Abstract
Replicative DNA polymerases, such as T4 polymerase, possess both elongation and 3′–5′ exonuclease proofreading catalytic activities. They arrest at the base preceding DNA damage on the coding DNA strand and specialized DNA polymerases have evolved to replicate across the lesion by a process known as TLS (translesion DNA synthesis). TLS is considered to take place in two steps that often require different enzymes, insertion of a nucleotide opposite the damaged template base followed by extension from the inserted nucleotide. We and others have observed that inactivation of the 3′–5′ exonuclease function of T4 polymerase enables TLS across a single site-specific abasic [AP (apurinic/apyrimidinic)] lesion. In the present study we report a role for auxiliary replicative factors in this reaction. When replication is performed with a large excess of DNA template over DNA polymerase in the absence of auxiliary factors, the exo− polymerase (T4 DNA polymerase deficient in the 3′–5′ exonuclease activity) inserts one nucleotide opposite the AP site but does not extend past the lesion. Addition of the clamp processivity factor and the clamp loader complex restores primer extension across an AP lesion on a circular AP-containing DNA substrate by the exo− polymerase, but has no effect on the wild-type enzyme. Hence T4 DNA polymerase exhibits a variety of responses to DNA damage. It can behave as a replicative polymerase or (in the absence of proofreading activity) as a specialized DNA polymerase and carry out TLS. As a specialized polymerase it can function either as an inserter or (with the help of accessory proteins) as an extender. The capacity to separate these distinct functions in a single DNA polymerase provides insight into the biochemical requirements for translesion DNA synthesis.
Keywords: abasic site, DNA minicircle, exonuclease-deficient polymerase, replication complex, T4 DNA polymerase, translesion DNA synthesis (TLS)
Abbreviations: AP, apurinic/apyrimidinic; exo−, T4 DNA polymerase deficient in 3′–5′ exonuclease activity; P/T, primer/template; TLS, translesion DNA synthesis; wt, wild-type
INTRODUCTION
Progression of replicative, high-fidelity DNA polymerases is usually arrested at lesions in the DNA template. In contrast, recently discovered specialized DNA polymerases are able to replicate past DNA damage [termed TLS (translesion DNA synthesis)]. These specialized polymerases can be accurate (error free) or mutagenic (error prone) during TLS. The prevailing model is that TLS is a multistep reaction requiring the presence of a DNA polymerase processivity factor and, in many cases, two types of specialized polymerases. One polymerase, called an inserter, efficiently inserts a nucleotide in front of the lesion; another, called an extender, is capable of elongating the primer after insertion (for recent reviews see [1,2]). However, specialized prokaryotic DNA polymerases have been reported that carry out both the insertion and extension reactions [1] and among the eukaryotic polymerases, POLQ (polymerase Q) can also efficiently perform both processes [3].
The incapacity of the replicative polymerases to accommodate damaged bases in their active sites, a capability that the specialized polymerases possess, is widely accepted as a major determinant of the different response of these enzymes to DNA damage, particularly to highly distorting DNA lesions [4]. In addition, replicative polymerases invariably have a 3′–5′ exonuclease activity to remove mismatched base-pairs that are often a consequence of TLS. In contrast, all of the specialized polymerases lack this ‘proofreading’ activity.
Many years ago, in vitro experiments led to the hypothesis that 3′–5′ exonuclease activity could prevent TLS by forcing the DNA polymerase into a futile cycle of incorporation/excision, a process referred to as polymerase ‘idling’ [5]. This notion was later supported by in vivo evidence showing that exonuclease-deficient mutants of Escherichia coli polymerase III can replicate past certain DNA lesions, even in the absence of its corresponding specialized polymerase, polymerase V [6–8]. Likewise, recent experiments have shown that inactivation of the proofreading reaction associated with replicative polymerase δ causes a mutator and cancer phenotype in mice [9].
Finally, a replicative polymerase that is unable to bypass a lesion on the template DNA will eventually dissociate from the DNA and terminate attempted elongation. Hence stabilizing the replication complex by DNA polymerase accessory proteins could be a third component that favours TLS.
The replicative T4 DNA polymerase exhibits both proofreading and elongation catalytic activities and we have studied this polymerase to understand TLS of P/T (primer/template) DNA containing one of the most frequent endogenous DNA lesions, an AP (apurinic/apyrimidinic) site. Elongation by the wt (wild-type) T4 DNA polymerase arrests at an AP site [10] and we have recently observed that inactivation of its 3′–5′ exonuclease activity allows in vitro TLS across this lesion, demonstrating that the absence of proofreading can contribute to the proficiency of TLS by T4 polymerase [11]. In the present study we present evidence for the role of accessory proteins in this process.
MATERIALS AND METHODS
Proteins and chemicals
Wt T4 DNA polymerase (gp43wt) and T4 DNA polymerase deficient in the 3′–5′ exonuclease activity (gp43D219A, exo−) were purified and stored as described previously [12,13]. The gp45 processivity clamp and the gp44/62 clamp loader were purified and characterized as described previously [14,15]. Purification and characterization of the gp32 single-stranded DNA-binding protein was performed as described in [16,17]. A gel of the proteins used in the present study is presented in Figure 1 of [11]. Protein concentrations were determined by UV absorbance at 280 nm, using molar absorption coefficients (ϵ) of 1.3×105 M−1·cm−1, 1.91×104 M−1·cm−1, 1.23×105 M−1·cm−1 and 3.7×104 M−1·cm−1, for monomeric gp43, monomeric gp45, gp44/62 complex and monomeric gp32 respectively. All protein concen-trations are reported in units of moles protein monomers.
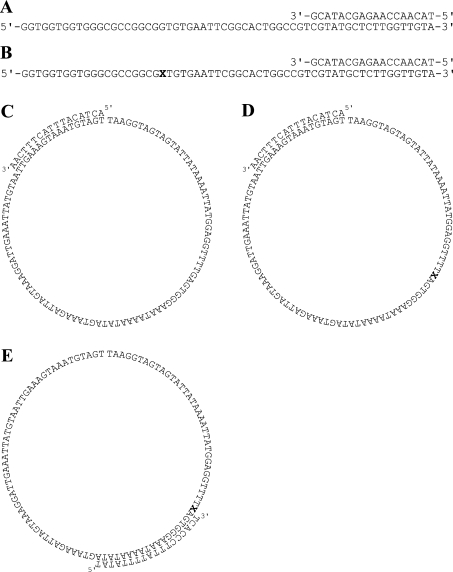
Figure 1. DNA substrates used in the present study.
