cognate receptors, as well as associated JAKs or SFKs [37,48]. All members of the STAT family contain a critical tyrosine near residue 700 that is phosphorylated by the activated JAKs or SFKs. Phosphorylation of this tyrosine activates the STAT monomers to dimerize via their SH2 (Src homology 2) domains and then translocate to the nucleus [49]. In the nucleus, STAT dimer and tetramers bind to specific DNA response elements to induce or repress gene transcription. In the present studies, Cr(VI) stimulated early, transient phosphorylation and nuclear translocation of STAT1 (Figure 3). In contrast, STAT3 was phosphorylated with a latent time course and pY-STAT3 remained in the nucleus for more that 72 h (Figures 4 and 5). JAK1/2 are the most well-described kinases that initiate STAT3 activation in response to cytokines such as IL-6 [48]. However, we found little JAK1 in the BEAS 2B cells (results not shown) and expression of dominant-negative JAK2 had no effect on Cr(VI)-stimulated STAT3 phosphorylation despite inhibiting IL-6-stimulated STAT3β phosphorylation.
The SFKs, especially c-Src and Lck, are major alternative kinases to the JAKs for activating STAT3 dimers [37,48]. Overexpression of active Lck in mouse T-cells causes constitutive activation of STAT3, STAT5, JAK1 and JAK2, but not STAT1 [37]. Since Cr(VI) activated Lck, but not Src in A549 airway cells [11] and lymphocytes [12], it was reasoned that Lck may be the alternative kinase mediating Cr(VI)-stimulated STAT3 activation. The results in Figure 7 confirm that eliminating Lck blocks Cr(VI)-stimulated phosphorylation of STAT3. Cr(VI) induction of IL-6 mRNA also required Lck protein, demonstrating that the Lck-mediated pathway functionally signals to induce an important inflammatory mediator. That Lck siRNA failed to block IL-6-stimulated STAT3 activation (Figure 7) indicated that Cr(VI) did not induce the cytokine first to stimulate a latent activation of STAT3. These novel observations suggest that activation of Lck was the initial upstream event in Cr(VI)-stimulated cell signalling cascades leading to prolonged STAT3 activation and transactivation of IL-6.
Cr(VI)-induced STAT3 transactivation of IL-6 in either primary airway cells or the BEAS 2B cell line (Figure 6) contrasts with the general belief that Cr(VI) only inhibits inducible genes [19,50,51]. This belief was based on earlier studies that mostly examined Cr(VI) effects on early gene inducibility. Transcriptional regulation by STAT3 can be stimulatory or inhibitory depending on the actions of the α and β splice variants [52]. STAT3α is essential in transmitting IL-6 signalling [52] and is the likely mediator of Cr(VI) stimulation of the IL-6 promoter (Figure 6). However, Cr(VI) stimulates equivalent activation of STAT3β, whose role is less clear. This isoform is essential for development, since this is the only isoform that can rescue the embryonic lethality of a STAT3 knockout [52]. Severgnini et al. [43] demonstrated that whole lung lysates of mice administered lipopolysaccharide had elevated levels of pY-STAT3β, which occurred earlier and persisted later than the activated STAT3α [43]. Mice deficient in STAT3β also had increased sensitivity to inflammation and STAT3β may assist in anti-inflammatory processes [43,52]. Thus it is possible that STAT3β either contributes to or modulates STAT3-dependent Cr(VI) induction of IL-6. Determining the relative roles of the two STAT3 isoforms in differential regulation of Cr(VI)-induced or -inhibited gene expression is beyond the scope of the present study. However, the observation that activation of both isoforms was prolonged in response to Cr(VI) exposure suggested that they may play a role in chronic phenotypic changes in the lung.
