Abstract
The glucose transporter GLUT4 and the aminopeptidase IRAP (insulin-responsive aminopeptidase) are the major cargo proteins of GSVs (GLUT4 storage vesicles) in adipocytes and myocytes. In the basal state, most GSVs are sequestered in perinuclear and other cytosolic compartments. Following insulin stimulation, GSVs undergo exocytic translocation to insert GLUT4 and IRAP into the plasma membrane. The mechanisms regulating GSV trafficking are not fully defined. In the present study, using 3T3-L1 adipocytes transfected with siRNAs (small interfering RNAs), we show that insulin-stimulated IRAP translocation remained intact despite substantial GLUT4 knockdown. By contrast, insulin-stimulated GLUT4 translocation was impaired upon IRAP knockdown, indicating that IRAP plays a role in GSV trafficking. We also show that knockdown of tankyrase, a Golgi-associated IRAP-binding protein that co-localizes with perinuclear GSVs, attenuated insulin-stimulated GSV translocation and glucose uptake without disrupting insulin-induced phosphorylation cascades. Moreover, iodixanol density gradient analyses revealed that tankyrase knockdown altered the basal-state partitioning of GLUT4 and IRAP within endosomal compartments, apparently by shifting both proteins toward less buoyant compartments. Importantly, the afore-mentioned effects of tankyrase knockdown were reproduced by treating adipocytes with PJ34, a general PARP (poly-ADP-ribose polymerase) inhibitor that abrogated tankyrase-mediated protein modification known as poly-ADP-ribosylation. Collectively, these findings suggest that physiological GSV trafficking depends in part on the presence of IRAP in these vesicles, and that this process is regulated by tankyrase and probably its PARP activity.
Keywords: adipocytes, GLUT4, GLUT1, insulin-responsive aminopeptidase (IRAP), iodixanol gradients, tankyrase
Abbreviations: AS160, Akt substrate of 160 kDa; DMEM, Dulbecco's modified Eagle's medium; GLUT, glucose transporter; GSK, glycogen synthase kinase; GST, glutathione S-transferase; GSV, glucose transporter 4 storage vesicles; HDM, heavy microsomes; IRAP, insulin-responsive aminopeptidase; IRS-1, insulin receptor substrate-1; LDM, light microsomes; PAR, polymers of ADP-ribose; PARP, poly-ADP-ribose polymerase; PARsylate, poly-ADP-ribosylate; PM, plasma membrane; PNS, post-nuclear supernatant; siRNA, small interfering RNA; TfR, transferrin receptor
INTRODUCTION
Insulin modulates the exocytosis of diverse membrane proteins from the endosomal compartments to the PM (plasma membrane) in adipocytes. This effect is most prominent on the glucose transporter GLUT4 and IRAP (insulin-responsive aminopeptidase) [1,2]. After de novo synthesis, both vesicular proteins are sorted by an incompletely defined process from the Golgi complex into the same pool of vesicles often referred to as the GSVs (GLUT4 storage vesicles) [3]. These vesicles continuously move to and from the PM in a regulated manner [1]. In the basal state, GSVs are sequestered primarily in the perinuclear space but also in the cytosolic periphery. In response to insulin, GSVs undergo robust exocytosis to deliver cargo proteins to the PM, enabling GLUT4 to import glucose and IRAP to proteolyse specific circulating hormones [2]. This regulated translocation requires insulin to activate both a PI3K (phosphoinositide 3-kinase)- and a Cbl-dependent signalling cascade [4]. The translocation is also stimulated by osmotic shock through a non-insulin signalling pathway mediated by the kinase FAK (focal adhesion kinase) [5].
Given that GLUT4 and IRAP co-localize extensively in GSVs, a possible explanation for their remarkably similar targeting is that one of them is directly regulated by insulin-sensitive trafficking machinery while the other merely tags along in the same vesicles. Several studies have addressed this using GLUT4 and IRAP knockout mice, but the results are somewhat mixed. First, in whole-body GLUT4 knockouts, the PM targeting of IRAP in fat and skeletal muscle cells becomes constitutive and does not respond to insulin stimulation, suggesting a role of GLUT4 in normal GSV trafficking [6]. In sharp contrast, tissue-specific GLUT4 knockout in mice does not impair insulin-stimulated IRAP translocation to the PM in cardiomyocytes or adipocytes [7,8], suggesting that GLUT4 is dispensable for normal GSV trafficking. Conversely, in whole-body IRAP knockouts, the insulin-stimulated PM translocation of GLUT4 is preserved [9], implying that GSV trafficking does not require the presence of IRAP either. Collectively, these murine models do not single out GLUT4 or IRAP as the cargo that governs GSV trafficking.
Circumstantial evidence involving cultured 3T3-L1 adipocytes suggests that the GSV trafficking machinery contacts GSVs by binding to IRAP. First, multiple components of this machinery were discovered on the basis of IRAP binding. They include AS160 (an Akt substrate of 160 kDa [10]), p115 (a vesicle-tethering factor implicated in ER-to-Golgi and post-Golgi movements [11]), FHOS (a formin homologue overexpressed in spleen [12]), and ACDs (acyl-CoA dehydrogenases [13]). All of these proteins interact with the IRAP cytosolic tail, a domain responsible for conferring insulin-stimulated translocation on IRAP and its co-localization with GLUT4 [2]. Interestingly, overexpression of certain fragments of this IRAP domain (amino acids 1–52 or 55–82) causes insulin-independent GLUT4 translocation [14], presumably by saturating the GSV targeting machinery and precluding its interaction with endogenous IRAP in GSVs.
A candidate component of the GSV trafficking machinery is the IRAP-binding protein tankyrase. Also known as TNKS-1 (tankyrase-1), this 165 kDa molecular scaffold resides in the Golgi region [15,16] and co-localizes with perinuclear GSVs in adipocytes [16]. Its ankyrin-repeat domain contains five IRAP-binding sites [17]. This domain also binds to additional partners such as Grb14 (growth-factor-receptor-bound protein 14) [17,18], a signalling adapter that modulates the glucose-lowering effect of insulin [19]. Most of these partners bind to the ankyrin-repeat domain of tankyrase using an RxxPDG sequence motif that corresponds to amino acids 96–101 in the IRAP cytosolic tail [16,17]. Overexpression of this tankyrase-binding region of IRAP, unlike the two aforementioned IRAP fragments, fails to cause insulin-independent GLUT4 translocation [14]. This prompted us to speculate that IRAP binding to tankyrase is not involved in sequestering GSVs but instead might modulate other aspects of GSV trafficking [16].
The functions of tankyrase likely overlap with the closely related tankyrase-2, which oligomerizes with tankyrase and also binds to IRAP [17,20]. Both tankyrases exhibit an unusual catalytic activity known as PARP (poly-ADP-ribose polymerase) activity [20,21], which can modify tankyrases themselves as well as IRAP and other partners through the addition of PAR (polymers of ADP-ribose) [16]. PAR formation (PARsylation) is readily reversed through hydrolysis. Circumstantial evidence suggests that cellular PARP activity is involved in glucose homoeostasis. First, nicotinamide, a vitamin that can inhibit PARPs at pharmacological levels, is known to inhibit glucose uptake in cultured adipocytes [22]. Moreover, in knockout mice lacking either the entire tankyrase-2 protein or just the PARP domain, the only obvious phenotype is the reduction in body weight (by up to 20%) and adiposity [23,24], leading to the speculation that tankyrase-2 might regulate energy metabolism [23].
