Abstract
The stress response gene IEX-1 (immediate early gene-X-1) is involved in the regulation of cell growth and cellular viability. To some extent, these effects include an interference with the proteasomal turnover of certain regulatory proteins. Here, we show that IEX-1 directly attenuates the activity and formation of the 26 S proteasome in HEK-293 cells (human embryonic kidney cells). We further demonstrate that IEX-1 reduces the overall expression levels of certain protein components of the 19 S proteasomal subunit such as S5a/Rpn10 and S1/Rpn2, whereas the expression of other proteasomal proteins was less or not affected. In contrast with direct apoptotic stimuli, such as the anti-cancer drug etoposide, leading to caspase-dependent degradation of S1 and S5a, the effect of IEX-1 is independent of proteolytic cleavage of these proteins. Furthermore, the decreasing effect of IEX-1 on S5a and S1 expression is still seen in the presence of cycloheximide, but not in the presence of actinomycin D, and quantitative real-time PCR revealed lower mRNA levels of S5a and S1 in IEX-1-overexpressing cells, suggesting an interference of IEX-1 with the gene transcription of S5a and S1. Additionally, luciferase assays confirmed an interference of IEX-1 with the activity of the S5a promoter. These findings indicate a role of IEX-1 in the maintenance and assembly of the 26 S proteasome, obviously involving an altered gene expression of certain proteasomal proteins. Thereby, IEX-1 may essentially modulate signalling pathways related to 26 S proteasome activity and involved in cellular growth control and apoptosis.
Keywords: apoptosis, gene transcription, immediate early gene-X-1 (IEX-1), polyubiquitination, proteasome assembly, regulated protein degradation
Abbreviations: ActD, actinomycin D; AMC, amido-4-methylcoumarin; DMEM, Dulbecco's modified Eagle's medium; HEK-293 cells, human embryonic kidney cells; Hsp90, heat-shock protein 90; hUMP1, human homologue of ubiquitin-mediated proteolysis 1; IEX-1, immediate early gene-X-1; IκB, inhibitory κB; MG-132, carbobenzoxy-L-leucyl-L-leucyl-leucinal; NF-κB, nuclear factor κB; Nrf2, NF-E2 (nuclear factor erythroid-derived 2)-related factor 2; POMP, proteasome maturation protein; PPARα, peroxisome-proliferator-activated receptorα; psmd, proteasome 26 S subunit non-ATPase; Rpn, regulatory particle non-ATPase; Rpt, regulatory particle triphosphatase; Suc-LLVY-AMC, N-succinyl-L-leucyl-L-leucyl-L-leucyl-7-AMC; Z-VAD-FMK, benzyloxycarbonylvalylalanyl-DL-aspartylfluoromethane
INTRODUCTION
The ubiquitin–proteasome pathway plays a pivotal role in signal transduction [1,2]. By mediating the tightly regulated turnover of a great variety of regulatory proteins, including cyclins, kinases, phosphatases or transcription factors, the ubiquitin–proteasome pathway essentially contributes to the orchestration of multiple cellular processes such as differentiation, proliferation and apoptosis [1–6]. One hallmark of this pathway is the polyubiquitination of the substrate protein that targets it to the proteolytic degradation complex, the 26 S proteasome. This high-molecular-mass multiprotein complex comprises a central catalytic part, the 20 S proteasome, and two axially positioned regulatory parts, the 19 S regulatory caps. The 20 S proteasome presents itself as a cylindrical structure composed of two pairs of ring-shaped subunits each consisting of seven non-identical α- (non-catalytic) and β- (catalytic) subunits respectively, and the 19 S subunit consists of approx. 20 non-identical proteins exhibiting ATPase [Rpt proteins (regulatory particle triphosphatase)] and non-ATPase [Rpn proteins (regulatory particle non-ATPase)] activity [7,8]. It is meanwhile known that the synthesis of certain proteasomal proteins as well as the formation and maturation of the 20 S and 19 S subunits and the assembly of the 26 S proteasome itself is also a tightly regulated process [9,10]. In yeast, it has been shown that the expression of various proteasomal proteins is under the control of a proteasome-sensitive transcription factor, Rpn4, that accounts for an adequate level of proteasome activity [11–13]. Any deregulation in proteasome activity leads to a loss in cellular homoeostasis and to severe cellular dysfunctions. Accordingly, an inadequate proteasome activity is involved in various pathological conditions including cancer, cachexia and neurodegenerative or autoimmune diseases [14–21].
One of the most prominent signal transduction pathways that are under control of the ubiquitin–proteasome module is the IκBα (inhibitory κBα)/NF-κB (nuclear factor κB) pathway. NF-κB represents a homo- or hetero-dimeric transcription factor of the Rel/NF-κB protein family and plays a fundamental role in various cellular processes, including protection from apoptosis. NF-κB activity is regulated by the κB inhibitor IκBα, which prevents its nuclear translocation and the site-specific transactivation of NF-κB target genes. In response to NF-κB-inducing conditions, polyubiquitination and subsequent proteasomal degradation of IκBα release NF-κB from its sequestration in the cytoplasm and promotes its transcriptional activity in the nucleus. As previously shown, the induced IκBα degradation is under control of the stress-responsive early response gene IEX-1 (immediate early gene-X-1) [22–24], also designated IER3 (intermediate early response 3). This widely expressed and inducible protein has been shown to block NF-κB activation by attenuating the signal-induced turnover of phospho-IκBα, an effect due to the inhibition of phospho-IκBα degradation by the 26 S proteasome [25]. Consequently, IEX-1 prevents the NF-κB-dependent protection from apoptosis. Furthermore, since IEX-1 itself is a target gene of NF-κB it might be part of the NF-κB self-termination process by preventing inappropriate IκBα degradation in the presence of stimuli like IL-1β or TNFα (tumour necrosis factor α) [25]. Such an exaggerated NF-κB induction gives rise to various severe pathological conditions, i.e. inflammation and cancer [26–29]. In accordance with the role of anti-apoptotic protection, the activity of the 26 S proteasome is subject to down-regulation by pro-apoptotic triggers, i.e. anti-cancer drug treatment. It has been recently shown that the major executor caspase, caspase 3, cleaves certain regulatory components of the 19 S proteasomal subunits [30], thereby efficiently consolidating the initiation and execution of apoptosis.
So, it is tempting to speculate that IEX-1 exerts its pro-apoptotic potential by affecting the proteasomal activity. We could show that IEX-1 affects the activity of the 26 S proteasome by down-regulating the expression of certain regulatory proteins, mainly those forming the 19 S subunit. Moreover, this effect of IEX-1 is caspase-independent but obviously dependent on altered gene transcription.