In summary, these results present a novel pathway through which a chronic exposure to low, non-toxic levels of Cr(VI) selectively activates important members of the STAT family in the lung. The results suggest that this pathway is initiated by Cr(VI) stimulation of Lck, causing increased phosphorylation and prolonged nuclear localization of STAT3. The full implications of this prolonged STAT3 activation in Cr(VI)-induced phenotypic change remain to be resolved. These studies, however, demonstrate that this Cr(VI)-stimulated pathway induces IL-6, an important inflammatory mediator in the lung. Thus this signalling pathway may play a role in regulating chronic pathological changes in the lung that result from environmental exposure to Cr(VI).
Acknowledgments
These studies were supported by grant ES10638 from the National Institute of Environmental Health Sciences. We thank Joseph Latoche (University of Pittsburgh), Dr Joseph Pilewski (University of Pittsburgh) and the Lung Transplant and Cystic Fibrosis programmes for providing primary human airway epithelial cells. Cell imaging support was provided by the University of Pittsburgh Center for Biological Imaging through the Center for the Environmental Basis of Human Disease.
References
- 1.Stearns D. M. Is chromium a trace essential metal? Biofactors. 2000;11:149–162. doi: 10.1002/biof.5520110301. [DOI] [PubMed] [Google Scholar]
- 2.Vincent J. B. Mechanisms of chromium action: low-molecular-weight chromium-binding substance. J. Am. Coll. Nutr. 1999;18:6–12. doi: 10.1080/07315724.1999.10718821. [DOI] [PubMed] [Google Scholar]
- 3.Antonini J. M., Taylor M. D., Zimmer A. T., Roberts J. R. Pulmonary responses to welding fumes: role of metal constituents. J. Toxicol. Environ. Health A. 2004;67:233–249. doi: 10.1080/15287390490266909. [DOI] [PubMed] [Google Scholar]
- 4.Bright P., Burge P. S., O'Hickey S. P., Gannon P. F. G., Robertson A. S., Boran A. Occupational asthma due to chrome and nickel electroplating. Thorax. 1997;52:28–32. doi: 10.1136/thx.52.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halasova E., Baska T., Kukura F., Mazurova D., Bukovska E., Dobrota D., Poliacek I., Halasa M. Lung cancer in relation to occupational and environmental chromium exposure and smoking. Neoplasma. 2005;52:287–291. [PubMed] [Google Scholar]
- 6.Park R. M., Bena J. F., Stayner L. T., Smith R. J., Gibb H. J., Lees P. S. Hexavalent chromium and lung cancer in the chromate industry: a quantitative risk assessment. Risk Anal. 2004;24:1099–1108. doi: 10.1111/j.0272-4332.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 7.Alcedo J. A., Wetterhahn K. E. Chromium toxicity and carcinogenesis. Int. Rev. Exp. Pathol. 1990;31:85–108. doi: 10.1016/b978-0-12-364931-7.50008-2. [DOI] [PubMed] [Google Scholar]
- 8.De Flora S., Serra D., Camoirano A., Zanacchi P. Metabolic reduction of chromium, as related to its carcinogenic properties. Biol. Trace Elem. Res. 1989;21:179–187. doi: 10.1007/BF02917250. [DOI] [PubMed] [Google Scholar]
- 9.Wetterhahn K. E., Hamilton J. W., Aiyar J., Borges K. M., Floyd R. Mechanism of chromium(VI) carcinogenesis. Reactive intermediates and effect on gene expression. Biol. Trace Elem. Res. 1989;21:405–411. doi: 10.1007/BF02917282. [DOI] [PubMed] [Google Scholar]
- 10.De Flora S. Threshold mechanisms and site specificity in chromium(VI) carcinogenesis. Carcinogenesis. 2000;21:533–541. doi: 10.1093/carcin/21.4.533. [DOI] [PubMed] [Google Scholar]