In the present study, we have investigated the role of GLUT4, IRAP and tankyrase in GSV trafficking in 3T3-L1 adipocytes. We found that GSV translocation was impaired by IRAP depletion but not by GLUT4 depletion. Interestingly, depletion of tankyrase or pharmacological inhibition of its PARP activity altered the intracellular distribution of GSVs and also attenuated their insulin-stimulated PM translocation.
MATERIALS AND METHODS
Adipocyte electroporation
3T3-L1 pre-adipocytes (American Type Culture Collection; Manassas, VA, U.S.A.) were maintained in DMEM (Dulbecco's modified Eagle's medium; Cellgro) containing 0.1% glucose and 10% fetal bovine serum (Omega Scientific) and differentiated as described in [25]. For electroporation, day 6 adipocytes were resuspended in serum-free DMEM (0.5 ml per T-75 flask) and electroporated in 650 μl aliquots with 2 nmol of siRNA (small interfering RNA) using a GenePulser Xcell (BioRad, 4-mm cuvettes, exponential protocol at 250 V and 950 μF). siRNAs were from Xeragon or IDT. The sense-strand GLUT4 siRNAs were 5′-GGTGATTGAACAGAGCTAC-3′ (G4-A), 5′-ACCCAAGGGCTGCTGTATT-3′ (G4-B) and 5′-CTGCCCGAAAGAGTCTAAA-3′ (G4-C), which began at nucleotides 309, 2451 and 875 of GenBank® NM_009204 respectively. The IRAP siRNAs were 5′-GCCCTGTTCCAGACAAACC-3′ (vp-A), 5′-GGCTGGTTGTTCCTCTTTA-3′ (vp-B), 5′-GGACGAGGA-TGAAGAGGAT-3′ (vp-C) and 5′-CCTGAGTCAGGATGTAAAT-3′ (vp-D), which began at nucleotides 2380, 2695, 267 and 1182 of GenBank® NM_172827 respectively. The tankyrase siRNAs were 5′-GAGATGCAGAGCACTATTC-3′ (T1A) and 5′-GTGCTGTCGACATGGCTCC-3′ (T1B), which began at nucleotides 3374 and 1413 of GenBank® NM_175091 respectively. The control sequences were 5′-GAGtTGCAGAGCACTAaTC-3′ (termed pmt, which differed from T1A at the two lower-cased nucleotides) and 5′-TTAGCTCGTGGGTCTCAGA-3′ (termed scr, a scrambled sequence showing no match to known genes).
Glucose uptake assays
Adipocytes were seeded in 24-well plates on day 6 and serum-starved on day 8 for 2 h as described in [26]. When indicated, PJ34 (Inotek) and sorbitol (Sigma) were added at 80 μM and 600 mM respectively for 45 min. After insulin stimulation (20 nM for 20 min), [3H]deoxy-D-glucose was added (0.1 μCi at 60 Ci/mmol; MP Biomedicals). After a 10 min incubation at 37 °C, cells were rinsed twice with PBS and solubilized in 1 M NaOH (400 μl/well). Aliquots (10 μl) were removed for protein analysis using a kit from BioRad. The remainder was subjected to scintillation counting, and the tracer uptake was normalized to the protein content.
Immunoadsorption of GLUT1-containing vesicles
Post-nuclear supernatants harvested as described below from two 10-cm plates of day 8–12 adipocytes were centrifuged at 42000 g for 75 min. The pellet was resuspended in 1 ml of HES buffer [20 mM Hepes (pH 7.5), 0.255 M sucrose and 1 mM EDTA] [27] supplemented with 120 mM NaCl, and precleared by incubating for 1 h with rabbit immunoglobulin (30 μg) and Protein A–Sepharose (240 μl, CL-4B; Pharmacia) at 4 °C. Aliquots (300 μl) of the supernatant were mixed for 1 h with a polyclonal anti-GLUT1 antibody (3 μl, Abcam No. 652, containing 30 μg of immunoglobulin) or control rabbit immunoglobulin (30 μg). After further incubation with Protein A–Sepharose (80 μl for 2 h), the immune complex was pelleted and washed four times using 120 mM NaCl/HES. The pellet was incubated for 10 min in the same NaCl/HES buffer supplemented with 2% C12E8 ([28], Fluka Biochemical), and the eluates were analysed by SDS/PAGE.
Differential centrifugation
Adipocytes electroporated on day 6 were serum-starved on day 8 for 2–3 h in DMEM/0.1% albumin (Sigma). When indicated, PJ34 (80 μM for 1 h) or insulin (20 nM for 20 min) was added prior to harvesting. Cells were scraped in HES buffer (2.2 ml/10-cm plate) at 4 °C and homogenized using a 7-ml Dounce homogenizer (Wheaton). Small aliquots were removed as whole-cell extracts. The remainder was centrifuged at 4 °C in an SW60 rotor at 19000 g for 20 min. From this spin, the pellet was processed as described in [27] to obtain the PM fraction, whereas the PNS (post-nuclear supernatant) was either separated by differential centrifugation into the cytosol fraction as well as LDMs (light microsomes) and HDMs (heavy microsomes) [27], or fractionated in iodixanol gradients (see below).
Iodixanol equilibrium sedimentation
The PNS of adipocytes described above was diluted with HES buffer to a protein concentration of 400 μg/ml. Iodixanol [4 g (60% w/v); Accurate Chemicals] was added to 10.25 g of PNS to obtain a final iodixanol concentration of 14% (w/v). Samples were loaded into OptiSeal tubes (Beckman No. 362181), and density gradients were formed at 57300 rev./min in an NVT65 rotor at 4 °C for 4 h. The tubes were then calibrated on the outside, and serial 600 μl fractions were collected from the top for immunoblotting and densitometry analysis. The area under each curve in Figures 6(B) and 7(D) was normalized to 100%. To compare basal with insulin-stimulated samples (Figure 6C), fractions (45 μl) from both samples were immunoblotted and quantified in parallel.
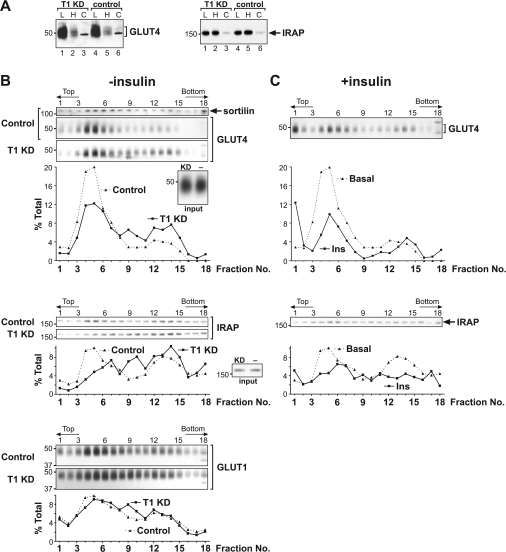
Figure 6. Effect of tankyrase knockdown on intracellular GLUT4 and IRAP distribution.