X represents the position of the artificial abasic site (tetrahydrofuran residue). (A) linear 60/17-mer substrate; (B) linear 60/17-mer substrate containing the abasic site; (C) circular 100/17-mer substrate; (D) circular 100/17-mer substrate containing an abasic site for running start experiments; (E) circular 100/18-mer substrate containing an abasic site for standing start experiments.
T4 polynucleotide kinase and DNA ligase were from New England Biolabs. dSpacer CE Phosphoramidite (tetrahydrofuran moiety) was purchased from Glen Research. [γ-32P]ATP (4500 Ci/mmol) was from ICN. Ultrapure dNTPs and ATP were from Amersham Bioscience.
DNA substrates
The sequences of the DNA templates/primers used in the present study are shown in Figure 1. The 60-mer linear templates A and B, either undamaged or containing a tetrahydrofuran moiety mimicking an abasic site (X), and the corresponding primers were chemically synthesized and purified on a 15% polyacrylamide/7M urea/30% formamide gel. After elution and ethanol precipitation, their concentrations were determined spectrophotometrically.
The 100-mer circular templates C and D were constructed as follows. The linear substrates C (intact) and D (containing the tetrahydrofuran moiety, indicated with an X) have the following sequence: 5′-TAAGGTAGTAGTATTATAAAATTATGGAGGTTTT(G/X)AGTGGGAAATAAAATATAGTAAAGATTAGTAAAGGATTGAAATTATGTAATTGAAAGTAAATGTAGT-3′. Each oligonucleotide (2 μM) was phosphorylated separately in 430 μl of polynucleotide kinase buffer containing 1 mM ATP and 40 units of T4 polynucleotide kinase for 30 min at 37 °C. Subsequently, a further 40 units of kinase was added and the incubation continued for an additional 30 min followed by 10 min at 75 °C. A 40-mer scaffold oligonucleotide (5′-TTTATAATACTACTACCTTAACTACATTTACTTTCAATTA-3′), complementary to the first 20 bases of the 5′-end and the last 20 bases of the 3′-end of both 100-mers, was then added to each reaction mixture at a concentration of 2 μM, together with NaCl to a final concentration of 50 mM. The mixtures were heated at 85 °C for 10 min and then slowly cooled to room temperature (20 °C). The reaction mixtures were brought to 1X DNA ligase buffer, supplemented with 50 mM ATP, in a final volume of 2 ml and 600 Weiss units of T4 DNA ligase were added to each reaction mixture. Samples were incubated at 37 °C for 1 h followed by 12 h at 16 °C. The reaction mixtures were ethanol precipitated and 100-mer minicircles were then purified from a 10% polyacrylamide/7 M urea/30% formamide gel after visualization of the material on fluorescent silica plates by UV shadowing. After elution from the gel and ethanol precipitation, the concentration of the substrates was determined spectrophotometrically. The templates were then hybridized to a 17-mer primer. Substrate E was obtained by hybridizing templates D to an 18-mer primer. All the primers were 32P-labelled at their 5′-end and hybridized to the respective templates at equimolar concentrations.
Primer extension assays
The DNA polymerase reaction (10 μl) contained 25 mM Hepes/HCl (pH 7.5), 5 mM MgCl2, 1 mM DTT (dithiothreitol), 200 μg/ml BSA, 5% glycerol, 40 mM NaCl and 100 μM of each of the four deoxynucleotides and the indicated concentrations of enzymes and substrates. When performed with the DNA polymerase alone, the reaction was initiated by the addition of the enzyme. When other replication proteins were present, 1 mM ATP was also added. gp45, gp44/66 and gp32 were first incubated for 1 min at 37 °C in the presence of the DNA substrate and polymerization reactions were then initiated by the addition of the indicated amounts of T4 DNA polymerase. Reactions were incubated at 37 °C for the indicated time and quenched by the addition of 3 μl of stop buffer (90% formamide, 0.1% xylene cyanol, 0.1% Bromophenol Blue and 1 mM EDTA). The reaction products were resolved on 15% polyacrylamide/7 M urea/30% formamide gels for the linear template and a 13% polyacrylamide gel for the circular template. Quantification of the results was performed using a Molecular Dynamics STORM 840 PhosphorImager and ImageQuant software. The extent of TLS was calculated as the ratio of the intensity of bands that lie in front or downstream of the lesion site to the intensity of all bands, excluding the primer. The extent of insertion in front of the lesion was calculated as the ratio of the intensity of the band incorporated in front of the lesion site to the intensity of all bands, excluding the primer. The extent of extension past the lesion was calculated as the ratio of the intensity of bands downstream of the lesion to the intensity of the band in front of the lesion.
RESULTS
Inactivation of the 3′–5′ exonuclease activity of the T4 DNA polymerase leads to either single nucleotide incorporation in front of an abasic site or extension past the lesion depending on the enzyme/template ratio used
We have previously shown that inactivation of the 3′–5′ exonuclease activity associated with the replicative DNA polymerase of the bacteriophage T4 allows both insertion and extension past an AP site when molar ratios of polymerase/DNA template are in the range of 0.1–3 [11]. These results show an intrinsic capacity of the exo− polymerase to replicate past an AP site which is not shared by its wt form. However, it seemed reasonable that a lower ratio of enzyme/template might correspond better to physiological conditions. We therefore decided to investigate the capacity of low concentrations of exonuclease-deficient and wt T4 polymerases to elongate a 5′-32P-labelled primer DNA annealed to templates containing an AP lesion. The substrates used in the present study are depicted in Figure 1. The two primers shown on the circular template are for different types of experiments, running start (Figure 1D, a 17-mer located 51 nucleotides downstream of the lesion) or a standing start (Figure 1E, an 18-mer, whose 3′-OH is located at the nucleotide that precedes the lesion). In the exonuclease-deficient polymerase the aspartic acid residue at position 219 that provides one of the four essential carboxylate residues for the two-metal ion exonuclease activity has been replaced by an alanine residue [18]. We examined our enzyme preparations for exonuclease activity; wt T4 gp43 polymerase readily digested the labelled primer of a 47/25-mer DNA/primer, while no digestion was seen with the exo− gp43D219A mutant (results not shown).