- 11.O'Hara K. A., Klei L. R., Barchowsky A. Selective activation of Src family kinases and JNK by low levels of chromium(VI) Toxicol. Appl. Pharmacol. 2003;190:214–223. doi: 10.1016/s0041-008x(03)00188-1. [DOI] [PubMed] [Google Scholar]
- 12.Vasant C., Rajaram R., Ramasami T. Apoptosis of lymphocytes induced by chromium(VI/V) is through ROS-mediated activation of Src-family kinases and caspase-3. Free Radical Biol. Med. 2003;35:1082–1100. doi: 10.1016/s0891-5849(03)00471-4. [DOI] [PubMed] [Google Scholar]
- 13.Wang S., Shi X. Mechanisms of Cr(VI)-induced p53 activation: the role of phosphorylation, mdm2 and ERK. Carcinogenesis. 2001;22:757–762. doi: 10.1093/carcin/22.5.757. [DOI] [PubMed] [Google Scholar]
- 14.Gao N., Jiang B. H., Leonard S. S., Corum L., Zhang Z., Roberts J. R., Antonini J., Zheng J. Z., Flynn D. C., Castranova V., Shi X. p38 Signaling-mediated hypoxia-inducible factor 1α and vascular endothelial growth factor induction by Cr(VI) in DU145 human prostate carcinoma cells. J. Biol. Chem. 2002;277:45041–45048. doi: 10.1074/jbc.M202775200. [DOI] [PubMed] [Google Scholar]
- 15.Kim G., Yurkow E. J. Chromium induces a persistent activation of mitogen-activated protein kinases by a redox-sensitive mechanism in H4 rat hepatoma cells. Cancer Res. 1996;56:2045–2051. [PubMed] [Google Scholar]
- 16.Kim Y. D., An S. C., Oyama T., Kawamoto T., Kim H. Oxidative stress, hogg1 expression and NF-κB activity in cells exposed to low level chromium. J. Occup. Health. 2003;45:271–277. doi: 10.1539/joh.45.271. [DOI] [PubMed] [Google Scholar]
- 17.Ye J., Zhang X., Young H. A., Mao Y., Shi X. Chromium(VI)-induced nuclear factor-κB activation in intact cells via free radical reactions. Carcinogenesis. 1995;16:2401–2405. doi: 10.1093/carcin/16.10.2401. [DOI] [PubMed] [Google Scholar]
- 18.Chen F., Bower J., Leonard S. S., Ding M., Lu Y., Rojanasakul Y., Kung H. F., Vallyathan V., Castranova V., Shi X. Protective roles of NF-κB for chromium(VI)-induced cytotoxicity is revealed by expression of IκB kinase-β mutant. J. Biol. Chem. 2002;277:3342–3349. doi: 10.1074/jbc.M101089200. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton J. W., Kaltreider R. C., Bajenova O. V., Ihnat M. A., McCaffrey J., Turpie B. W., Rowell E. E., Oh J., Nemeth M. J., Pesce C. A., Lariviere J. P. Molecular basis for effects of carcinogenic heavy metals on inducible gene expression. Environ. Health Perspect. 1998;106(Suppl. 4):1005–1015. doi: 10.1289/ehp.98106s41005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaltreider R. C., Pesce C. A., Ihnat M. A., Lariviere J. P., Hamilton J. W. Differential effects of arsenic(III) and chromium(VI) on nuclear transcription factor binding. Mol. Carcinog. 1999;25:219–229. [PubMed] [Google Scholar]
- 21.Shumilla J. A., Broderick R. J., Wang Y., Barchowsky A. Chromium(VI) inhibits the transcriptinal activity of NF-κB by decreasing the interaction of p65 with CBP. J. Biol. Chem. 1999;274:36207–36212. doi: 10.1074/jbc.274.51.36207. [DOI] [PubMed] [Google Scholar]
- 22.Shumilla J. A., Wetterhahn K. E., Barchowsky A. Inhibition of NF-κB binding to DNA by chromium, cadmium, mercury, zinc, and arsenite in vitro: evidence of a thiol mechanism. Arch. Biochem. Biophys. 1998;349:356–362. doi: 10.1006/abbi.1997.0470. [DOI] [PubMed] [Google Scholar]