(A) PNSs of adipocytes electroporated on day 6 with T1A (lanes 1–3) or the point-mutation control pmt (lanes 4–6) and serum-starved on day 8 were subjected to differential centrifugation. From each 10 cm plate of adipocytes, 1% of the LDM (lanes 1 and 4), 5% of the HDM (lanes 2 and 5) and 1.5% of the cytosolic fraction (lanes 3 and 6) were loaded on to SDS gels for immunoblotting. The blots shown are representative of five independent experiments. (B) Day 6 adipocytes were electroporated with T1A (solid curves) or the control pmt (dotted curves) and harvested on day 8 after a 2 h serum starvation. PNS of controls and knockdowns were resolved in parallel in 14% self-forming iodixanol density gradients, and fractions were immunoblotted for the indicated proteins. The effect of the knockdown shown is representative of four batches of electroporated cells. The insets compare knockdowns (left-hand lane) with controls (right-hand lane) (20 μg of protein/lane) for the amount of GLUT4 and IRAP in the input PNS. (C) Adipocytes electroporated with the control siRNA pmt as in (B) were stimulated with 20 nM insulin for 20 min. PNS were fractionated in 14% iodixanol gradients, immunoblotted for GLUT4 and IRAP in parallel with (B), and plotted as solid curves. The dotted curves representing basal controls were taken from (B). The insulin effect shown is representative of two independent experiments. T1 KD, tankyrase 1 knockdown.
Figure 7. Effect of PJ34 on tankyrase autoPARsylation and GLUT4/IRAP trafficking.
(A) Day 8 adipocytes were treated with PJ34 (lane 1; 80 μM for 45 min) or without (lane 2). Detergent-soluble extracts were incubated with GST–IRAPaa78-109 resins to pull down tankyrase. The precipitates were sequentially immunoblotted for poly(ADP-ribose) epitopes (left-hand panel) and tankyrase (right-hand panel). (B) Serum-starved day 8 adipocytes were pretreated with 80 μM PJ34 (open bars) or without (solid bars) for 45 min. Basal and insulin-stimulated [3H]deoxy-D-glucose uptake was determined (means±S.E.M. of four wells). The result shown is representative of three other experiments using similar PJ34 concentrations for various durations. (C) Serum-starved day 8 adipocytes were treated with PJ34 (80 μM for 45 min) and then with insulin (20 nM for 20 min) as indicated. The PM content of the indicated proteins was determined as in Figure 1(C). The bar graph shows the means±S.E.M. of two basal and four insulin-stimulated samples combined from two experiments. A representative set of immunoblots is shown in the right-hand panel. (D) Serum-starved day 8 adipocytes were treated with PJ34 (80 μM for 1 h, solid curves) or without (dotted curves). PNS were fractionated in 14% iodixanol gradients as in Figure 6, and the immunoblots were quantified also as in Figure 6. The PJ34 effect shown is representative of three independent experiments. PJ, PJ34; Ctrl, control.
Affinity precipitation of tankyrase
To concentrate tankyrase-1 and -2 for immunoblotting (Figures 3A and 7A), adipocytes were lysed in buffer A as described in [16] containing 1% Triton X-100 and clarified at 13000 g for 10 min. Equal protein amounts were incubated with GST (glutathione S-transferase)–IRAPaa78-109 resins (15 μg, [16]) at 4 °C for at least 2 h to pull down both tankyrases.
Figure 3. Tankyrase knockdown impairs glucose uptake reversibly and dose-dependently.
Adipocytes were electroporated on day 6 with a tankyrase siRNA (T1A or T1B), a point-mutation control (Pmt) or buffer alone (Buff). (A) Extracts prepared on day 8 (lanes 1–6) or day 9 (lanes 7–9) were immunoblotted for tankyrase (upper panels) and tankyrase-2 (lower panels). (B) [3H]Deoxy-D-glucose uptake in the basal state and after insulin stimulation (20 nM for 20 min) was determined on day 8 (left-hand panel) or day 9 (right-hand panel) as in Figure 1(B). The results shown are representative of four (day 8) or two (day 9) experiments. TNKS-1, tankyrase-1; TNKS-2, tankyrase-2.
Immunoblotting and statistical analyses
Primary antibodies were directed against tankyrase (1 μg/ml H-350; Santa Cruz); tankyrase-2 (10 μg/ml, T12; [16]); IRAP (1:12000 [16]); GLUT4 (5 μg/ml, 1F8; [16]); GLUT1 (1:1000, ab652; Abcam); TfR (transferrin receptor; 1:1000; Zymed); caveolin-1 and phospho-tyrosine (both at 1:1000; Transduction Lab); ras (0.5 μg/ml; Transduction Lab); AS160 (1:500; Abcam); poly(ADP-ribose) (1μg IgY/ml; Tulip BioLabs); IRS-1 (insulin receptor substrate-1; Upstate); IRβ (insulin receptor β; C19; Santa Cruz); actin (1:2000; Sigma); sortilin (1:500; [29]); GSK (glycogen synthase kinase)-3 (Ser9), phospho-Akt S473, and PAS (phospho-specific Akt substrate), the latter three were from Cell Signaling and used at 1:1000. The immunoblots were quantified by densitometry using Kodak 1D image analysis software. The intensity of each band was normalized to the lane representing the control (adipocytes not subjected to knockdown, or insulin or PJ34 treatment) prior to being pooled between experiments for statistical analyses. P values were calculated using the Student's unpaired two-tailed t test.
RESULTS
IRAP translocation remains intact despite substantial GLUT4 depletion
To explore how GSVs interact with their targeting machinery, we electroporated 3T3-L1 adipocytes with siRNA to knock down GLUT4 and examined the impact on GSV trafficking. The electroporation was performed typically on day 6 of adipogenesis, since more mature adipocytes exhibited a lower viability during the procedure. We found that two independent siRNAs, G4-A and G4-B, caused ∼80% GLUT4 depletion in whole-cell extracts harvested on day 8, but had no effect on GLUT1, caveolin-1 or actin (Figure 1A). Interestingly, both G4-A and G4-B as well as a third GLUT4 siRNA (G4-C, results not shown) caused a mild decrease in IRAP expression, a trend that mirrored the IRAP down-regulation in GLUT4 knockout mice [6–8] and presumably reflected IRAP destabilization due to GLUT4 deficiency. As expected, GLUT4 knockdown decreased insulin-stimulated glucose uptake by 35–40% while having little effect on basal glucose uptake (Figure 1B), consistent with GLUT4 mediating only a minor portion of basal uptake [30].
Figure 1. GLUT4 knockdown does not impair IRAP translocation.