MATERIALS AND METHODS
Materials
MG-132 (the proteasome inhibitor carbobenzoxy-L-leucyl-L-leucyl-leucinal) and Suc-LLVY-AMC [N-succinyl-L-leucyl-L-leucyl-L-leucyl-7-AMC (amido-4-methylcoumarin)] were from Biomol (München, Germany). Etoposide (Vepesid) was from Bistol-Myers-Squibb (München, Germany). Z-VAD-FMK (benzyloxycarbonyl-valylalanyl-DL-aspartylfluoromethane) was from R&D Systems (Wiesbaden, Germany). Cycloheximide and ActD (actinomycin D) were obtained from Sigma (Munich, Germany).
Cell lines and their handling
Retrovirally transduced HEK-293 cells (human embryonic kidney cells) stably expressing tetrameric and trimeric anti-IEX-1 hammerhead ribozymes or not – designated 293-Ri2/10, 293-Ri2/9 and 293-babe respectively [31] – were cultured in DMEM (Dulbecco's modified Eagle's medium) low-glucose medium (PAA Laboratories, Cölbe, Germany) supplemented with 1% (w/v) glutamine (Life Technologies, Eggenstein, Germany), 10% (v/v) fetal calf serum (Biochrom KG, Berlin, Germany) and 1 μg/ml puromycin (BD-Clontech, Heidelberg, Germany). Stably mock/pTO-LacZ or pTO-IEX-1 transfected TRex-293 cells (Invitrogen, Schelp, The Netherlands) – designated Tet-293-Mock and Tet-293-IEX-1 respectively – were kept in culture using DMEM low-glucose medium supplemented with 1% glutamine, 10% fetal calf serum, 5 μg/ml blasticidin and 400 μg/ml zeocin (both from Invitrogen). All cells were kept at 37 °C in a 5% CO2 atmosphere at 85% humidity. Prior to the experiments, cells were kept for at least 24 h without selection antibiotics.
Western blotting
Cells were lysed in 500 μl of lysis buffer [50 mM Tris/HCl, pH 7.4, 1% (v/v) Triton X-100, 150 mM NaCl, 1 mM EDTA, 0.1 mM PMSF and 0.1 mM Na3VO4] supplemented with protease inhibitor cocktail (Complete™-Mini/EDTA free; Roche) and further treated three times with sonication for 10 s. After adjusting to equal protein concentrations (using the Dc-Protein assay; Bio-Rad, München, Germany), the samples were supplemented with a 0.5 vol. of 3×SDS/PAGE sample buffer, heated (95 °C) for 5 min and submitted to 8–20% PAA (polyacrylamide agarose)-gradient electrophoresis. Upon semi-dry transfer on to a PVDF membrane, blots were blocked for 2 h with 5% non-fat milk powder and 0.05% Tween 20 in PBS (blocking solution) at room temperature. Afterwards, blots were exposed to the primary antibodies against S5a/Rpn10, S1/Rpn2, S6′/Rpt5, α6/C2, or hUMP1 (human homologue of ubiquitin-mediated proteolysis 1)/POMP (proteasome maturation protein) (Affiniti/Biomol, Hamburg, Germany), against caspase 3 (Cell Signaling Technology, Frankfurt, Germany), against α-tubulin (Sigma, Deisenhofen, Germany) and against Hsp90 (heat-shock protein 90; Santa Cruz Biotechnology, Heidelberg, Germany) at a 200- to 1000-fold dilution in primary antibody diluent [5% (w/v) BSA in 0.05% Tween 20 in TBS (Tris-buffered saline; 50 mM Tris/HCl, pH 7.6, and 150 mM NaCl)] overnight at 4 °C. After extensive washing with 0.05% Tween 20 in TBS, blots were exposed to the appropriate horse-radish peroxidase-conjugated secondary antibody (Cell Signaling Technology) diluted (1:1000) in blocking buffer and were developed using the LumiGlo peroxidase detection kit (Cell Signaling Technology).
Fluorimetric proteasome assay
For the determination of proteasomal activity in HEK-293 cell-derived clones, a flourimetric assay protocol was performed as described in [32]. In brief, cells were harvested by mild trypsinization. Then, 5×105 cells were resuspended in 500 μl medium and incubated with the proteasome inhibitor MG-132 (5 μg/ml) or not. After 30 min, 10 μl of Suc-LLVY-AMC (10 mM dissolved in DMSO) was added, and at various time points the formation of the fluorescent dye AMC was measured at λ=460 nm using a spectralfluorimeter (PerkinElmer). Specific chymotryptic proteasomal activity was determined by subtraction of the mostly proteasome-independent fluorescence, measured in the presence of MG-132.
Sucrose-density-gradient centrifugation
Identical amounts (2–3 mg of protein) of cellular extracts (in 25 mM Hepes/HCl, pH 7.4, 40 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.2 mM PMSF, 0.5 mM Na3VO4, 250 mM sucrose and 0.5% Triton X-100 supplemented with protease inhibitor cocktail) were loaded on top of 10–30% linear sucrose gradients (in lysis buffer) and the material was centrifuged using a SW28 rotor (Beckman) at 28000 rev./min for 30 h at 4 °C. After centrifugation, 0.5 ml fractions were collected and 20 μl of each fraction was run on an 8–20% gradient gel and immunoblotted.
RNA preparation and real-time PCR
Total RNA was isolated using the RNeasy kit from Qiagen (Hilden, Germany) and reverse-transcribed into single-stranded cDNA, as described previously [33]. Two microlitres of cDNA were subjected to semiquantitative real-time PCR (iCycler; BioRad) using the SYBR Green assay with gene-specific primers at a final concentration of 0.2 μM. The primer sequences and the PCR conditions for the detection of the proteasomal subunits were: 5′-gaaggtttccggagtcggcagtt-3′ (S1/Rpn2, sense) and 5′-ctccagagcaatgccaatagcct-3′ (S1/Rpn2, antisense); 5′-aggcactgtcaccagatgagaac-3′ (S5a/Rpn10, sense) and 5′-aggcactgtcaccagatgagaac-3′ (S5a/Rpn10, antisense); 5′-cacagtcaccttggaggacatc-3′ (S3/Rpn3, sense) and 5′-gaagatgaccacgaggagttgg-3′ (S3/Rpn3, antisense); 5′-gtcaagagtcgggatttgtggg-3′ (S6′/Rpt5, sense) and 5′-ttggagctcatgggtgactctc-3′ (S6′/Rpt5, antisense); 5′-ggcttactgctgatgctagactg-3′ (α6/C2, sense) and 5′-agtcaggtcctgttctgcaggaa-3′ (α6/C2, antisense); 94 °C for 5 min; 35 cycles 94 °C for 30 s, 60 °C for 30 s, 72 °C for 90 s; 72 °C for 10 min. For control, β-actin was amplified in parallel using primers from BD Biosciences Clontech. The real-time PCR was performed with a MyiQ Single Color Real-time PCR Detection System (Bio-Rad). Data were collected during annealing steps and were further analysed by using the iCycler iQ optical system software (Bio-Rad). All samples were analysed in duplicate and data are expressed as amount of mRNA in arbitrary units. The quality of PCR products was checked by melting curve analysis following the amplification as well as by analytical RT (reverse transcriptase)–PCR and subsequent gel electrophoresis.