After primer extension, the newly synthesized DNA products were resolved by denaturing PAGE and visualized on a Phosphor-Imager. Lanes 1–4 of Figure 2(A) show representative data for the replication of linear templates A and B by the wt and exo− forms of T4 DNA polymerase. As can be seen, both DNA polymerases catalysed similar elongation of the undamaged substrate A at the lowest concentrations of enzyme investigated; we also found no difference in activity at higher concentrations of enzymes (results not shown).
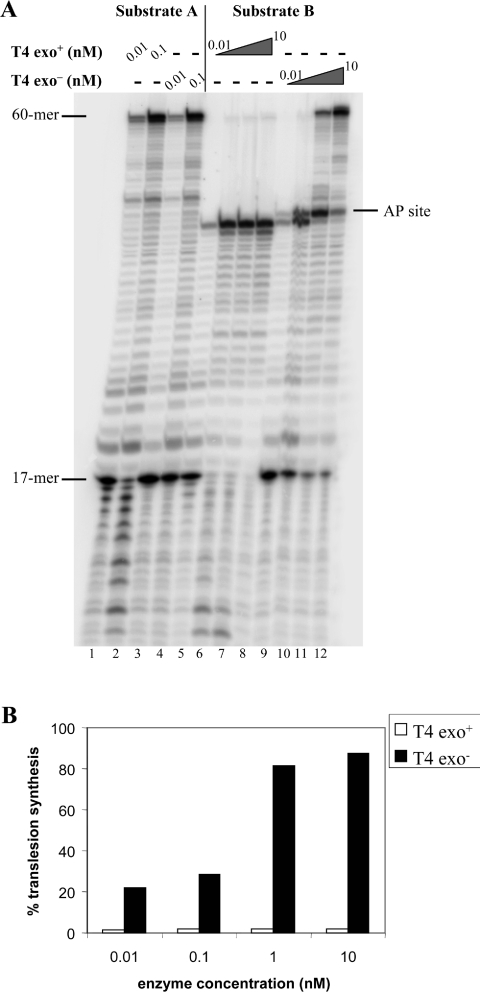
Figure 2. Replication of linear DNA substrates by gp43 wt and gp43D219A exo− DNA polymerases.
(A) PhosphorImage of the reaction products for undamaged substrate, A, or substrate that contains an AP lesion. Reactions were performed as described in the Materials and methods section with 30 nM substrates A or B for 30 min at 37 °C at the indicated concentrations of polymerases. In the case of lanes 5–8 and 9–12, concentrations of the enzymes were 0.01 nM, 0.1 nM, 1 nM and 10 nM respectively. The position of the 17-mer (primer) and of the 60-mer (full-length product) are indicated on the left-hand side of the gel, while the location of the abasic site is indicated on the right-hand side. (B) Quantification of the data shown in (A) for the damaged substrate.
Replication of the AP-containing linear DNA template B by the wt polymerase showed that the enzyme was blocked at the base preceding the lesion at all of the concentrations used (Figure 2A, lanes 5–8). Quantification of the data indicates that the extent of blockage as a fraction of total DNA synthesis did not vary with the amount of enzyme (Figure 2B). The faint band observed at the position of the full-length product is the result of a small amount of undamaged DNA in our preparation of substrate B [11].
Replication of substrate B with the exonuclease-deficient polymerase gave a different result (Figure 2A, lanes 9–12). Two distinct patterns were apparent. At the higher concentrations of enzyme (lanes 11 and 12) we observed both incorporation in front of the lesion and extension beyond it, which reached up to 80% of the total elongated primers (Figure 2B), in agreement with our previous findings [11]. At the lower concentrations however, the polymerase catalysed incorporation of nucleotide opposite the AP site but was unable to extend the resulting primer, even though we note an increase in the capacity to insert in front of the lesion as a function of the polymerase concentration (Figure 2A, lanes 9 and 10 and Figure 2B). The data in lanes 9 and 10 of Figure 2A were obtained with a molar ratio of template/enzyme of 3000 and 300 respectively, and similar results were also observed for ratios of template/enzyme between these values (results not shown).
The circular or linear structure of the template DNA can affect the efficiency of TLS [19]. We therefore examined the capacities of increasing concentrations of wt and exo− T4 DNA polymerases to replicate past an AP site on a circular DNA substrate (Figures 1C and 1D). Both polymerases efficiently replicated the undamaged circular substrate at enzyme concentrations of 0.1 and 1 nM (Figure 3A, lanes 2–5). In all cases, the length of the product synthesized by wt enzyme was the same as the template strand, i.e. 100 nucleotides (lanes 2 and 3). In contrast, the exonuclease-deficient polymerase synthesized longer products (lanes 4 and 5), thereby showing an increased capacity of strand displacement compared with the wt enzyme, as previously reported for the T4 polymerase with a shorter circular template and for other exonuclease-deficient polymerases [11].
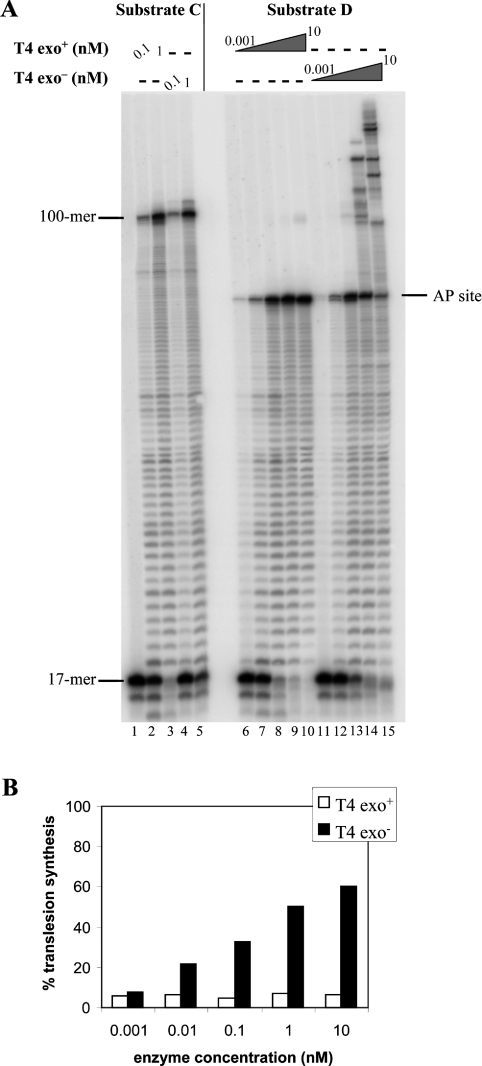
Figure 3. Replication of circular DNA substrates by gp43 wt and gp43D219A exo− DNA polymerases in a running start reaction.