3T3-L1 adipocytes were electroporated on day 6 of differentiation with either a GLUT4 siRNA (G4A or G4B), a scrambled siRNA (Scr) or buffer alone (buf). Cells were insulin-stimulated as indicated (20 nM for 20 min) on day 8 prior to analysis. (A) Whole-cell extracts were immunoblotted (right-hand panel, 30 μg protein/lane) for the indicated proteins. The bar graph to the left shows densitometry quantification (means±S.E.M.) of the knockdown samples (mean of lanes 2, 4 and 6) normalized against the control (mean of lanes 1, 3 and 5). (B) [3H]Deoxy-D-glucose uptake in the basal state and after insulin stimulation was normalized against the protein content and shown in arbitrary units (a.u.). Each data point represents the means±S.E.M. of four replicates. (C) The PM fractions were immunoblotted for the indicated proteins (right-hand panel, 15 μg protein/lane), quantified by densitometry, and normalized against the unstimulated control (lane 3). The bar graph to the left shows the mean of insulin-stimulated knockdowns (lanes 2 and 6) and the mean of insulin-stimulated controls (lanes 1 and 5). *P<0.05 from the control; N.S., not significantly different. Each panel was repeated once (A and C) or twice (B) with similar results. Ctrl, control; KD, knockdown.
Next, to assess GSV translocation, we immunoblotted the PM fraction for IRAP, an established GSV marker [2]. Despite the tendency to express less IRAP than control, GLUT4-knockdown adipocytes recruited significantly more IRAP to the PM after insulin stimulation (Figure 1C, lanes 2 and 6 compared with lanes 1 and 5). A similar trend has been observed in fat-specific GLUT4 knockout mice, where adipocytes stimulated ex vivo with insulin recruit more IRAP to the PM than wild-type controls (50% compared with 31% of total IRAP) [8]. Figure 1(C) also shows that GLUT4 knockdown did not impair the PM targeting of GLUT1 or caveolin-1. Thus substantial GLUT4 depletion did not attenuate insulin-stimulated PM translocation of IRAP and, by inference, GSVs.
IRAP knockdown impairs GLUT4 translocation
In a reciprocal experiment, we explored whether GSV trafficking is dependent on the presence of IRAP. Of the four siRNAs designed against IRAP (also known as vp-165), vp-A and vp-B achieved knockdown most effectively (Figure 2A lanes 2 and 3, and results not shown). Compared with a scrambled siRNA (Scr, lane 4) or buffer alone (buff, lane 1), the glucose uptake after vp-A or vp-B electroporation was normal in the basal state but was reduced in the insulin-stimulated state by ∼45% (Figure 2A, graph). A modest reduction of insulin-stimulated glucose uptake was also observed using two other less effective IRAP siRNAs (vp-C and -D, results not shown). Therefore IRAP knockdown attenuated insulin-stimulated glucose uptake in adipocytes. This effect was not due to decreased GLUT4 or GLUT1 expression (Figure 2B).
Figure 2. IRAP knockdown attenuates insulin-stimulated translocation of GLUT4 and GLUT1.
(A) Adipocytes were electroporated on day 6 with buffer alone (Buff), vp-A, vp-B or a scrambled control (Scr) (lanes 1–4 respectively). Whole-cell extracts (30 μg of protein) on day 8 were immunoblotted for IRAP and GLUT4 (upper panels). The bar graph shows basal and insulin-stimulated glucose uptake determined as in Figure 1(B). The experiment was repeated three times with similar results. (B) Whole-cell extracts of adipocytes electroporated with a scrambled siRNA (Scr, lane 1) or vp-A (lane 2) were immunoblotted (30 μg protein/lane) for the indicated proteins, quantified by densitometry and normalized to lane 1. The bar graph shows the means±S.E.M. of triplicates. *P<0.05. (C) Adipocytes electroporated with a scrambled siRNA (Scr, lanes 1 and 3) or vp-A (lanes 2 and 4) were stimulated with insulin (20 nM for 20 min) as indicated. The PM fractions (15 μg protein/lane) were immunoblotted for the indicated proteins. The bar graph shows the means±S.E.M. of two sets of independent experiments, each normalized to lane 1 prior to being pooled for analysis. (D) Membrane fractions of adipocytes prepared as described in the Materials and methods section were immunoblotted for IRAP and TfR (2.5, 1.5 and 1 μl in lanes 3–5 respectively), and 300 μl aliquots were immunoprecipitated (IP) using GLUT1 antiserum (lanes 1 and 6) or control immunoglobulin (lanes 2 and 7). The supernatants were immunoblotted for GLUT1 and tubulin (30 μl, lanes 1 and 2), whereas the precipitates were eluted with 300 μl of 2% C12E8 and immunoblotted for IRAP and TfR (20 μl eluate/lane). The results shown are representative of four independent experiments performed at slightly varied stoichiometry. (E) Adipocytes electroporated with vp-A (lanes 1 and 2) or a scrambled siRNA (Scr, lanes 3 and 4) were stimulated with insulin (20 nM for 20 min) as indicated. Whole-cell extracts were immunoblotted using antibodies against AS160 (left-hand panel) or phospho-Akt substrates (right-hand panel). Ctrl, control; KD, knockdown.
To explore how IRAP knockdown affected glucose uptake, we purified the PM fraction of adipocytes transfected with vp-A or a scrambled siRNA. Figure 2(C) shows that IRAP knockdown attenuated insulin-stimulated GLUT4 translocation to the PM (lane 3 compared with 4), indicating that the translocation depended in part on the presence of IRAP. This effect was specific, since the PM targeting of the TfR was not affected (lane 3 compared with 4). Unexpectedly, IRAP knockdown did impair GLUT1 translocation to the PM (Figure 2C), an effect that probably contributed to the decreased glucose uptake in these cells (Figure 2A).
A plausible explanation for why IRAP knockdown impaired the translocation of GLUT1 but not TfR would be preferential IRAP–GLUT1 co-localization in vesicular compartments, leading to partially linked trafficking of the two proteins. This notion is consistent with the report that insulin-stimulated increase of GLUT1 in PM (6-fold) approaches the 8–14-fold increase in GLUT4/IRAP and exceeds the approx. 3-fold increase in TfR (for references, see [29]). The co-localization of GLUT1 with GLUT4/IRAP is also supported by the reported recovery of 85% of cellular GLUT1 in vesicles immunoadsorbed using anti-GLUT4 antibodies [31]. To directly assess IRAP–GLUT1 co-localization, we immunoadsorbed GLUT1-containing vesicles using an antibody that recognized the GLUT1 N-terminal cytosolic tail. This resulted in substantial depletion of GLUT1 from the input material (Figure 2D, lane 1 compared with lane 2). (We were unable to directly demonstrate immunoprecipitated GLUT1 due to interference from the immunoglobulin heavy chain on Western blots.) Next, we used the non-ionic detergent C12E8 [28] to elute co-purified proteins from immunoadsorbed GLUT1 vesicles. By comparing the IRAP-to-TfR ratio between the eluates and the input, Figure 2(D) also shows that GLUT1 vesicles were enriched for IRAP by approx. 2.5-fold over TfR. Thus the substantial presence of IRAP in GLUT1 vesicles might underlie to some extent the impaired GLUT1 translocation following IRAP knockdown.
To show that the decreased translocation of GLUT4 and GLUT1 upon IRAP knockdown was not due to inhibition of insulin signalling, we assessed the signalling using Akt-mediated phosphorylation as a readout. Of particular interest is AS160, an IRAP-binding protein [10] whose phosphorylation by Akt is implicated in GSV translocation [32]. Figure 2(E) shows that IRAP knockdown did not affect insulin-induced gel-mobility shift of AS160 (left-hand panel), nor did it affect insulin-stimulated phosphorylation of prominent Akt substrates, including a band that co-migrated with AS160, and a 250 kDa band that was presumably the Akt substrate AS250 [33] (right-hand panel). Therefore IRAP knockdown apparently did not affect insulin-Akt signalling.