Cloning of the Rpn10/S5a [psmd4 (proteasome 26 S subunit non-ATPase 4)] promoter and luciferase reporter gene assay
Using the 5′-primers tccgggctctgtactgggctta (pos. 147722) or ccaactgagaactgcaatggc (pos. 148630) and the 3′-primer caagcttcaccaatcacagacctc (pos. 148812) containing a HindIII restriction site, 1090 and 186 bp fragments of the 5′-flanking region of the Rpn10 gene (psmd4, GenBank® accession no. AL592424; reported transcription start pos. 148806) were amplified and cloned into the pGL3basic luciferase vector (Promega) via KpnI/HindIII sites. Both constructs were verified by automated BigDye (PerkinElmer) DNA cycle sequencing using an ABI 3770 instrument (PerkinElmer). These psmd4 vectors psmd4[−186] and psmd4[−1090] or the empty pGL3 vector (0.6 μg each) were co-transfected with 0.2 μg of pTKRL (pTK-Renilla) used as transfection control, as described recently [25]. After treatment, transfected cells were lysed with dual lysis buffer (Promega) and firefly or Renilla luciferase activities were determined using the dual-luciferase reporter kit (Promega) on a luminometer (Bertholdt). Optical units determined for firefly luciferase were normalized for Renilla luciferase chemiluminescence.
Statistics
Data are presented as means±S.D. and analysed by Student's t test. A P value <0.05 (indicated by an asterisk in the Figures) or <0.02 (indicated by a double asterisk in the Figures) was considered as statistically significant.
RESULTS
IEX-1 attenuates the activity of the 26 S proteasome in HEK-293 cells
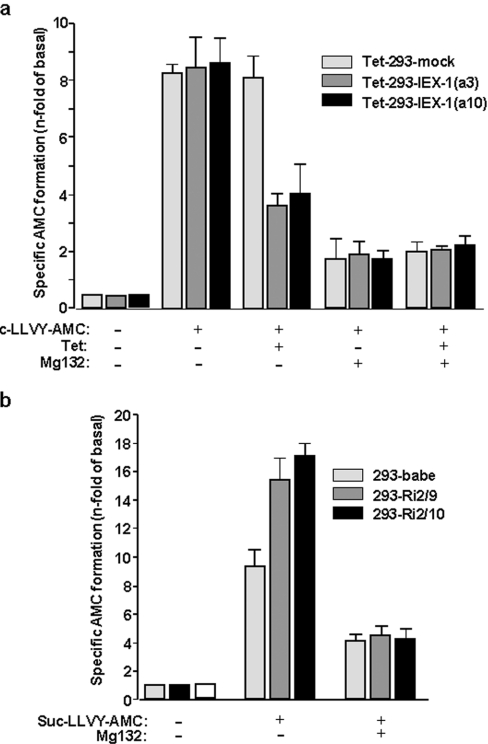
For the determination of the activity of the 26 S proteasome in IEX-1-overexpressing cells, the formation of the fluorescent AMC product from cleavage of the proteasomal Suc-LLVY-AMC substrate was measured in mock- or IEX-1-transfected Tet-293 cells treated with tetracycline, or not. As shown in Figure 1(a), in Tet-293-mock cells, the activity of the proteasome was not altered by tetracycline treatment, whereas in Tet-293-IEX-1 cells, here represented by the two independent clones Tet-293-IEX-1(a3) and Tet-293-IEX-1(a10), the formation of the AMC substrate was greatly reduced if IEX-1 was overexpressed in response to tetracyline administration. To verify the specificity of this effect, the proteasome inhibitor MG-132 was added prior to tetracycline stimulation. Under these conditions, the residual activity of AMC formation that is independent of the proteasome was not affected by IEX-1 overexpression.
Figure 1. IEX-1 inhibits the activity of the 26 S proteasome.
(a) Tet-293-mock and Tet-293-IEX-1 cells (clones a3 and a10) treated with or without tetracycline (16 h, 1 μM), or (b) 293-babe, 293-Ri2/10 and 293-Ri2/9 cells were submitted to a fluorimetric 26 S proteasome assay. For determination of residual non-proteasomal activity accounting for AMC formation, the specific 26 S proteasome inhibitor MG-132 was added in parallel. Results are expressed as n-fold AMC formation in comparison with control cells without Suc-LLVY-AMC and express the means±S.D. for five independent experiments performed in duplicate and adjusted to equal cell numbers.
In contrast, an increase in AMC formation was observed in ribozyme-transduced HEK-293 cells with disrupted IEX-1 expression (Figure 1b). In 293-Ri2/10 cells expressing a tetrameric anti-IEX-1 hammerhead ribozyme and exhibiting the most efficient knockdown of IEX-1 expression [31], the cleavage of Suc-LLVY-AMC was more pronounced as compared with control cells (293-babe). A similar but slightly lower effect was seen in 293-Ri2/9 cells expressing a trimeric anti-IEX-1 hammerhead ribozyme and exhibiting a slightly less efficient IEX-1 knockdown [31]. Again, these IEX-1-related effects in 293-Ri2/10 and 293-Ri2/9 cells were not seen in the presence of MG-132, hence indicating proteasome specificity.
Altered protein levels of S5a and S1 by IEX-1 in HEK-293 cells
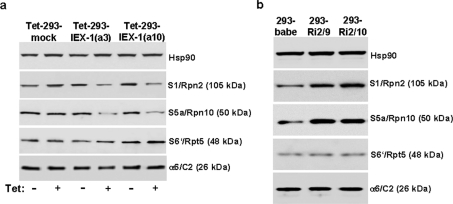
In order to elucidate whether the decrease in proteasomal activity could be attributed to an altered expression of certain proteasomal protein components [10], total lysates from IEX-1-overexpressing or IEX-1-deficient HEK-293 cells were submitted to Western-blot analysis detecting the 19 S subunit components S5a/Rpn10, S1/Rpn2 and S6′/Rpt-5 as well as of the 20 S subunit component α6/C2.
In comparison with mock-transfected Tet-293 cells, the protein levels of S5a and S1 were strongly reduced in the two clones (a3 and a10) of Tet-293-IEX-1 cells when IEX-1 was overexpressed in a tetracycline-dependent fashion (Figure 2a). By contrast, the expression of Rpt-5 and α6/C2 was not significantly affected. Conversely, the knockdown of IEX-1 expression in ribozyme-transduced HEK-293 cells, as seen in 293-Ri2-9 and 293-Ri2-10 cells, significantly increased the expression levels of S5a and S1, but not of S6′ and α6/C2 (Figure 2b).
