Abstract
Although resistin was first suggested as a possible link between obesity and diabetes, we have demonstrated previously that expression of resistin is induced by LPS (lipopolysaccharide). In the present study, we showed that LPS increased levels of resistin mRNA and promoter activity in murine RAW264.7 macrophages. Investigation of cis-regulatory elements in the mouse resistin promoter required for LPS-mediated induction showed that an Octamer (ATTTGCAT) element, located at −914 to −907, was required for maximal promoter activity in response to LPS stimulation. Co-transfection of RAW264.7 cells with a resistin promoter–luciferase construct and an Oct-1 or Oct-2 expression plasmid (pCG-Oct-1 or pCG-Oct-2) showed that Oct-2, but not Oct-1, activated the resistin promoter upon LPS treatment. Binding of Oct-2 to the Octamer element was demonstrated by supershift DNA-affinity precipitation and chromatin immunoprecipitation assays. Reverse transcription–PCR and Western blot results showed that levels of Oct-2 mRNA and protein were both up-regulated by LPS in RAW264.7 cells. The LPS-induced increase in Oct-2 protein was inhibited by LY294002 (a phosphoinositide 3-kinase inhibitor) post-transcriptionally, and the inhibition also resulted in a lower response of both resistin mRNA and promoter activity to LPS treatment. Moreover, specific knockdown of Oct-2 by RNA interference impaired the LPS-induced increase in resistin mRNA and promoter activity. Together, these results indicate that Oct-2 is involved in the LPS-mediated induction of resistin gene expression in macrophages and suggest that activation of Oct-2 is a part of LPS signalling pathways in macrophages.
Keywords: lipopolysaccharide (LPS), macrophage, obesity, Octamer, Oct-2, resistin
Abbreviations: Ab, antibody; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; Ets, E twenty-six; FBS, foetal bovine serum; FIZZ, found in inflammatory zone; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IL, interleukin; iNOS, inducible nitric oxide synthase; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MEK, MAPK/extracellular-signal-regulated kinase kinase; NF-κB, nuclear factor κB; PBMC, peripheral blood mononuclear cell; PI3K, phosphoinositide 3-kinase; RNAi, RNA interference; RT, reverse transcription; TNF-α, tumour necrosis factor α
INTRODUCTION
Resistin, a 12.5 kDa cysteine-rich protein predominantly expressed in adipocytes, has been shown to contribute to insulin resistance in diet-induced or genetically obese mice [1]. An association of resistin with obesity and insulin resistance has also been demonstrated in humans [2,3]. Several lines of evidence suggest that the metabolic effect of resistin on impaired glucose tolerance and insulin action can be attributed to its enhancement of glucose production by the liver [4–6]. However, resistin has also been shown to inhibit insulin-stimulated glycogenesis and glucose oxidation in skeletal muscle, leading to impaired glucose tolerance in rats [7]. In addition, adenovirus-mediated chronic hyper-resistinaemia leads to whole-body insulin resistance involving impaired insulin signalling in skeletal muscle, liver and adipose tissue, resulting in glucose intolerance, hyperinsulinaemia and hypertriglyceridaemia [8]. Despite the accumulated evidence showing that resistin can inhibit the effects of insulin on glucose metabolism, the role of resistin in obesity and diabetes remains controversial. A lower level of resistin expression in adipose tissue has been reported in several different obese mice models compared with their lean counterparts [9], and no association has been found between plasma resistin levels or adipocyte resistin mRNA levels and Type 2 diabetes or insulin resistance in humans [10–13].
Sequence analysis revealed that resistin is identical with FIZZ3, which belongs to the FIZZ (found in inflammatory zone) protein family. FIZZ1, the first member of the FIZZ family to be identified, is overexpressed in allergic inflammation [14]. Because of this, Gomez-Ambrosi and Fruhbeck [15] proposed that resistin might be involved in obesity-related inflammatory processes. Our group was the first to provide evidence supporting this idea by demonstrating that LPS (lipopolysaccharide) up-regulates resistin levels in rat adipose tissue and white blood cells and in human PBMCs (peripheral blood mononuclear cells) [16]. Kaser et al. [17] demonstrated that resistin expression in human PBMCs is markedly increased by pro-inflammatory cytokines, including IL (interleukin)-1, IL-6 and TNF-α (tumour necrosis factor α), as well as by LPS. In human studies, plasma resistin levels have been shown to be highly associated with levels of inflammatory markers, such as CRP (C-reactive protein), IL-6 and ICAM-1 (intercellular adhesion molecule 1) [18–20]. Lehrke et al. [21] have shown that LPS can activate resistin expression in PBMC-derived primary human macrophages and induce hyper-resistinaemia in healthy volunteers 6–8 h after intravenous injection. These results strongly suggest a link between resistin and inflammation; however, the regulation of the resistin gene during inflammatory processes remains unclear. The goal of the present study was therefore to investigate the molecular mechanism of resistin expression induced by LPS in macrophages.