(A) PhosphorImage of the reaction products. Reactions were performed as described in the Materials and methods section with 15 nM of running start substrates C (undamaged) or D (with an AP lesion) for 30 min at 37 °C at the indicated concentrations of polymerases. In the case of lanes 6–10 and 11–15, concentrations of the enzymes were 0.001 nM, 0.01 nM, 0.1 nM, 1 nM and 10 nM respectively. The position of the 17-mer (primer) and of the 100-mer (full-length product) are indicated on the left-hand side of the gel, while the location of the abasic site is indicated on the right-hand side. (B) Quantification of the data shown in (A) for the damaged substrate.
Replication of the damaged circular template D by the wt enzyme showed arrest at the base preceding the lesion, similar to results found with the linear template B. No incorporation in front of the AP site was observed with any of the concentrations of wt polymerase used (Figure 3A, lanes 6–10, and Figure 3B). Replication of substrate D by the exonuclease-deficient enzyme gave substantial TLS products at the highest concentrations of enzyme (Figure 3A, lanes 14 and 15, and Figure 3B); these products are longer than the full-length template, as observed for the undamaged substrate. However, the lowest enzyme concentrations were unable to extend from the primer 3′-OH terminus, even though the polymerase incorporated increasing amounts of nucleotide in front of the AP site in a concentration-dependent fashion (Figure 3A, lanes 11–13, and Figure 3B), thus reproducing the results observed with the linear template.
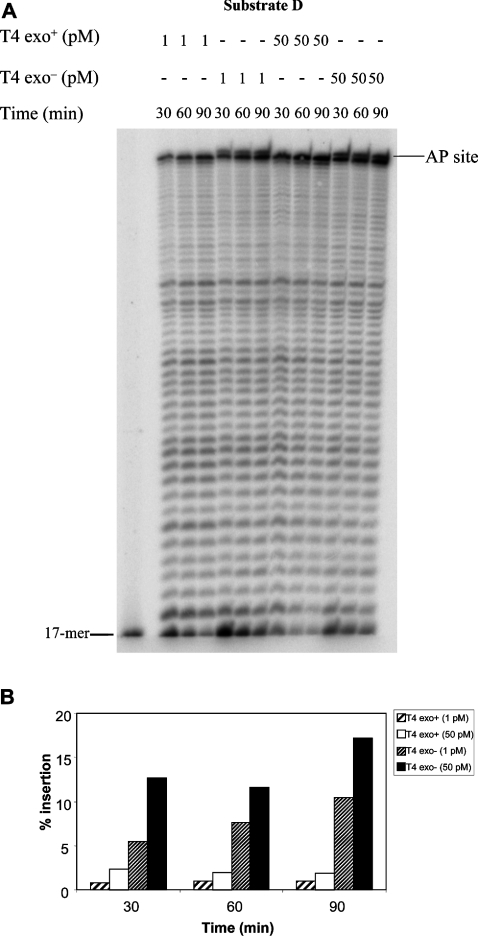
It is possible that primer extension past the lesion is kinetically inhibited at low concentrations of enzyme. We therefore asked whether the exonuclease-deficient T4 polymerase could eventually extend from the nucleotide inserted in front of the lesion at longer incubation times. Figure 4(A) shows that two concentrations of exo− polymerase that were unable to perform TLS after 30 min incubation did not extend the lesion-inserted nucleotide during up to 90 min of incubation. We observed a moderate augmentation in the amount of nucleotide incorporated in front of the lesion during these experiments (Figure 4B), showing that the enzyme was active throughout the incubation. As expected, no insertion in front of the AP site was detected with the wt polymerase. Taken together these results show that at low protein concentrations, the exonuclease-deficient form of the T4 polymerase can insert one nucleotide in front of an abasic site but cannot further extend the primer from its 3′-OH.
Figure 4. Insertion of a nucleotide in front of the lesion of an AP-containing circular DNA substrate in a running start reaction.
(A) PhosphorImage of the reaction products. Running start reactions were performed as described in the Materials and methods section with 15 nM substrate D at the indicated times and concentrations of polymerases. The position of the 17-mer (primer) is indicated on the left-hand side of the gel while the location of the abasic site is indicated on the right-hand side. (B) Quantification of the data shown in (A).
Addition of the replication factors gp45 and gp44/62 allows extension by the exonuclease-deficient T4 DNA polymerase past the abasic site
During DNA replication, the T4 DNA polymerase associates with auxiliary replicative factors [20]. These factors include a homotrimeric processivity factor (gp45), a heterodimeric clamp loader (gp44/gp66) and single-stranded DNA-binding protein (gp32); their combined action increases binding of the enzyme to DNA and processivity of replication, and could thereby influence the capacity of the T4 polymerase to replicate past a lesion. We have previously reported that addition of these factors did not modify the intrinsic capacity of high concentrations of wt or exo− T4 DNA polymerase to replicate past an abasic site on linear or circular DNA substrates [11].
In the present study we have performed experiments with low concentrations of enzyme where the exo− polymerase inserts one nucleotide in front of the lesion but is unable to proceed beyond. Representative gels obtained with circular running start or standing start damaged templates D and E are shown in Figures 5(A) and 6(A). Lane 8 of Figure 5(A) shows a reaction with all the replicative factors but in the absence of the polymerase. A minor contaminating polymerase activity was detected, but it appears to be negligible compared with the activity shown by the polymerase alone in lanes 1 or 9 of Figure 5(A), taking into account that a nearly 10000 times molar excess of auxiliary proteins is present in lane 8.