Figure 2. IEX-1 decreases protein levels of S1 and S5a in HEK-293 cells.
Total lysates from (a) Tet-293-mock and Tet-293-IEX-1 cells (clones a3 and a10) treated with or without tetracycline (16 h, 1 μM), or from (b) 293-babe, 293-Ri2/10 and 293-Ri2/9 cells were submitted to Western-blot analysis detecting the proteasomal proteins a6/C2 (20 S proteasome component), S1, S5a and S6′ (all 19 S proteasome components) or, as the control, Hsp90. A representative result from five independent experiments is shown.
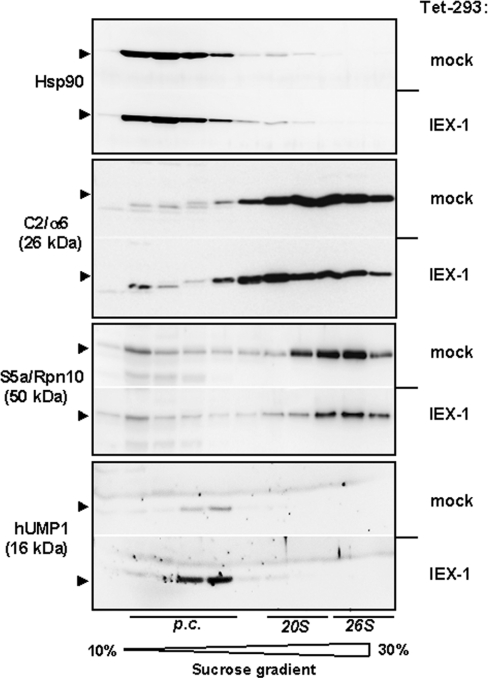
To further address the impact of IEX-1 expression on proteasome formation, sucrose-density-gradient centrifugation was performed and fractions thereof were analysed by Western blot. As compared with tetracycline-treated mock-Tet-293 cells, IEX-1-overexpressing HEK-293 cells, here shown for the clone Tet-293-IEX-1(a3), again exhibited much lower levels of the S5a protein that is sedimented in higher density fractions corresponding to the 26 S proteasome (Figure 3). In contrast, the C2 protein detected over a broader range in the density gradient was not affected in its overall amount by IEX-1 overexpression. However, C2-positive sediments in higher density fractions (containing material of the 26 S and 20 S proteasome) were less pronounced, whereas those in lower density fractions (containing material of the 20 S proteasome precursors) were enriched in Tet-293-IEX-1 cells. Furthermore, expression of the proteasomal maturation factor hUMP1/POMP was strongly increased in IEX-1-overexpressing cells, probably reflecting accumulation of undigested hUMP1 protein due to an impaired proteasome assembly (Figure 3, bottom).
Figure 3. Impaired protein assembly of the 26 S proteasome in IEX-1-expressing HEK-293 cells.
Cellular lysates from Tet-293-mock and Tet-293-IEX-1 cells (clone a3) treated with tetracycline (16 h, 1 μM) were submitted to 10–30% (w/v) sucrose-density-gradient ultracentrifugation (28 000 rev./min; 30 h; SW28 rotor) and fractions thereof were analysed by Western blot detecting the proteasomal proteins α6/C2 and S5a, the maturation factor hUMP1 or, as the control, Hsp90. A representative result from three independent experiments is shown. The positions of sedimented proteasomal subcomplexes and precursor complexes (p.c.) are indicated.
The attenuating effect of IEX-1 on S5a and S1 expression enhances the reduction of S5a and S1 protein levels resulting from etoposide-induced apoptosis and is caspase-independent
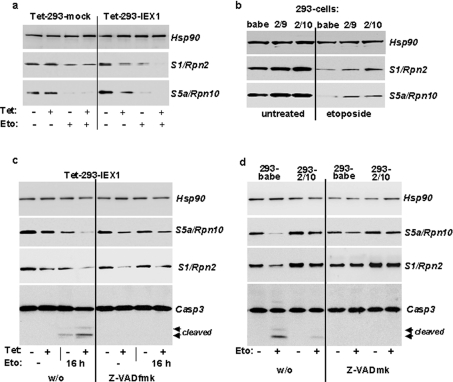
Next we looked whether down-regulation of S5a and S1 is related to apoptosis-dependent and caspase 3-mediated degradation of these proteins. Upon administration of the anti-cancer drug etoposide (20 μg/ml) and in the absence of tetracycline, the protein levels of S5a and S1 quite similarly declined in mock- and IEX-1-transfected Tet-293 cells within 24 h in comparison with untreated cells. If tetracycline was added 16 h prior to etoposide administration the drug-induced decrease in S5a and S1 protein levels was much more pronounced in Tet-293-IEX-1 cells, here the clone Tet-293-IEX-1(a3), as compared with Tet-293-mock cells (Figure 4a). Conversely, in HEK-293 cells subjected to ribozyme-mediated knockdown of IEX-1 expression (293-Ri2-10 and 293-Ri2-9), the etoposide-dependent decrease in S5a and S1 protein levels was much less pronounced as compared with 293-babe control transfectants (Figure 4b).
Figure 4. The decreasing effect of IEX-1, but not of etoposide, on the protein levels of S1 and S5a is caspase-independent.
(a) Tet-293-mock and Tet-293-IEX-1 cells (clone a3) pretreated with or without tetracycline (8 h, 1 μM), or (b) 293-babe, 293-Ri2/10 and 293-Ri2/9 cells were subjected to treatment with or without etoposide (10 μM) for 24 h. Then, cell lysates were submitted to Western blot analysis detecting the proteasomal proteins S1 and S5a or, as the control, Hsp90. (c) Tet-293-IEX-1 cells (clone a3) pretreated with or without tetracycline (8 h, 1 μM), or (d) 293-babe and 293-Ri2/10 cells were subjected to treatment with etoposide (10 μM) for 24 h or not, either in the absence (left four lanes) or presence (right four lanes) of Z-VAD-FMK. Then, cell lysates were submitted to Western blot analysis detecting the proteasomal proteins S1 and S5a or, as the control, Hsp90. In all panels, one representative result from three independent experiments is shown.