Tankyrase knockdown attenuates insulin-stimulated glucose uptake
Next, we investigated whether GSV translocation was modulated by the IRAP-binding protein tankyrase [16]. To knockdown tankyrase, we compared eight siRNAs and found T1A and T1B to be the most effective. We also designed the control siRNA, pmt, by introducing point mutations into T1A. Figure 3(A) shows that in adipocytes electroporated on day 6 and assayed on day 8, T1A achieved >90% tankyrase depletion (lane 2, upper panel) whereas the control siRNA pmt had no effect (lane 3). The knockdown by T1A was stable through to at least day 9 (results not shown). By comparison, T1B achieved only a modest knockdown on day 8 (lane 5), an effect essentially lost by day 9 (lane 8). Neither T1A nor T1B affected the expression of tankyrase-2 (lower panel). Importantly, insulin-stimulated glucose uptake on day 8 was attenuated robustly by T1A and modestly by T1B (Figure 3B, left-hand panel), indicating a dose-dependent tankyrase effect on glucose uptake.
Tankyrase knockdown conceivably could have attenuated glucose uptake by blocking adipogenic differentiation, since our adipocytes were electroporated on day 6, prior to completing adipogenesis. Arguing against this possibility is that neither T1A nor T1B affected the expression of the adipogenesis markers PPARγ (peroxisome-proliferator-activated receptor γ) and adiponectin (results not shown). Moreover, as tankyrase recovered from the knockdown by day 9 in T1B-electroporated cells (Figure 3A, lane 8), glucose uptake normalized concomitantly (Figure 3B, right-hand panel). This phenotypic reversibility suggested that tankyrase knockdown did not interfere with adipogenesis.
Tankyrase knockdown inhibits GLUT4 and IRAP translocation
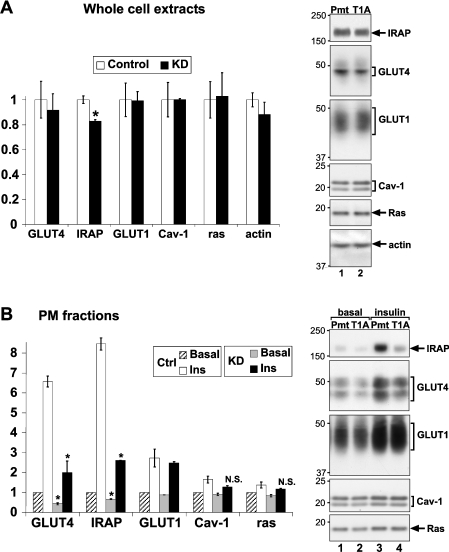
To investigate how tankyrase knockdown impaired glucose uptake, we first examined the overall expression of GLUT4 and GLUT1. Figure 4(A) shows that the expression of neither transporter was affected by tankyrase knockdown. Next, we purified the PM fraction of basal and insulin-stimulated adipocytes, and assessed GLUT4 and GLUT1 translocation by immunoblotting. Figure 4(B) shows that tankyrase knockdown impaired the insulin-stimulated PM translocation of GLUT4 (lane 3 compared with 4) but not GLUT1, caveolin-1 or ras. (The translocation of caveolin and ras as previously reported [34,35] was not always discernible in our hands.) As for IRAP, the PM content after insulin stimulation was robustly decreased by tankyrase knockdown (Figure 4B, lane 4 compared with lane 3) while the overall expression was only slightly decreased (Figure 4A), confirming that IRAP translocation was impaired. Therefore tankyrase knockdown specifically impaired the translocation of the major GSV cargo proteins, GLUT4 and IRAP.
Figure 4. Tankyrase knockdown impairs the translocation of IRAP and GLUT4 but not GLUT1.
Day 6 adipocytes were electroporated with T1A siRNA or a point-mutation control (Pmt). On day 8, (A) whole-cell extracts (30 μg protein/lane) and (B) the PM fractions (15 μg protein/lane) were immunoblotted and quantified as described in Figure 2(C). The bar graph to the left shows the means±S.E.M. of two basal samples and of four insulin-stimulated samples. *P<0.05. This experiment was repeated twice with similar results.
Tankyrase knockdown does not affect insulin-induced phosphorylation
A potential mechanism whereby tankyrase knockdown could impair GLUT4/IRAP translocation is through inhibiting insulin signalling, since tankyrase interacts with Grb14 [18], an insulin receptor adapter implicated in glucose homoeostasis [19], and also with PP1 (protein phosphatase 1) [17], a phosphatase that could conceivably modulate signalling. Arguing against this mechanism is that the knockdown did not prevent insulin signalling from recruiting GLUT1, ras and caveolin-1 to the PM (Figure 4B), nor did the knockdown attenuate insulin-induced phosphorylation of the IR, IRS-1, Akt, GSK3 and p42/p44 ERKs (extracellular-signal-regulated kinases) (Figure 5A, lane 4 compared with lane 2). Moreover, the induction of glucose uptake by osmotic shock, which signals through a non-insulin pathway [5], was also impaired by the knockdown (Figure 5B), supporting the notion that tankyrase knockdown impaired GSV formation or trafficking rather than blocking upstream insulin signalling.
Figure 5. Insulin-induced phosphorylation is preserved but hypertonicity-induced glucose uptake is impaired in tankyrase-depleted adipocytes.
(A) Day 6 adipocytes were electroporated with T1A (lanes 3 and 4) or the point-mutation control Pmt (lanes 1 and 2). After insulin stimulation (20 nM for 30 min) as indicated (lanes 2 and 4) on day 8, extracts were immunoblotted for the IR, other signalling molecules as indicated, and their phosphorylated forms (30 μg protein/lane). (B) Adipocytes electroporated on day 6 with T1A (open bars) or the point-mutation control Pmt (solid bars) were assayed on day 8 for [3H]deoxy-D-glucose uptake in the basal state and after osmotic shock (600 mM sorbitol for 20 min). The average uptake of 6 wells (means±S.E.M.) is shown. This study was repeated once with similar results. ERK, extracellular-signal-regulated kinase.
Tankyrase knockdown alters intracellular GLUT4/IRAP distribution
We suspected that the Golgi-associated tankyrase modulated GLUT4/IRAP sorting into exocytosis-competent compartments, rather than directly affecting the exocytosis from these compartments. This is because tankyrase does not move with GSVs to the PM upon insulin stimulation [16] and because vesicular sorting at the Golgi complex and the trans-Golgi network is an integral step of intracellular GSV itinerary [1,36–38]. We therefore hypothesized that tankyrase knockdown altered the basal-state distribution of GLUT4 and IRAP within endosomal compartments. To test this idea, we used differential centrifugation to separate PNSs of serum-starved adipocytes into the cytosolic fraction as well as HDMs and LDMs. We found that tankyrase knockdown apparently did not alter the partitioning of GLUT4 and IRAP among these fractions (Figure 6A); regardless of the knockdown, both proteins were highly enriched in LDMs. (For this reason, a lower percentage of LDMs was loaded in Figure 6A than the other fractions.)