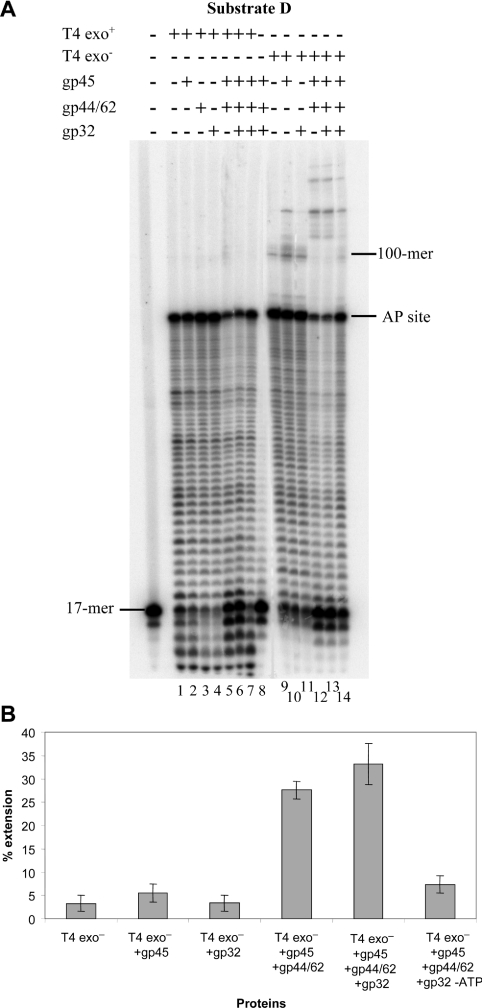
Figure 5. Replication of the AP-containing circular DNA substrate by gp43wt and gp43D219A exo− DNA polymerases, in combination with other replicative proteins, in a running start reaction.
(A) PhosphorImage of the reaction products. Reactions were performed as described in the Materials and methods section with 15 nM running start substrate D, 0.05 nM of either gp43 wt or gp43D219A exo− polymerases, 100 nM of gp45, 200 nM of gp44/62 and 150 nM of gp32. The incubation time was 30 min at 37 °C. The position of the 17-mer primer is indicated on the left-hand side of the gel while the location of the abasic site and of the full-length 100-mer product are indicated on the right-hand side. Lane 14 is the same as lane 13, except that ATP has been omitted from the reaction mixture. (B) Quantification of the extension from the AP site by the D219A exo− polymerase, either alone or in combination with the replicative proteins. Data are the average of three independent experiments±S.E.M.
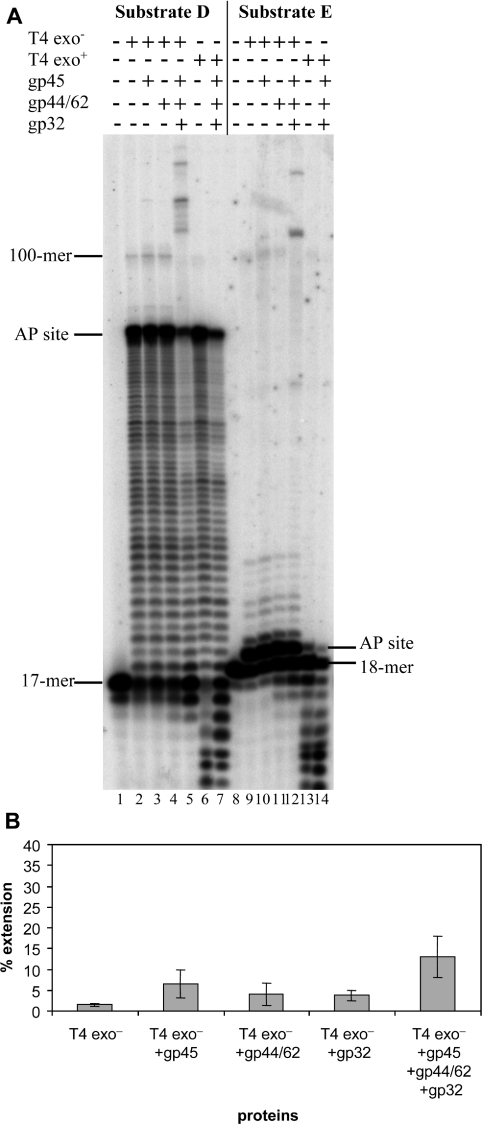
Figure 6. Replication of the AP-containing circular DNA substrate by gp43wt and gp43D219A exo− DNA polymerases, in combination with other replicative proteins, in both running and standing start reactions.
(A) Reactions were performed as described in the Materials and methods section with 15 nM running start or standing start substrate D, 0.05 nM of either gp43 wt or gp43D219A exo− polymerases, 100 nM of gp45, 200 nM of gp44/62 and 150 nM of gp32. The incubation time was 30 min at 37 °C. The position of the 17-mer primer, the AP site and the 100-mer full-length product for the running start reaction are indicated on the left-hand side of the gel, while the location of the 18-mer primer and of the abasic site for the standing start reaction are indicated on the right-hand side. (B) Quantification of the extension from the AP site by the D219A exo− polymerase, either alone or in combination with the replicative proteins. Data are the average of four independent experiments±S.E.M.
Addition of gp45, gp44/62 and gp32 separately or in combination did not substantially modify the replication of undamaged substrate C by the wt or exo− polymerases alone (results not shown and Figure 6 of [11]), although a reduced utilization of the primer was noticed in the presence of gp45 and gp44/62 (see also data in Figures 5 and 6). Auxiliary factors, either separately or together, did not enable replication by wt polymerase past the abasic site in a running start reaction (Figure 5A, lanes 1–7 and Figure 6, lanes 6 and 7). Moreover, shorter exposures of the gel indicated that DNA synthesis was always blocked at the base preceding the lesion, even in the presence of the factors (results not shown).
Addition of gp45, gp44/62 and gp32 to the reaction with exonuclease-deficient polymerase gave a different picture. Although a low level of extension past the AP site was detected even when the polymerase alone was used (lane 9 of Figure 5A, quantified in Figure 5B, and lane 2 of Figure 6), a lower exposure of the gel revealed that the vast majority of the products ended at both the base preceding and in front of the lesion, as seen in Figures 3 and 4. Neither the single-stranded DNA-binding protein, gp32, alone at a ratio of one molecule of protein per six nucleotides (lane 11 of Figure 5A) nor the clamp loader, gp44/62, alone (lane 4 of Figure 6) altered the polymerase behaviour (quantified in Figure 5B). However, a slight increase in the polymerase capacity to extend past the lesion was observed upon addition of the processivity clamp, gp45, alone (lane 10 of Figure 5A and lane 3 of Figure 6, quantified in 5B and 6B). This was somewhat unexpected since a clamp loading factor is required to assemble the processivity clamp on circular double-stranded DNA. This result might be explained by the observation that the gp45 has both open and closed subunit interfaces in solution [21]; hence some of the molecules are open trimers and may not need to be charged by gp44/62 to exert their stimulatory action.