In the presence of the caspase inhibitor Z-VAD-FMK (Figures 4c and 4d), the etoposide-dependent decrease in S5a and S1 protein levels in all transfected HEK-293 cells was completely blocked, indicating that this decreasing effect by etoposide was indeed due to caspase-mediated degradation of S5a and S1. Interestingly, caspase inhibition by Z-VAD-FMK did not affect the tetracycline-induced decrease in S5a and S1 protein levels in IEX-1-overexpressing 293-Tet cells (Figure 4c), neither in the absence nor in the presence of etoposide. Moreover, in ribozyme-transduced HEK-293 cells with disrupted IEX-1 expression, the higher protein levels of S5a and S1 were still seen in the presence of Z-VAD-FMK (Figure 4d). These findings demonstrate that the effect of IEX-1 on S5a and S1 expression itself is independent of caspase activity, but significantly enhances the caspase-dependent decrease in S1 and S5a protein levels during etoposide-induced apoptosis. Along with these additive actions of etoposide and IEX-1, apoptotic cell death increases with IEX-1 overexpression as indicated by the stronger caspase 3 activation seen in Figure 4(c), but decreases with IEX-1 knockdown, as indicated by the reduced caspase 3 activation seen in Figure 4(d).
IEX-1 interferes with S5a and S1 gene transcription
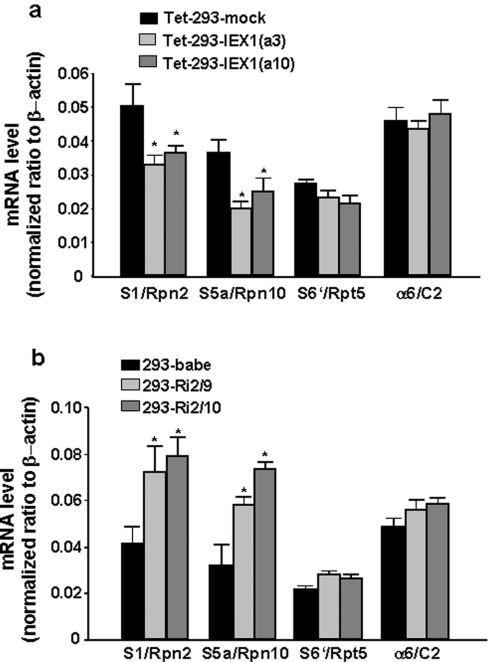
It was therefore tempting to speculate that IEX-1 may interfere with gene transcription of S5a and S1, particularly since these two proteins are subject to tight transcriptional regulation [10]. For this purpose, quantitative real-time PCR was conducted in order to determine the mRNA levels of S5a and S1 as well as of S6′ and C2 for comparison. As shown in Figure 5(a), after normalization to the amount of β-actin mRNA used as internal control, the levels of S5a and S1 mRNA were significantly reduced in tetracycline-treated Tet-293 cells overexpressing IEX-1. Compared with Tet-293-mock cells, the mRNA levels of S5a and S1 decreased by 32 and 47% respectively in Tet-293-IEX-1(a10) cells, and by 35 and 53% respectively in Tet-293-IEX-1(a3) cells. In contrast, mRNA levels of S6′ and C2 were not significantly affected by tetracycline treatment in mock- or IEX-1-transfected Tet-293 cells. When analysing HEK-293 cells with disrupted IEX-1 expression, it turned out that in these cells (293-Ri2/9 and 293-Ri2/10) the amount of S5a mRNA (increased by 80 and 120% respectively) and S1 mRNA (increased by 70 and 90% respectively) were significantly higher than in 293-babe control transfectants (Figure 5b), whereas the mRNA levels of S6′ and C2 did not differ in this way.
Figure 5. IEX-1 suppresses gene expression of S1 and S5a on the transcriptional level.
Total RNA from (a) Tet-293-mock and Tet-293-IEX-1 cells (clones a3 and a10) pretreated with tetracycline (16 h, 1 μM) or from (b) 293-babe, 293-Ri2/9 and 293-Ri2/10 cells was reverse-transcribed and then submitted to semi-quantitative real-time PCR using specific primers for S5a, S6′, S1, C2 and, as the control, β-actin. The amount of mRNA normalized to β-actin is shown and expressed as means±S.D. (n=5); asterisk indicates statistically significant (P<0.05).
The negative interference of IEX-1 with S1 and S5a is abrogated during transcriptional but not during translational inhibition
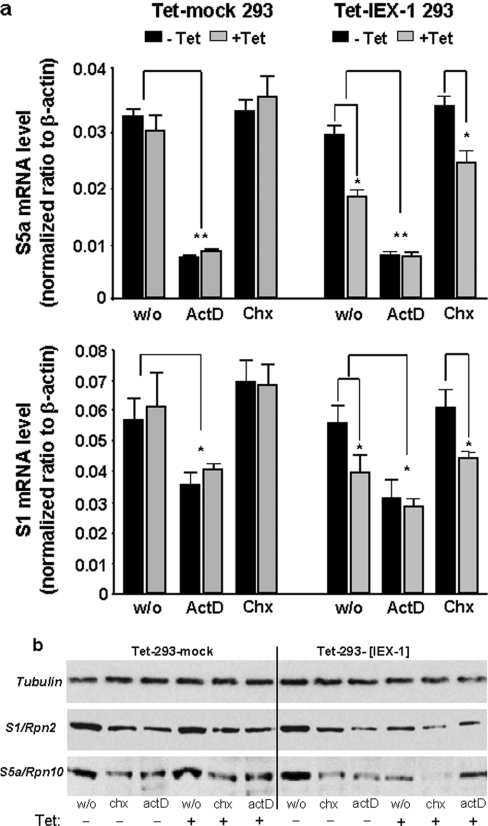
In order to show whether the attenuating effect of IEX-1 on S1 and S5a expression depends on de novo mRNA synthesis, the transcriptional inhibitor ActD (0.2 μg/ml, 24 h) was added 16 h after induction of IEX-1 overexpression. As shown by real-time PCR (Figure 6a), mRNA levels of S1 and S5a were significantly suppressed in the presence of ActD reflecting blockade of RNA de novo synthesis. Under these conditions, the overexpression of IEX-1 did not further decrease the expression levels of S1 or S5a mRNA, thus indicating a non-additive action of IEX-1 and ActD. In contrast, cycloheximide (0.5 μg/ml, 24 h) that blocks de novo protein synthesis did not interfere with the levels of S1 or S5a mRNA and IEX-1 overexpression still decreased the amount of both mRNAs in the presence of cycloheximide, indicating that neither the S1 nor S5a gene transcription under these conditions nor the negative effect of IEX-1 involves de novo protein synthesis.
Figure 6. The decreasing effect of IEX-1 on S1 and S5a expression is blocked by transcriptional but not by translational inhibition.
Tet-293-mock and Tet-293-IEX-1 cells (clone a3), pretreated with tetracycline (16 h, 1 μM), or not, were further incubated with the protein translation inhibitor cycloheximide (Chx; 0.5 μg/ml) and the RNA transcription inhibitor ActD (0.2 μg/ml), or without (w/o) for 24 h. Then (a) total RNA was isolated and submitted to reverse transcription and subsequent semi-quantitative real-time PCR detecting S1, S5a and, as the control, β-actin mRNAs. The amount of mRNA normalized to β-actin is shown and expressed as means±S.D. (n=4); asterisk indicates statistically significant (P<0.05); double asterisk indicates statistically significant (P<0.02). In parallel (b), cell lysates were submitted to Western-blot analysis detecting the proteasomal proteins S1 and S5a or, as the control, α-tubulin. In all panels, one representative result from three independent experiments is shown.