Because the differential centrifugation shown in Figure 6(A) yielded relatively few fractions on the basis of differences in sedimentation rate, it might not detect a subtle redistribution of proteins. We therefore turned to iodixanol (OptiPrep) equilibrium density gradients, which resolved microsomes into 18 fractions on the basis of buoyant density instead of sedimentation rate. This method has been used to separate GLUT4 in LDMs into a denser ‘peak 1’ and a lighter ‘peak 2’, each containing 44% and 39% respectively of the input GLUT4 [39,40]. Peak 1 shows a greater depletion of GLUT4 upon insulin stimulation (44% compared with 25%), whereas peak 2 is characterized by a higher sortilin content [39]. In the present study, the iodixanol gradients of serum-starved control adipocytes showed a major GLUT4 peak near the top of the gradient and a minor peak near the bottom (Figure 6B, top panel, dotted curve). The major peak (centred about fraction 5) was sortilin-rich and thus presumably corresponded to peak 2 of previous studies [39,40]. Whether our minor GLUT4 peak (centred about fraction 13) corresponded to peak 1 of previous studies was unclear, since its GLUT4 content was much lower than would have been expected of peak 1. Figure 6(B) also shows that the IRAP distribution in control adipocytes, albeit more even than GLUT4, also peaked at fractions 5 and 13 (middle panel, dotted curve). Importantly, after tankyrase knockdown, both GLUT4 and IRAP were substantially depleted from the lighter peak and shifted toward the middle and the bottom of the gradients (Figure 6B, top two panels, solid curves). These changes were specific, since the GLUT1 profile in the same gradient was largely unaffected (Figure 6B, lower panel). Thus tankyrase knockdown specifically affected the density profile of compartments that harboured GLUT4 and IRAP in the basal state, resulting in an overall increase in the buoyant density of these compartments. This tankyrase effect was highly reproducible in four batches of electroporated cells; however, the profiles themselves were somewhat variable between experiments.
Since tankyrase knockdown shifted GLUT4 and IRAP away from the lighter fractions of iodixanol gradients (Figure 6B) and also impaired their insulin-stimulated translocation (Figure 4B), we suspected that the lighter fractions might contain GLUT4 and IRAP that were highly insulin-responsive and exocytosis-competent. Indeed, when control adipocytes were stimulated with insulin, the greatest depletion of intracellular GLUT4 and IRAP was from the lighter fractions and particularly fraction 5 (Figure 6C, solid curves compared with dotted curves), consistent with these fractions harbouring highly exocytosis-competent GSVs. A caveat is that intracellular redistribution, rather than exocytosis, could also deplete a protein from a given fraction. Nevertheless, our findings are consistent with the notion that altered basal-state GLUT4/IRAP partitioning within endosomal compartments (Figure 6B) contributed to their impaired translocation upon insulin stimulation (Figure 4B).
PARP inhibition affects GSV trafficking
Tankyrase has a catalytic activity that can PARsylate (poly-ADP-ribosylate) itself and, at least in vitro, IRAP [16], raising the possibility that tankyrase might modulate GSV trafficking by PARsylating either the cargo or the trafficking machinery. This was evaluated by treating adipocytes with the general PARP inhibitor PJ34 [41,42]. To confirm that PJ34 inhibited tankyrase-mediated PARsylation, we used GST–IRAP resins to affinity-precipitate tankyrase for immunoblotting with anti-PAR antibodies. The immunoblot in Figure 7(A) shows that PJ34 (80 μM for 1 h) abolished tankyrase autoPARsylation as expected (lane 1 compared with lane 2, left-hand panel). Of note, tankyrase typically exhibited a slower gel mobility on anti-PAR blots than on anti-tankyrase blots due to the bulky nature of PAR modification (Figure 7A, lane 2, left-hand panel compared with right-hand panel). Importantly, we found that PJ34 substantially attenuated insulin-stimulated glucose uptake (Figure 7B) and the PM translocation of both GLUT4 and IRAP (Figure 7C). This latter effect was specific, since GLUT1 translocation remained intact (Figure 7C). To draw additional analogy between PARP inhibition and tankyrase knockdown, we used iodixanol gradients to examine the effect of PJ34 treatment (80 μM for 1 h) on serum-starved adipocytes. Figure 7(D) shows that this treatment partially depleted GLUT4 (left-hand panel) and IRAP (right-hand panel) from the lighter peak at fraction 5 and redistributed them toward the denser portion of the gradients.
DISCUSSION
We have investigated the regulation of GLUT4 and IRAP trafficking in 3T3-L1 adipocytes by combining siRNA-mediated knockdown with subcellular fractionation. Our data indicate that substantial depletion of GLUT4 does not affect the insulin-stimulated translocation of IRAP. In contrast, the presence of IRAP is important to insulin-stimulated GLUT4 translocation and glucose uptake. Moreover, the intracellular distribution of GLUT4/IRAP and their insulin-stimulated translocation are regulated by the IRAP-binding protein tankyrase.
In our GLUT4-depleted adipocytes, the intact IRAP translocation (Figure 1C) is in agreement with the robust IRAP translocation reported in tissue-specific GLUT4 knockout mice [7,8] as well as in adipocytes derived from NIH-3T3 cells without inducing GLUT4 expression [43]. These findings collectively suggest that GLUT4 is dispensable for physiological trafficking of IRAP and, by inference, GSVs. Although GLUT4 is known to interact with components of the GSV targeting machinery [44–46], we suspect that GSVs lacking GLUT4 can nevertheless interact with the targeting machinery by utilizing IRAP as a handle.
In IRAP-depleted adipocytes, the impaired insulin-stimulated GLUT4 translocation and glucose uptake (Figures 2A and 2C) raise the possibility that IRAP contributes to the insulin responsiveness of GSVs, whereas GLUT4 may be merely a passenger therein. This notion is supported by direct IRAP interaction with multiple regulators of GSV trafficking [10–13]. However, the impaired GLUT4 translocation in IRAP-knockdown adipocytes (Figure 2C) is in sharp contrast with the normal GLUT4 translocation in adipocytes isolated from IRAP-null mice [9]. This phenotypic difference may have several explanations. First, unlike transient knockdown in 3T3-L1 adipocytes, life-long IRAP deficiency may allow mice to develop compensatory mechanism(s) that normalize GLUT4 trafficking and glucose homoeostasis. Secondly, the development of this putative compensatory mechanism might require complete absence of IRAP, which occurs only in the mouse model.