Addition of both gp45 and gp44/62 significantly increased the capacity of the exo− polymerase to extend past the AP site (lane 12 of Figure 5A, quantified in 5B). Neither the amount nor the distribution of these extended products varied when gp32 was added (lane 13 of Figure 5A and lane 5 of Figure 6, quantified in 5B). Omitting ATP from the reaction reduced the amount of the products extended (lane 14 of Figure 5A, quantified in 5B). As mentioned before, addition of gp45 and gp44/62 with or without gp32 resulted in a reduced utilization of the primer by the polymerase in the running start reaction (lanes 5, 6, 12 and 13 of Figure 5A and lane 5 of Figure 6) in a fashion that appeared to be at least in part ATP-dependent (lane 14 of Figure 5A).
We also investigated the effect of the auxiliary proteins on the extension past an abasic site by the wt and exo− forms of the T4 polymerase in a standing start reaction, where the 3′-OH of the primer is situated just before the lesion. The wt enzyme was unable to incorporate in front of the lesion, with only a faint band revealed by overexposure of the gel (lane 13 of Figure 6). Addition of the accessory protein did not change the intensity of this band or allow any extension (lane 14 of Figure 6). For low concentrations of the exo− polymerase alone, the vast majority of the products ended in front of the AP site with the presence of a few extended products of reduced length (lane 9 of Figure 6), similar to that which was observed with the running start reaction. However, significant extension of these products was obtained only upon simultaneous addition of gp45, gp44/62 and gp32 (lane 12 of Figure 6, quantified in Figure 6B). The stimulatory effect of the accessory proteins appears to be reduced in the standing start reaction compared with the running start (compare Figure 5 with Figure 6); this may be due to an increased difficulty in charging the processivity factor when the 3′-OH of the primer is so close to the lesion.
Taken together, these results show that addition of both the processivity factor gp45 and of the clamp loader gp44/62 in the presence of ATP favours primer extension by low concentrations of an exonuclease-deficient form of T4 polymerase across an AP site placed on a circular DNA template. In contrast, addition of accessory factors does not permit wt DNA polymerase to either incorporate a nucleotide in front of the lesion or to extend the primer beyond it.
DISCUSSION
A number of laboratories, including ours, have reported that the 3′–5′ exonuclease-deficient T4 DNA polymerase mutant D219A can extend beyond an abasic lesion when replicating synthetic linear or circular DNA templates and that dAMP is the deoxyribonucleoside monophosphate preferentially incorporated in front of the lesion [11,22,23]. Although these observations clearly reflect an intrinsic capacity of the mutant polymerase to replicate past an AP site, they were obtained at relatively high molar ratios of enzyme/DNA template. These experimental conditions may differ from the physiological situation in vivo, where the concentration of the polymerase is probably much lower than that of the DNA. In the case of bacteriophage T4, roughly 600 molecules of polymerase are estimated to be engaged in the replication of the 1.7×105 base-long phage genome [24]. During replication the polymerase/DNA ratio is probably even lower since a T4-infected cell has been calculated to contain, in addition to the genomic DNA, 60 replication forks or 120 growing chains [25]. This reasoning led us to investigate the capacity of the exonuclease-deficient form of the T4 polymerase to replicate past an abasic site on template DNA at low enzyme/DNA ratios.
TLS by exo− T4 polymerase is considered to take place in two steps, insertion of a nucleotide opposite the damaged template base followed by extension from the inserted nucleotide. Data reported in Figures 2 and 3 show that, depending on the polymerase/DNA ratio, these steps are affected differently by the presence of the AP lesion. At high polymerase/DNA ratios (from 0.03 to 0.3), the exo− polymerase is able to synthesize past the lesion while at low polymerase/DNA ratios (from 3×10−4 to 3×10−3), the exo− polymerase inserts a nucleotide in front of the AP site but is unable to extend from the resulting 3′-OH. Taken together, these data indicate that, under experimental conditions likely to be closer to a physiological situation than those employed in previous studies, the exonuclease-deficient T4 polymerase can insert one nucleotide in front of an abasic site but cannot further elongate.
No extension is observed after 90 min of reaction with low polymerase/DNA ratios, even though the enzyme continues to catalyse insertion across from the lesion (Figure 4). Significant unreplicated substrate remains in these experiments compared with experiments with higher protein concentrations because the TLS by low protein concentrations is incomplete (compare Figures 2B, 3B and 4B). The equilibrium binding constant for the formation of the ternary complex of T4 polymerase with undamaged P/T DNA and non-hydrolysable nucleotide triphosphate has been estimated, from analytical ultracentrifugation experiments, as Kd=koff/kon=5 nM [26]. The inability of low concentrations of exo− mutant to extend from the mispaired terminal base pair of the primer may reflect a diminished affinity of the enzyme for the P/T junction after TLS which can be overcome by sufficiently high protein concentrations.
During DNA replication, the T4 DNA polymerase associates with replicative auxiliary factors that increase the DNA binding capacity and processivity of the enzyme and could modulate the TLS activity of the enzyme. We have previously reported that addition of auxiliary replicative proteins did not modify the intrinsic bypass capacity of wt or exo− T4 DNA polymerase to replicate linear or circular DNA substrates containing an abasic site [11]. However, the concentration of exonuclease-deficient polymerase used in those experiments was high enough to catalyse substantial translesion replication on its own, as shown in the present study, and this could have masked effects of the accessory proteins. We therefore decided to investigate the effect of replicative auxiliary factors under conditions where the mutant polymerase is not able to extend past the lesion.