When analysing the effects of these inhibitors on protein levels as well (Figure 6b), it turned out that both ActD and cycloheximide decrease the expression of S1 and S5a. But while no additive effect was seen with IEX-1 and ActD, the decrease in S1 and S5a protein levels seen with cycloheximide was much more pronounced in IEX-1-overexpressing cells, indicating an additive action. These data suggest that the suppressing effect of IEX-1 on S1 and S5a expression is due to an interference with de novo mRNA synthesis of these genes. This effect does not involve post-transcriptional or co-translational events that control mRNA stability because this would be additive to the effect of ActD.
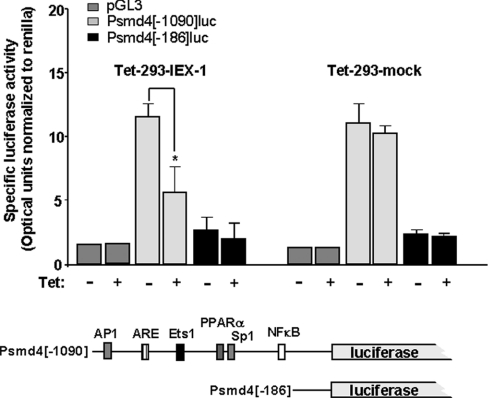
Suppression of S5a promoter activity by IEX-1
Since S5a appears to be the most affected 19 S subunit gene by IEX-1 (see above), we chose the promoter of this gene for analysing the effect of IEX-1 on promoter activity of proteasomal subunit genes. For this purpose, parts of the S5a promoter, which has not been characterized so far, were fused to the firefly luciferase reporter gene and used for luciferase assays in Tet-293 cells. From a database search we generated two promoter fragments (Figure 7, lower panel): one [psmd4 (−1090)] containing a potential antioxidant response element and binding site for Nrf2 [NF-E2 (nuclear factor erythroid-derived 2)-related factor 2] as well as potential binding sites for AP1 (activator protein 1), PPARα (peroxisome-proliferator-activated receptor α), Sp1 (stimulating protein 1), Ets-1 (avian erythroblastosis virus E2 oncogene homolog 1) and, most notably, NF-κB, and one lacking these sites (psmd4[−186]). As shown in Figure 7, significant promoter activity as measured by firefly luciferase optical units (normalized to Renilla luciferase) was detected in Tet-293 cells transfected with psmd4[−1090] compared with cells transfected with promoterless pGL3, but only marginal promoter activity was seen in cells transfected with psmd4[−186]. When overexpressing IEX-1 by the addition of tetracycline in Tet-293-IEX-1 cells (clone a3), but not in Tet-293-mock cells, the psmd4[−1090]-driven luciferase expression declined significantly (Figure 7), whereas the residual promoter activities seen with psmd4-[−186] and pGL3 were not affected. These findings strongly support our conclusions drawn from the preceding experiments that IEX-1 specifically interferes with the expression of the 19 S subunit genes on the transcriptional level, obviously by exerting an inhibitory effect on the promoter activity, obviously by exerting an inhibitory effect on the promoter activity, as shown here for S5a.
Figure 7. IEX-1 overexpression attenuates S5a promoter activity.
Tet-293-IEX-1 (clone a3) or Tet-293-mock cells were co-transfected with pTKRL (for normalization) and firefly luciferase reporter gene constructs containing two fragments (psmd4[−1090] and psmd4[−186]) of the S5a promoter as depicted in the lower part of the Figure or with the promoterless pGL3 vector. Potential binding sites were identified from the genomic DNA sequence containing the psmd4/S5a gene (accession no. AL592424) using the Internet-based Transcription Element Search Software (http://www.cbil.upenn.edu/tess). After transfection, cells were incubated with 1 μM tetracycline or not for 24 h. Then, cells were lysed and submitted to dual luciferase assay using Renilla luciferase activity (pTKRL) for normalization of optical units obtained with firefly luciferase and expressed as means±S.D. (n=4); asterisk indicates statistically significant (P<0.05).
DISCUSSION
Under certain cellular conditions (i.e. serum starvation, DNA-damaging insults), the stress response gene IEX-1 exerts a strong pro-apoptotic effect [31,33–35], which to some extent is related to its attenuating effect on NF-κB activation [25]. This involves an interference of IEX-1 with the signal-induced degradation of IκBα by the 26 S proteasome. Whereas it is known that certain pro-apoptotic triggers, as shown above for the anti-cancer drug etoposide, decrease the levels of certain proteasomal protein components by caspase-mediated cleavage and thereby consolidate the cell death committing apoptotic programme [30], we could show that the effect of IEX-1 does not involve this process. In fact, IEX-1 affects the expression of these proteins independently of caspase activity and, thereby, acts additive to the decreasing effect of apoptotic stimuli on protein levels of proteasomal components which is caspase-dependent.
Intriguingly, IEX-1 directly attenuates the expression of certain proteasomal components, particularly those that are part of the 19 S regulatory subunit of the 26 S proteasome [10] and conferring specificity for polyubiquitinated substrates such as IκBα [36,37]. This decreased expression of S5a or S1 in response to IEX-1 is not due to a direct protein interaction as previously described for α-synuclein [38], but obviously results from a suppressed gene transcription as shown by the declined mRNA levels of both genes in HEK-293 cells if IEX-1 is overexpressed. In support of this assumption, conditions of transcriptional blockade by ActD abrogated this negative interference of IEX-1 with S1 and S5a expression, suggesting that the effect of IEX-1 does not depend on post-transcriptional events, i.e. a decreased mRNA stabilization or an increased mRNA turnover, but rather is related to the transcription process and de novo RNA synthesis. Furthermore, treatment with the translational inhibitor cycloheximide did not alter the effect of IEX-1 on S1 and S5a expression, also indicating that IEX-1 expression does not affect a post-transcriptional or a co-translational event. In further support of this assumption, the activity of the putative S5a promoter as determined by luciferase reporter gene assay was attenuated by IEX-1.