- 23.Majumder S., Ghoshal K., Summers D., Bai S., Datta J., Jacob S. T. Chromium(VI) down-regulates heavy metal-induced metallothionein gene transcription by modifying transactivation potential of the key transcription factor, metal-responsive transcription factor 1. J. Biol. Chem. 2003;278:26216–26226. doi: 10.1074/jbc.M302887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye J., Wang S., Leonard S. S., Sun Y., Butterworth L., Antonini J., Ding M., Rojanasakul Y., Vallyathan V., Castranova V., Shi X. Role of reactive oxygen species and p53 in chromium(VI)-induced apoptosis. J. Biol. Chem. 1999;274:34974–34980. doi: 10.1074/jbc.274.49.34974. [DOI] [PubMed] [Google Scholar]
- 25.Barchowsky A., Soucy N. V., O'Hara K. A., Hwa J., Noreault T. L., Andrew A. S. A novel pathway for nickel-induced interleukin-8 expression. J. Biol. Chem. 2002;277:24225–24231. doi: 10.1074/jbc.M202941200. [DOI] [PubMed] [Google Scholar]
- 26.Devor D. C., Bridges R. J., Pilewski J. M. Pharmacological modulation of ion transport across wild-type and ΔF508 CFTR-expressing human bronchial epithelia. Am. J. Physiol. Cell Physiol. 2000;279:C461–C479. doi: 10.1152/ajpcell.2000.279.2.C461. [DOI] [PubMed] [Google Scholar]
- 27.Myerburg M. M., Butterworth M. B., McKenna E. E., Peters K. W., Frizzell R. A., Kleyman T. R., Pilewski J. M. Airway surface liquid volume regulates ENaC by altering the serine protease–protease inhibitor balance: a mechanism for sodium hypersabsorption in cystic fibrosis. J. Biol. Chem. 2006;281:27942–27949. doi: 10.1074/jbc.M606449200. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X., Norman M., Roth L., Li X. Protein–DNA array-based identification of transcription factor activities regulated by interaction with the glucocorticoid receptor. J. Biol. Chem. 2004;279:38480–38485. doi: 10.1074/jbc.M403948200. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao F., Barchowsky A., Nemec A. A., Fabisiak J. P. Microbial stimulation by mycoplasma fermentans synergistically amplifies IL-6 release by human lung fibroblasts in response to residual oil fly ash (ROFA) and nickel. Toxicol. Sci. 2004;81:467–479. doi: 10.1093/toxsci/kfh205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds M., Peterson E., Quievryn G., Zhitkovich A. Human nucleotide excision repair efficiently removes chromium–DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien T. J., Ceryak S., Patierno S. R. Complexities of chromium carcinogenesis: role of cellular response, repair and recovery mechanisms. Mutat. Res. 2003;533:3–36. doi: 10.1016/j.mrfmmm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Laskin D. L., Fakhrzadeh L., Heck D. E., Gerecke D., Laskin J. D. Upregulation of phosphoinositide 3-kinase and protein kinase B in alveolar macrophages following ozone inhalation. Role of NF-κB and STAT-1 in ozone-induced nitric oxide production and toxicity. Mol. Cell. Biochem. 2002;234–235:91–98. [PubMed] [Google Scholar]
- 34.Di Stefano A., Caramori G., Capelli A., Gnemmi I., Ricciardolo F. L., Oates T., Donner C. F., Chung K. F., Barnes P. J., Adcock I. M. STAT4 activation in smokers and patients with chronic obstructive pulmonary disease. Eur. Respir. J. 2004;24:78–85. doi: 10.1183/09031936.04.00080303. [DOI] [PubMed] [Google Scholar]
- 35.Becker S., Groner B., Muller C. W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 36.Kim O. S., Park E. J., Joe E. H., Jou I. JAK-STAT signaling mediates gangliosides-induced inflammatory responses in brain microglial cells. J. Biol. Chem. 2002;277:40594–40601. doi: 10.1074/jbc.M203885200. [DOI] [PubMed] [Google Scholar]