Given that GSV translocation apparently depended on IRAP (Figure 2C), we investigated whether the translocation was modulated by the IRAP partner tankyrase [16]. We found that upon tankyrase knockdown, the insulin-stimulated translocation of IRAP and GLUT4 as well as glucose uptake were attenuated (Figures 3B and 4B). We therefore speculate that GLUT4/IRAP sorting into exocytosis-competent compartments is promoted by the Golgi-associated tankyrase. This notion is in line with the observations that the Golgi constitutes part of the recycling path of GLUT4/IRAP and that the Golgi-associated coat protein GGA promotes the entry of nascent GLUT4/IRAP into exocytosis-competent compartments [1,36–38]. Although the exact sorting step(s) regulated by tankyrase remains unclear, it might involve GLUT4/IRAP movement between compartments that exhibit different buoyant densities in iodixanol gradients. This would explain why tankyrase depletion apparently traps GLUT4/IRAP in compartments of relatively high buoyant densities (Figure 6B), presumably by blocking their entry into lower-density compartments that are highly insulin responsive (Figure 6C). Whether tankyrase modulates GSV trafficking by binding to IRAP remains to be demonstrated. Besides tankyrase, several known regulators of GSV trafficking also bind to IRAP [10–13]. Thus IRAP probably has additional roles in vesicular trafficking that are tankyrase-independent. Supporting this notion is that GLUT1 translocation is impaired by IRAP knockdown (Figure 2C) but not by tankyrase knockdown (Figure 4B).
Since the effects of tankyrase knockdown on intracellular GLUT4/IRAP distribution (Figure 6B) and PM translocation (Figure 4B) could be reproduced using the PARP inhibitor PJ34 (Figure 7), we speculate that tankyrase uses its PARP activity to modulate GSV trafficking. This activity can modify the IRAP cytosolic domain in vitro [16], raising the possibility that tankyrase PARsylates IRAP to generate a reversible tag on GSVs that guides their sorting, much like phosphorylation- and ubiquitin-directed sorting of other cargo proteins [47].
Acknowledgments
We are grateful to Dr Susanna Keller (Department of Medicine, University of Virginia, Charlottesville, VA, U.S.A.) for insightful comments, and to Dr Gustav Lienhard (Department of Biochemistry, Dartmouth Medical School, Hanover, NH, U.S.A.) for the sortilin antibody. This work was supported by an American Diabetes Association award (7-05-CD-05) to N.-W. C.
References
- 1.Bryant N. J., Govers R., James D. E. Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell. Biol. 2002;3:267–277. doi: 10.1038/nrm782. [DOI] [PubMed] [Google Scholar]
- 2.Keller S. R. The insulin-regulated aminopeptidase: a companion and regulator of GLUT4. Front. Biosci. 2003;8:S410–S420. doi: 10.2741/1078. [DOI] [PubMed] [Google Scholar]
- 3.Ducluzeau P. H., Fletcher L. M., Vidal H., Laville M., Tavare J. M. Molecular mechanisms of insulin-stimulated glucose uptake in adipocytes. Diabetes Metab. 2002;28:85–92. [PubMed] [Google Scholar]
- 4.Mora S., Pessin J. E. An adipocentric view of signaling and intracellular trafficking. Diabetes Metab. Res. Rev. 2002;18:345–356. doi: 10.1002/dmrr.321. [DOI] [PubMed] [Google Scholar]
- 5.Chen D., Elmendorf J. S., Olson A. L., Li X., Earp H. S., Pessin J. E. Osmotic shock stimulates GLUT4 translocation in 3T3L1 adipocytes by a novel tyrosine kinase pathway. J. Biol. Chem. 1997;272:27401–27410. doi: 10.1074/jbc.272.43.27401. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H., Li J., Katz E. B., Charron M. J. GLUT4 ablation in mice results in redistribution of IRAP to the plasma membrane. Biochem. Biophys. Res. Commun. 2001;284:519–525. doi: 10.1006/bbrc.2001.4994. [DOI] [PubMed] [Google Scholar]
- 7.Abel E. D., Graveleau C., Betuing S., Pham M., Reay P. A., Kandror V., Kupriyanova T., Xu Z., Kandror K. V. Regulation of insulin-responsive aminopeptidase expression and targeting in the insulin-responsive vesicle compartment of glucose transporter isoform 4-deficient cardiomyocytes. Mol. Endocrinol. 2004;18:2491–2501. doi: 10.1210/me.2004-0175. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho E., Schellhorn S. E., Zabolotny J. M., Martin S., Tozzo E., Peroni O. D., Houseknecht K. L., Mundt A., James D. E., Kahn B. B. GLUT4 overexpression or deficiency in adipocytes of transgenic mice alters the composition of GLUT4 vesicles and the subcellular localization of GLUT4 and insulin-responsive aminopeptidase. J. Biol. Chem. 2004;279:21598–21605. doi: 10.1074/jbc.M312269200. [DOI] [PubMed] [Google Scholar]
- 9.Keller S. R., Davis A. C., Clairmont K. B. Mice deficient in the insulin- regulated membrane aminopeptidase show substantial decreases in glucose transporter GLUT4 levels but maintain normal glucose homeostasis. J. Biol. Chem. 2002;277:17677–17686. doi: 10.1074/jbc.M202037200. [DOI] [PubMed] [Google Scholar]
- 10.Larance M., Ramm G., Stockli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- 11.Hosaka T., Brooks C. C., Presman E., Kim S. K., Zhang Z., Breen M., Gross D. N., Sztul E., Pilch P. F. p115 interacts with the GLUT4 vesicle protein, IRAP, and plays a critical role in insulin-stimulated GLUT4 translocation. Mol. Biol. Cell. 2005;16:2882–2890. doi: 10.1091/mbc.E05-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tojo H., Kaieda I., Hattori H., Katayama N., Yoshimura K., Kakimoto S., Fujisawa Y., Presman E., Brooks C. C., Pilch P. F. The Formin family protein, formin homolog overexpressed in spleen, interacts with the insulin-responsive aminopeptidase and profilin IIa. Mol. Endocrinol. 2003;17:1216–1229. doi: 10.1210/me.2003-0056. [DOI] [PubMed] [Google Scholar]
- 13.Katagiri H., Asano T., Yamada T., Aoyama T., Fukushima Y., Kikuchi M., Kodama T., Oka Y. Acyl-coenzyme A dehydrogenases are localized on GLUT4-containing vesicles via association with insulin-regulated aminopeptidase in a manner dependent on its dileucine motif. Mol. Endocrinol. 2002;16:1049–1059. doi: 10.1210/mend.16.5.0831. [DOI] [PubMed] [Google Scholar]
- 14.Waters S. B., D'Auria M., Martin S. S., Nguyen C., Kozma L. M., Luskey K. L. The amino terminus of insulin-responsive aminopeptidase causes GLUT4 translocation in 3T3-L1 adipocytes. J. Biol. Chem. 1997;272:23323–23327. doi: 10.1074/jbc.272.37.23323. [DOI] [PubMed] [Google Scholar]
- 15.Altan-Bonnet N., Sougrat R., Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Chi N. W., Lodish H. F. Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 17.Sbodio J. I., Chi N. W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RxxPDG motif and is a novel tankyrase partner. J. Biol. Chem. 2002;277:31887–31892. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- 18.Lyons R. J., Deane R., Lynch D. K., Ye Z. S., Sanderson G. M., Eyre H. J., Sutherland G. R., Daly R. J. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 2001;276:17172–17180. doi: 10.1074/jbc.M009756200. [DOI] [PubMed] [Google Scholar]