Still, it could be just speculated how IEX-1 interferes with gene transcription of S1 and S5a. In yeast, the transcription of proteasomal proteins is under control of the transcription factor Rpn4, which is mainly regulated by its proteasomal degradation [11–13]. Accordingly, during conditions with high proteasomal activity the de novo synthesis of protein components is restrained, whereas the lack of proteasome activity favours the increased expression of particularly those proteins compensating for the declined proteasomal activity [10]. Although the human counterpart of Rpn4 has not been identified yet, recent reports indicate that in mammalian cells the antioxidant-related transcription factor Nrf2, which is known to be involved into cytoprotection during oxidative stress, may control the expression of these proteins [39,40]. Kwak and Kensler [40] recently described a crucial role of Nrf2 in the induced expression of the proteasomal subunit protein β5/PSMB5 in response to oxidative stress, and other conditions are known – including proteasome inhibition, PI3K (phosphoinositide 3-kinase)/Akt or ERK (extracellular-signal-regulated kinase) activation – leading to increased Nrf2 activity and, thereby, stimulating expression of proteasomal proteins [41,42]. However, it still has to be elucidated whether or not the effect of IEX-1 on the gene expression of certain proteasomal proteins is indeed related to the action of this transcription factor as could be expected from S5a promoter analysis and luciferase assays, and whether other transcription factors, i.e. PPARα [43] or NF-κB, also play a role.
A major consequence of the blocking effect of IEX-1 on the expression of certain subcomponents of the 26 S proteasome is a marked impairment of its assembly (Figure 3) that is already blocked on the level of the precursor particles [10,44,45]. Moreover, along with the decreased formation of the 26 S proteasome, the maturation of the 20 S proteasome seems also to be affected as indicated by the accumulation of the maturation factor hUMP1 and by the altered distribution of sedimented 20 S protein, i.e. α6/C2, in the sucrose density gradients (Figure 3). It is not clear yet how the reduced de novo expression of 19 S proteins and consequently the 19 S subunit depletion influences the assembly of the 20 S proteasome. Due to the reduced levels of certain protein components, the immature 19 S precursors may disintegrate and assemble improperly with 20 S precursors, thereby blocking further 20 S subunit maturation. Another explanation could be that the inappropriate assembly of the 19 S subunit excessively consumes certain proteasome-related chaperones [45] that are then unavailable for the completion of the 20 S subunit.
Irrespective of the exact mechanisms, proteasome activity targeted to polyubiquitinated proteins (i.e. IκBα) is significantly compromised by IEX-1, giving rise, at least in part, to an increased sensitivity to apoptotic triggers. This is underlined by the finding that many conditions affecting proteasome activity enhance apoptosis. In this way, proteasome inhibitors and anti-cancer drugs and, as can be assumed, IEX-1 expression sensitize various cell types to diverse apoptotic stimuli [46,47]. Thus it can be speculated that IEX-1 is part of a cellular regulation process that dampens the exaggerated processing and turnover of regulatory proteins. Thereby, IEX-1 may contribute to the maintenance of cellular homoeostasis and integrity which, for example, is a prerequisite in the elimination of pre-malignant cells. Accordingly, IEX-1 expression has been found to be reduced in certain types of tumour indicating a potential role of IEX-1 in tumour suppression [48–50].
Acknowledgments
This work forms part of the Ph.D. Thesis of J.M. and was supported by a grant (no. SFB415/A13 to H.S.) from the German Research Society DFG (Deutsche Forschungsgemeinschaft). We thank Professor O. Janssen from the Institute for Immunology, UKSH-Campus Kiel, for help with the gradient centrifugation and helpful discussion.
References
- 1.Welchman R., Gordon C., Mayer R. J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 2.Dalton W. The proteasome. Semin. Oncol. 2004;6:3–9. doi: 10.1053/j.seminoncol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z. J. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhananjayan S. C., Ismail A., Nawaz Z. Ubiquitin and control of transcription. Essays Biochem. 2005;41:69–80. doi: 10.1042/EB0410069. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H. G., Wang J., Yang X., Hsu H. C., Mountz J. D. Regulation of apoptosis proteins in cancer cells by ubiquitin. Oncogene. 2004;23:2009–2015. doi: 10.1038/sj.onc.1207373. [DOI] [PubMed] [Google Scholar]
- 6.Naujokat C., Hoffmann S. Role and function of the 26 S proteasome in proliferation and apoptosis. Lab. Invest. 2002;82:965–980. doi: 10.1097/01.lab.0000022226.23741.37. [DOI] [PubMed] [Google Scholar]
- 7.Voges D., Zwickl P., Baumeister W. The 26 S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Baumeister W., Walz J., Zuhl F., Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 10.Meiners S., Heyken D., Weller A., Ludwig A., Stangl K., Kloetzel P. M., Krüger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 11.Xie Y., Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26 S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju D., Wang L., Mao X., Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- 13.London M. K., Keck B. I., Ramos P. C., Dohmen R. J. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 2004;567:259–264. doi: 10.1016/j.febslet.2004.04.078. [DOI] [PubMed] [Google Scholar]
- 14.Devo A., Soane T., Welchman R., Mayer R. J. The ubiquitin–proteasome system and cancer. Essays Biochem. 2005;41:187–203. doi: 10.1042/EB0410187. [DOI] [PubMed] [Google Scholar]
- 15.Khal J., Wyke S. M., Russell S. T., Hine A. V., Tisdale M. J. Expression of the ubiquitin–proteasome pathway and muscle loss in experimental cancer cachexia. Br. J. Cancer. 2005;93:774–780. doi: 10.1038/sj.bjc.6602780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani A., Gelmann E. P. The ubiquitin–proteasome pathway and its role in cancer. J. Clin. Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 17.Spataro V., Norbury C., Harris A. L. The ubiquitin–proteasome pathway in cancer. Br. J. Cancer. 1998;77:448–455. doi: 10.1038/bjc.1998.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layfield R., Lowe J., Bedford L. The ubiquitin–proteasome system and neurodegenerative disorders. Essays Biochem. 2005;41:157–171. doi: 10.1042/EB0410157. [DOI] [PubMed] [Google Scholar]
- 19.Mountz J. D. Significance of increased circulating proteasome in autoimmune disease. J. Rheumatol. 2002;10:2027–2030. [PubMed] [Google Scholar]
- 20.Shringarpure R., Davies K. J. Protein turnover by the proteasome in aging and disease. Free Radical Biol. Med. 2002;32:1084–1089. doi: 10.1016/s0891-5849(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi T., Faustman D. L. Implications of altered apoptosis in diabetes mellitus and autoimmune disease. Apoptosis. 2001;6:31–45. doi: 10.1023/a:1009667926296. [DOI] [PubMed] [Google Scholar]