Results in Figures 5 and 6 show that addition of both the processivity clamp factor gp45 and of the clamp loader gp44/62 allows exo− T4 polymerase to extend primer DNA across an abasic site on a circular template. In contrast with our data, a previous study has reported that enhancement of wt or exo − T4 polymerase processivity did not affect its ability to bypass an abasic site [27]. The origin for these conflicting observations is unclear, but probably reflects the different DNA substrates used. We observed that auxiliary factors enhance extension in these reaction conditions for both running and standing start reactions. Results also indicate that the presence of the single-stranded DNA-binding protein, gp32, does not enhance or reduce the amount of extended products. We found that the presence of 5–10 times molar excess of gp45 and gp44/62 over the DNA template-primer concentration was necessary to obtain a significant extension past the lesion (results not shown). Similar experimental conditions are often reported in the literature [23,28] and may reflect a relative inefficacy of the in vitro loading process. We estimate the amount of elongation past the lesion in these experiments to be approx. 30% of the total reaction products in the case of the running start reaction. This value could be an underestimation, since, for reasons that we do not presently understand, we also observe a reduced utilization of the primer by both wt and exo− polymerases in the presence of gp45 and gp44/62 in a reaction that requires ATP. Addition of auxiliary factors did not permit either insertion in front of the lesion or extension beyond it by the wt DNA polymerase.
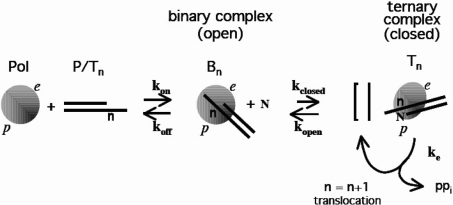
X-ray crystallography studies of RB69 DNA polymerase, a Family B DNA polymerase related to T4 polymerase [28–32], are useful in understanding the behaviour of T4 DNA polymerase [29]. The polymerase forms a binary complex with the P/T junction (Bn) in which the primer strand is located in the exonuclease site of the enzyme (e) [30,31] (Figure 7). Addition of the nucleotide substrate rotates the P/T DNA by 40° and induces the formation of a closed complex [28–32] (Tn, designated as an ellipse in Figure 7) in which the coding base (n) and complementary nucleotide substrate (N) are held tightly in a pocket at the polymerization catalytic site of the ternary complex (p). This closed conformation is maintained without electrostatic or hydrogen bonds between the bases (n and N) and the protein, indicating that the steric constraints of the binding pocket are sufficient for selection of the correct Watson–Crick base pair [31,32]. Once formed, this closed ternary complex is poised for catalysis and the primer can be extended by one nucleotide. When T4 polymerase encounters an AP lesion on template DNA, ‘replication’ of the AP site produces a mismatched 3′ primer terminus whose steric characteristics are no longer those of a Watson–Crick base pair [33,34]. This terminal mispair fits poorly in the catalytic site p and is expected to destabilize binding. P/T DNA returns to the proofreading site where the exonuclease activity will remove the 3′ terminal base [31]. As a result, wt T4 DNA polymerase is invariably arrested at the base preceding an AP lesion in the template strand. In the case of an exonuclease-deficient mutant, the base incorporated across from the AP site is not removed and a fraction of the P/T DNA can return to the polymerization site where chain extension past this lesion could resume, although at a lower rate (Figure 7). Polymerization is in kinetic competition with dissociation of P/T DNA from the complex [36]. In the event that P/T DNA containing mispair is released, a mismatch at the 3′ end of the primer may inhibit reassociation of free DNA to the polymerase. This inhibition, coupled with dissociation of the replication complex at low protein concentration, could explain the failure of the exo− DNA polymerase to elongate past the lesion (Figure 4).
Figure 7. Model for DNA replication by T4 polymerase based on structural data for the homologous DNA polymerase RB69 [28–32].
In this scheme T4 DNA polymerase (Pol), which has a polymerase catalytic site (p) and an exonuclease catalytic site (e), binds to an undamaged P/T DNA with base n at the coding position on the template strand (P/Tn) to form a binary complex (Bn). Pol and Bn are in an open conformation indicated as a sphere and P/Tn is situated in the exonuclease site of the binary complex (see text). Binding of complementary substrate dNTP (N) to Bn induces a closed ternary complex (Tn) depicted as an ellipse. During this conformational change the P/T DNA rotates 40 ° and its extremity moves 40 Å (1 Å=0.1 nm) so that N, n and the 3′-OH terminus of the primer strand are at p and the enzyme catalyses polymerization (ke). Addition of N to the primer strand is followed by pyrophosphate (PPi) release and translocation. Subsequent nucleotide addition that leads to elongation probably occurs via a closed polymerase complex, designated [], whose structure has not yet been determined. The polymerase complex attains thermodynamic equilibrium between each nucleotide addition step [36] and consequently can either dissociate or bind the following dNTP and enter the next elongation cycle.
In the present study we report that the efficiency of chain extension past an AP lesion in a circular DNA substrate at low polymerase/DNA ratio is enhanced by the clamp processivity factor, gp45. The processivity clamp is tethered to the polymerase by the flexible C-terminus of the polymerase. An X-ray structure of the RB69 clamp has been reported previously and model building of its interactions with RB69 polymerase shows that the clamp can remain attached to the polymerase as the P/T DNA swings between the polymerization and exonuclease active sites [30,31]. In addition, Hingorani and O'Donnell [35] have argued that the apparently flexible connection and the rather small area of contact between the polymerase and its processivity factor might allow the polymerase to transiently release its grip from the DNA, while it still remains bound to the clamp. If these ideas are correct, the processivity factor promotes extension past an AP lesion in these conditions by maintaining a high enough local concentration of DNA polymerase and DNA to push the equilibrium toward the binary and ternary complexes (Figure 7).
In conclusion, in vitro bypass of an AP site can be a ‘two polymerase affair’ in which DNA polymerases δ, η, ι and Rev1, for example, work as ‘inserters’ and DNA polymerase ξ works as an ‘extender’ (see Table 1 of [1]). However, polymerases also exist, such as polymerase V or POLQ (polymerase Q), that have the ability to efficiently carry out both the insertion and extension reactions [1,2,4]. T4 DNA polymerase presents a novel situation in which a single DNA polymerase can behave first as an inserter, and then as an extender with the help of replicative accessory proteins.
Acknowledgments
This work was partially supported by the CNRS (Centre National de la Recherche Scientifique) and by grant 3477 to G.V. from the Association pour la Recherche sur le Cancer. G.B. was supported by a grant from the Fondation pour la Recherche Médicale.