- 19.Cooney G. J., Lyons R. J., Crew A. J., Jensen T. E., Molero J. C., Mitchell C. J., Biden T. J., Ormandy C. J., James D. E., Daly R. J. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J. 2004;23:582–593. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sbodio J. I., Lodish H. F., Chi N. W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem. J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 22.Grunfeld C., Shigenaga J. K. Nicotinamide and other inhibitors of ADP-ribosylation block deoxyglucose uptake in cultured cells. Biochem. Biophys. Res. Commun. 1984;123:785–791. doi: 10.1016/0006-291x(84)90298-5. [DOI] [PubMed] [Google Scholar]
- 23.Chiang Y. J., Nguyen M. L., Gurunathan S., Kaminker P., Tessarollo L., Campisi J., Hodes R. J. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol. Cell. Biol. 2006;26:2037–2043. doi: 10.1128/MCB.26.6.2037-2043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao S. J., Poitras M. F., Cook B. D., Liu Y., Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol. Cell. Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen M. T., Satoh H., Favelyukis S., Babendure J. L., Imamura T., Sbodio J. I., Zalevsky J., Dahiyat B. I., Chi N. W., Olefsky J. M. JNK and tumor necrosis factor-α mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:35361–35371. doi: 10.1074/jbc.M504611200. [DOI] [PubMed] [Google Scholar]
- 26.Janez A., Worrall D. S., Olefsky J. M. Insulin-mediated cellular insulin resistance decreases osmotic shock-induced glucose transport in 3T3-L1 adipocytes. Endocrinology. 2000;141:4657–4663. doi: 10.1210/endo.141.12.7801. [DOI] [PubMed] [Google Scholar]
- 27.Piper R. C., Hess L. J., James D. E. Differential sorting of two glucose transporters expressed in insulin-sensitive cells. Am. J. Physiol. 1991;260:C570–C580. doi: 10.1152/ajpcell.1991.260.3.C570. [DOI] [PubMed] [Google Scholar]
- 28.Mastick C. C., Aebersold R., Lienhard G. E. Characterization of a major protein in GLUT4 vesicles. Concentration in the vesicles and insulin-stimulated translocation to the plasma membrane. J. Biol. Chem. 1994;269:6089–6092. [PubMed] [Google Scholar]
- 29.Morris N. J., Ross S. A., Lane W. S., Moestrup S. K., Petersen C. M., Keller S. R., Lienhard G. E. Sortilin is the major 110-kDa protein in GLUT4 vesicles from adipocytes. J. Biol. Chem. 1998;273:3582–3587. doi: 10.1074/jbc.273.6.3582. [DOI] [PubMed] [Google Scholar]
- 30.Liao W., Nguyen M. T., Imamura T., Singer O., Verma I. M., Olefsky J. M. Lentiviral shRNA-mediated knockdown of GLUT4 in 3T3-L1 adipocytes. Endocrinology. 2006;47:2245–2252. doi: 10.1210/en.2005-1638. [DOI] [PubMed] [Google Scholar]
- 31.Calderhead D. M., Kitagawa K., Tanner L. I., Holman G. D., Lienhard G. E. Insulin regulation of the two glucose transporters in 3T3-L1 adipocytes. J. Biol. Chem. 1990;265:13801–13808. [PubMed] [Google Scholar]
- 32.Sano H., Kane S., Sano E., Miinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 33.Gridley S., Chavez J. A., Lane W. S., Lienhard G. E. Adipocytes contain a novel complex similar to the tuberous sclerosis complex. Cell. Signalling. 2006;18:1626–1632. doi: 10.1016/j.cellsig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Kandror K. V., Stephens J. M., Pilch P. F. Expression and compartmentalization of caveolin in adipose cells: coordinate regulation with and structural segregation from GLUT4. J. Cell. Biol. 1995;129:999–1006. doi: 10.1083/jcb.129.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goalstone M. L., Draznin B. Effect of insulin on farnesyltransferase activity in 3T3-L1 adipocytes. J. Biol. Chem. 1996;271:27585–27589. doi: 10.1074/jbc.271.44.27585. [DOI] [PubMed] [Google Scholar]
- 36.Slot J. W., Garruti G., Martin S., Oorschot V., Posthuma G., Kraegen E. W., Laybutt R., Thibault G., James D. E. Glucose transporter (GLUT-4) is targeted to secretory granules in rat atrial cardiomyocytes. J. Cell. Biol. 1997;137:1243–1254. doi: 10.1083/jcb.137.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shewan A. M., van Dam E. M., Martin S., Luen T. B., Hong W., Bryant N. J., James D. E. GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in syntaxins 6 and 16 but not TGN38: involvement of an acidic targeting motif. Mol. Biol. Cell. 2003;14:973–986. doi: 10.1091/mbc.E02-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson R. T., Khan A. H., Furukawa M., Hou J. C., Li L., Kanzaki M., Okada S., Kandror K. V., Pessin J. E. Entry of newly synthesized GLUT4 into the insulin-responsive storage compartment is GGA dependent. EMBO J. 2004;23:2059–2070. doi: 10.1038/sj.emboj.7600159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashiramoto M., James D. E. Characterization of insulin-responsive GLUT4 storage vesicles isolated from 3T3-L1 adipocytes. Mol. Cell. Biol. 2000;20:416–427. doi: 10.1128/mcb.20.1.416-427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perera H. K., Clarke M., Morris N. J., Hong W., Chamberlain L. H., Gould G. W. Syntaxin 6 regulates GLUT4 trafficking in 3T3-L1 adipocytes. Mol. Biol. Cell. 2003;14:2946–2958. doi: 10.1091/mbc.E02-11-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seimiya H., Muramatsu Y., Ohishi T., Tsuruo T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell. 2005;7:25–37. doi: 10.1016/j.ccr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 42.Suarez-Pinzon W. L., Mabley J. G., Power R., Szabo C., Rabinovitch A. Poly (ADP-ribose) polymerase inhibition prevents spontaneous and recurrent autoimmune diabetes in NOD mice by inducing apoptosis of islet-infiltrating leukocytes. Diabetes. 2003;52:1683–1688. doi: 10.2337/diabetes.52.7.1683. [DOI] [PubMed] [Google Scholar]
- 43.Gross D. N., Farmer S. R., Pilch P. F. GLUT4 storage vesicles without GLUT4: transcriptional regulation of insulin-dependent vesicular traffic. Mol. Cell. Biol. 2004;24:7151–7162. doi: 10.1128/MCB.24.16.7151-7162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giorgino F., de Robertis O., Laviola L., Montrone C., Perrini S., McCowen K. C., Smith R. J. The sentrin-conjugating enzyme mUbc9 interacts with GLUT4 and GLUT1 glucose transporters and regulates transporter levels in skeletal muscle cells. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1125–1130. doi: 10.1073/pnas.97.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bogan J. S., Hendon N., McKee A. E., Tsao T. S., Lodish H. F. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature. 2003;425:727–733. doi: 10.1038/nature01989. [DOI] [PubMed] [Google Scholar]
- 46.Jung C. Y. Proteins that interact with facilitative glucose transporters: implication for function. Exp. Physiol. 1998;83:267–273. doi: 10.1113/expphysiol.1998.sp004112. [DOI] [PubMed] [Google Scholar]
- 47.Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 2004;6:252–259. doi: 10.1038/ncb1107. [DOI] [PubMed] [Google Scholar]