- 22.Kondratyev A. D., Chung K. N., Jung M. O. Identification and characterization of a radiation-inducible glycosylated human early-response gene. Cancer Res. 1996;56:1498–1502. [PubMed] [Google Scholar]
- 23.Schäfer H., Trauzold A., Siegel E. G., Fölsch U. R., Schmidt W. E. PRG1: a novel early-response gene transcriptionally induced by pituitary adenylate cyclase activating polypeptide in a pancreatic carcinoma cell line. Cancer Res. 1996;56:2641–2648. [PubMed] [Google Scholar]
- 24.Wu M. X. Roles of the stress-induced gene IEX-1 in regulation of cell death and oncogenesis (Review) Apoptosis. 2003;8:11–18. doi: 10.1023/a:1021688600370. [DOI] [PubMed] [Google Scholar]
- 25.Arlt A., Kruse M. L., Breitenbroich M., Gehrz A., Kocs B., Minkenberg J., Fölsch U. R., Schäfer H. The early response gene IEX-1 attenuates NFκB activation in 293 cells, a possible counterregulatory process leading to enhanced cell death. Oncogene. 2003;22:3343–3351. doi: 10.1038/sj.onc.1206524. [DOI] [PubMed] [Google Scholar]
- 26.Karin M., Greten F. R. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 27.Pande V., Ramos M. J. NFκB in human disease: current inhibitors and prospects for de novo structure based design of inhibitors. Curr. Med. Chem. 2005;12:357–374. doi: 10.2174/0929867053363180. [DOI] [PubMed] [Google Scholar]
- 28.Karin M., Cao Y., Greten F. R., Li Z. W. NFκB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto Y., Gaynor R. B. Role of the NFκB pathway in the pathogenesis of human disease states. Curr. Mol. Med. 2001;1:287–296. doi: 10.2174/1566524013363816. [DOI] [PubMed] [Google Scholar]
- 30.Sun X. M., Butterworth M., MacFarlane M., Dubiel W., Ciechanover A., Cohen G. M. Caspase activation inhibits proteasome function during apoptosis. Mol. Cell. 2004;14:81–93. doi: 10.1016/s1097-2765(04)00156-x. [DOI] [PubMed] [Google Scholar]
- 31.Grobe O., Arlt A., Krupp G., Ungefroren H., Fölsch U. R., Schmidt W. E., Schäfer H. Functional disruption of IEX-1 expression by concatameric hammer-head ribozymes alters growth properties of 293 cells. FEBS Lett. 2001;494:196–200. doi: 10.1016/s0014-5793(01)02344-4. [DOI] [PubMed] [Google Scholar]
- 32.Masdehors P., Merle-Beral H., Maloum K., Omura S., Magdelenat H., Deli J. Deregulation of the ubiquitin system and p53 proteolysis modify the apoptotic response in B-CLL lymphocytes. Blood. 2000;96:269–274. [PubMed] [Google Scholar]
- 33.Schäfer H., Arlt A., Trauzold A., Hünermann-Jansen A., Schmidt W. E. The putative apoptosis inhibitor IEX-1L is a mutant non-spliced variant of p22PRG1/IEK−1 and is not expressed in vivo. Biochem. Biophys. Res. Commun. 1999;262:139–145. doi: 10.1006/bbrc.1999.1131. [DOI] [PubMed] [Google Scholar]
- 34.Segev D. L., Hoshiya Y., Hoshiya M., Tran T. T., Carey J. L., Stephen A. E., MacLaughlin D. T., Donahoe P. K., Maheswaran S. Mullerian-inhibiting substance regulates NF-kappa B signaling in the prostate in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 2002;99:239–244. doi: 10.1073/pnas.221599298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osawa Y., Nagaki M., Banno Y., Brenner D. A., Nozawa Y., Moriwaki H., Nakashima S. Expression of the NF-kappa B target gene X-ray-inducible immediate early response factor-1 short enhances TNF-alpha-induced hepatocyte apoptosis by inhibiting Akt activation. J. Immunol. 2003;170:4053–4060. doi: 10.4049/jimmunol.170.8.4053. [DOI] [PubMed] [Google Scholar]
- 36.Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T. The ubiquitin–proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappaB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z., Hagle J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin–proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 38.Snyder H., Mensah K., Theisler C., Lee J., Matouschek A., Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J. Biol. Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 39.Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. Antioxidants enhance mammalian proteasome expression through the Keap1–Nrf2 signaling pathway. Mol. Cell. Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwak M. K., Kensler T. W. Induction of 26 S proteasome subunit PSMB5 by the bifunctional inducer 3-methylcholanthrene through the Nrf2–ARE, but not the AhR/Arnt–XRE, pathway. Biochem. Biophys. Res. Commun. 2006;354:1350–1357. doi: 10.1016/j.bbrc.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 41.Kang K. W., Lee S. J., Park J. W., Kim S. G. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol. Pharmacol. 2002;62:1001–1010. doi: 10.1124/mol.62.5.1001. [DOI] [PubMed] [Google Scholar]
- 42.Numazawa S., Yoshida T. Nrf2-dependent gene expressions: a molecular–toxicological aspect. J. Toxicol. Sci. 2004;29:81–89. doi: 10.2131/jts.29.81. [DOI] [PubMed] [Google Scholar]
- 43.Anderson S. P., Howroyd P., Liu J., Qian X., Bahnemann R., Swanson C., Kwak M. K., Kensler T. W., Corton J. C. The transcriptional response to a peroxisome proliferator-activated receptor alpha agonist includes increased expression of proteome maintenance genes. J. Biol. Chem. 2004;279:52390–52398. doi: 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- 44.Witt E., Zantopf D., Schmidt M., Kraft R., Kloetzel P. M., Krüger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. J. Mol. Biol. 2000;301:1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- 45.Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 46.Schilling D., Pittelkow M. R., Kumar R. IEX-1, an immediate early gene, increases the rate of apoptosis in keratinocytes. Oncogene. 2001;20:7992–7997. doi: 10.1038/sj.onc.1204965. [DOI] [PubMed] [Google Scholar]
- 47.Arlt A., Grobe O., Sieke A., Kruse M. L., Fölsch U. R., Schmidt W. E., Schäfer H. Expression of the NF-κB target gene p22PRG1/IEX−1 does not prevent cell death but instead triggers apoptosis in HeLa cells. Oncogene. 2001;20:69–77. doi: 10.1038/sj.onc.1204061. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y., Sun H., Drake J., Kittrell F., Abba M. C., Deng L., Gaddis S., Sahin A., Baggerly K., Medina D., et al. From mice to humans: identification of commonly deregulated genes in mammary cancer via comparative SAGE studies. Cancer Res. 2004;64:7748–7755. doi: 10.1158/0008-5472.CAN-04-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nambiar P. R., Nakanishi M., Gupta R., Cheung E., Firouzi A., Ma X. J., Flynn C., Dong M., Guda K., Levine J., et al. Genetic signatures of high- and low-risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res. 2004;64:6394–6401. doi: 10.1158/0008-5472.CAN-04-0933. [DOI] [PubMed] [Google Scholar]
- 50.Dilley W. G., Kalyanaraman S., Verma S., Cobb J. P., Laramie J. M., Lairmore T. C. Global gene expression in neuroendocrine tumors from patients with the MEN1 syndrome. Mol. Cancer. 2005;4:9. doi: 10.1186/1476-4598-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]