References
- 1.Friedberg E. C., Lehmann A. R., Fuchs R. P. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell. Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Seki M., Masutani C., Yang L. W., Schuffert A., Iwai S., Bahar I., Wood R. D. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash S., Johnson R. E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Villani G., Boiteux S., Radman M. Mechanism of ultraviolet-induced mutagenesis: extent and fidelity of in vitro DNA synthesis on irradiated templates. Proc. Natl. Acad. Sci. U.S.A. 1978;75:3037–3041. doi: 10.1073/pnas.75.7.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandewiele D., Borden A., O'Grady P. I., Woodgate R., Lawrence C. W. Efficient translesion replication in the absence of Escherichia coli Umu proteins and 3′–5′ exonuclease proofreading function. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15519–15524. doi: 10.1073/pnas.95.26.15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs R. P., Napolitano R. L. Inactivation of DNA proofreading obviates the need for SOS induction in frameshift mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13114–13119. doi: 10.1073/pnas.95.22.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borden A., O'Grady P. I., Vandewiele D., Fernandez de Henestrosa A. R., Lawrence C. W., Woodgate R. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J. Bacteriol. 2002;184:2674–2681. doi: 10.1128/JB.184.10.2674-2681.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsby R. E., Hays L. E., Chen X., Olmsted E. A., Slayton W. B., Spangrude G. J., Preston B. D. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagher D., Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 11.Tanguy Le Gac N., Delagoutte E., Germain M., Villani G. Inactivation of the 3′–5′ exonuclease of the replicative T4 DNA polymerase allows translesion DNA synthesis at an abasic site. J. Mol. Biol. 2004;336:1023–1034. doi: 10.1016/j.jmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Morris C. F., Hama-Inaba H., Mace D., Sinha N. K., Alberts B. Purification of the gene 43, 44, 45, and 62 proteins of the bacteriophage T4 DNA replication apparatus. J. Biol. Chem. 1979;254:6787–6796. [PubMed] [Google Scholar]
- 13.Nossal N. G., Hinton D. M., Hobbs L. J., Spacciapoli P. Purification of bacteriophage T4 DNA replication proteins. Methods Enzymol. 1995;262:560–584. doi: 10.1016/0076-6879(95)62045-1. [DOI] [PubMed] [Google Scholar]
- 14.Pietroni P., Young M. C., Latham G. J., von Hippel P. H. Dissection of the ATP-driven reaction cycle of the bacteriophage T4 DNA replication processivity clamp loading system. J. Mol. Biol. 2001;309:869–891. doi: 10.1006/jmbi.2001.4687. [DOI] [PubMed] [Google Scholar]
- 15.Reddy M. K., Weitzel S. E., von Hippel P. H. Processive proofreading is intrinsic to T4 DNA polymerase. J. Biol. Chem. 1992;267:14157–14166. [PubMed] [Google Scholar]
- 16.Nossal N. G. DNA synthesis on a double-stranded DNA template by the T4 bacteriophage DNA polymerase and the T4 gene 32 DNA unwinding protein. J. Biol. Chem. 1974;249:5668–5676. [PubMed] [Google Scholar]
- 17.Young M. C., Kuhl S. B., von Hippel P. H. Kinetic theory of ATP-driven translocases on one-dimensional polymer lattices. J. Mol. Biol. 1994;235:1436–1446. doi: 10.1006/jmbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- 18.Frey M. W., Nossal N. G., Capson T. L., Benkovic S. J. Construction and characterization of a bacteriophage T4 DNA polymerase deficient in 3′–5′ exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomer G., Livneh Z. Analysis of unassisted translesion replication by the DNA polymerase III holoenzyme. Biochemistry. 1999;38:5948–5958. doi: 10.1021/bi982599+. [DOI] [PubMed] [Google Scholar]
- 20.Trakselis M. A., Mayer M. U., Ishmael F. T., Roccasecca R. M., Benkovic S. J. Dynamic protein interactions in the bacteriophage T4 replisome. Trends Biochem. Sci. 2001;26:566–572. doi: 10.1016/s0968-0004(01)01929-6. [DOI] [PubMed] [Google Scholar]
- 21.Alley S. C., Shier V. K., Abel-Santos E., Sexton D. J., Soumillion P., Benkovic S. J. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry. 1999;38:7696–7709. doi: 10.1021/bi9827971. [DOI] [PubMed] [Google Scholar]
- 22.Berdis A. J. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 23.Kadyrov F. A., Drake J. W. Characterization of DNA synthesis catalyzed by bacteriophage T4 replication complexes reconstituted on synthetic circular substrates. Nucleic Acids Res. 2002;30:4387–4397. doi: 10.1093/nar/gkf576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg A. San Francisco: Freeman and Company; 1980. DNA Replication; p. 187. [Google Scholar]
- 25.Werner R. Distribution of growing points in DNA of bacteriophage T4. J. Mol. Biol. 1968;33:679–692. doi: 10.1016/0022-2836(68)90313-6. [DOI] [PubMed] [Google Scholar]
- 26.Delagoutte E., Von Hippel P. H. Function and assembly of the bacteriophage T4 DNA replication complex: interactions of the T4 polymerase with various model DNA constructs. J. Biol. Chem. 2003;278:25435–25447. doi: 10.1074/jbc.M303370200. [DOI] [PubMed] [Google Scholar]
- 27.Reineks E. Z., Berdis A. J. Evaluating the effects of enhanced processivity and metal ions on translesion DNA replication catalyzed by the bacteriophage T4 DNA polymerase. J. Mol. Biol. 2003;328:1027–1045. doi: 10.1016/s0022-2836(03)00370-x. [DOI] [PubMed] [Google Scholar]
- 28.Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 29.Hogg M., Cooper W., Reha-Krantz L., Wallace S. S. Kinetics of error generation in homologous B-family DNA polymerases. Nucleic Acids Res. 2006;34:2528–2535. doi: 10.1093/nar/gkl300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shamoo Y., Steitz T. A. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 31.Franklin M. C., Wang J., Steitz T. A. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 32.Reineks E. Z., Berdis A. J. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 33.Hogg M., Wallace S. S., Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freisinger E., Grollman A. P., Miller H., Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hingorani M. M., O'Donnell M. Sliding clamps: a (tail)ored fit. Curr. Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 36.Greive S. J., von Hippel P. H. Thinking quantitatively about transcriptional regulation. Nat. Rev. Mol. Cell. Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]