- 37.Yu C. L., Jove R., Burakoff S. J. Constitutive activation of the Janus kinase-STAT pathway in T lymphoma overexpressing the Lck protein tyrosine kinase. J. Immunol. 1997;159:5206–5210. [PubMed] [Google Scholar]
- 38.Andrew A. S., Warren A. J., Barchowsky A., Temple K. A., Klei L., Soucy N. V., O'Hara K. A., Hamilton J. W. Genomic and proteomic profiling of responses to toxic metals in human lung cells. Environ. Health Perspect. 2003;111:825–835. doi: 10.1289/ehp.111-1241504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye J., Shi X. Gene expression profile in response to chromium-induced cell stress in A549 cells. Mol. Cell. Biochem. 2001;222:189–197. [PubMed] [Google Scholar]
- 40.Levy D. E., Darnell J. E., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 41.Sampath D., Castro M., Look D. C., Holtzman M. J. Constitutive activation of an epithelial signal transducer and activator of transcription (STAT) pathway in asthma. J. Clin. Invest. 1999;103:1353–1361. doi: 10.1172/JCI6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao H., Guo R. F., Speyer C. L., Reuben J., Neff T. A., Hoesel L. M., Riedemann N. C., McClintock S. D., Sarma J. V., Van Rooijen N., et al. Stat3 activation in acute lung injury. J. Immunol. 2004;172:7703–7712. doi: 10.4049/jimmunol.172.12.7703. [DOI] [PubMed] [Google Scholar]
- 43.Severgnini M., Takahashi S., Rozo L. M., Homer R. J., Kuhn C., Jhung J. W., Perides G., Steer M., Hassoun P. M., Fanburg B. L., et al. Activation of the STAT pathway in acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;286:L1282–L1292. doi: 10.1152/ajplung.00349.2003. [DOI] [PubMed] [Google Scholar]
- 44.Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hokuto I., Ikegami M., Yoshida M., Takeda K., Akira S., Perl A. K., Hull W. M., Wert S. E., Whitsett J. A. Stat-3 is required for pulmonary homeostasis during hyperoxia. J. Clin. Invest. 2004;113:28–37. doi: 10.1172/JCI200419491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doganci A., Sauer K., Karwot R., Finotto S. Pathological role of IL-6 in the experimental allergic bronchial asthma in mice. Clin. Rev. Allergy Immunol. 2005;28:257–270. doi: 10.1385/CRIAI:28:3:257. [DOI] [PubMed] [Google Scholar]
- 47.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J. Clin. Invest. 2003;112:978–980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rane S. G., Reddy E. P. Janus kinases: components of multiple signaling pathways. Oncogene. 2000;19:5662–5679. doi: 10.1038/sj.onc.1203925. [DOI] [PubMed] [Google Scholar]
- 49.Schindler C., Shuai K., Prezioso V. R., Darnell J. E., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 50.Hamilton J. W., Wetterhahn K. E. Differential effects of chromium(VI) on constitutive and inducible gene expression in chick embryo liver in vivo and correlation with chromium(VI)-induced DNA damage. Mol. Carcinog. 1989;2:274–286. doi: 10.1002/mc.2940020508. [DOI] [PubMed] [Google Scholar]
- 51.Wei Y. D., Tepperman K., Huang M. Y., Sartor M. A., Puga A. Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin. J. Biol. Chem. 2004;279:4110–4119. doi: 10.1074/jbc.M310800200. [DOI] [PubMed] [Google Scholar]
- 52.Maritano D., Sugrue M. L., Tininini S., Dewilde S., Strobl B., Fu X., Murray-Tait V., Chiarle R., Poli V. The STAT3 isoforms α and β have unique and specific functions. Nat. Immunol. 2004;5:401–409. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]