EXPERIMENTAL
Materials
LPS from Escherichia coli (serotype 0111:B4) was purchased from Sigma–Aldrich. DNA recombination enzymes were obtained from New England Biolabs. The pGL3-basic and phRL-TK plasmids and the luciferase assay system were from Promega. DMEM (Dulbecco's modified Eagle's medium) and FBS (foetal bovine serum) were obtained from Gibco BRL, Life Technologies. SuperFect transfection reagent was purchased from Qiagen. [α-32P]dCTP was purchased from NEN Life Science Products. Octamer consensus binding oligonucleotides and Ab (antibody) against Oct-1, Oct-2 and β-actin were from Santa Cruz Biotechnology. LY294002 [PI3K (phosphoinositide 3-kinase) inhibitor], SB203580 [p38 MAPK (mitogen-activated protein kinase) inhibitor] and U0126 [a MEK (MAPK/extracellular-signal-regulated kinase kinase) inhibitor] were from Calbiochem; all were dissolved in DMSO and used at a maximum concentration of 0.1% in culture medium. Plasmids pCG-Oct-1 and pCG-Oct-2, and their parent plasmid pCG were gifts from Dr W. Herr (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.) [22].
Cell culture and LPS treatment
RAW264.7 cells, a murine macrophage cell line, were obtained from the American Type Culture Collection (Manassas, VA, U.S.A.) and were maintained in DMEM supplemented with 10% (v/v) FBS, 2 mM glutamine and 1000 units/ml penicillin/streptomycin. In these experiments, resistin mRNA levels and promoter activity were compared in untreated cells and cells treated with LPS; unless otherwise specified, treatment was with 10 ng/ml LPS for 4 h.
RNA isolation and analyses of resistin mRNA
Total cellular RNA was isolated from RAW264.7 cells according to the method of Chomczynski and Sacchi [23]. RNA concentrations were estimated from the absorbance at 260 nm. First-strand cDNA was synthesized from total RNA using the SuperScript II cDNA Synthesis method (Invitrogen) with oligo-dT primer. The mRNA levels of resistin and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were determined by real-time PCR with an iCycler instrument (Bio-Rad) or by semi-quantitative RT (reverse transcription)–PCR as indicated. Fold induction of mRNA expression level was determined by the ΔΔCt-method as described by Livak and Schmittgen [24] or quantified by densitometry. The specific primer sets were designed by Beacon designer 5.0 (Premier Biosoft International) and are listed in Table 1. The mRNA levels of Oct-1, Oct-2, iNOS (inducible nitric oxide synthase) and GAPDH were determined by semi-quantitative RT–PCR using specific primers as listed in Table 1. The amplified DNA fragments were sequenced to confirm their identity.
Table 1. Sequences of the oligonucleotides used.
Cis-elements are underlined. The lower-case letters indicate the mutated sequences.
| Oligonucleotide | Sequence (5′→3′) | |
|---|---|---|
| Oligonucleotides used for real-time PCR | ||
| Resistin | Forward | CTGATGTCGGGGAAGTGAGC |
| Reverse | GTGCAGGTGCCTGTAGAGAC | |
| GAPDH | Forward | ACCCACTCGTCCACCTTTGA |
| Reverse | CATACCAGGAAATGAGCTT | |
| Oligonucleotides used for RT–PCR | ||
| Oct-1 | Forward | CCTGCAACCAGCACAGTTTA |
| Reverse | CTACGATTCAAGCCCTCAGC | |
| Oct-2 | Forward | AATGGACCCGACATTAACCA |
| Reverse | AAATGGTCGTTTGGCTGAAG | |
| iNOS | Forward | AGGAACATCTGGCCAGGCTG |
| Reverse | ACTTGGGATGCTCCATGGTC | |
| GAPDH | Forward | AAAGGATCCACTGGCGTCTTCACCACC |
| Reverse | GAATTCGTCATGGATGACCTTGGCCAG | |
| Oligonucleotides used for mutagenesis | ||
| C/EBP-mt | Forward | ACGCGTCTTTTCATTTGTaCgcTATAAACTTCCTCTGCTA |
| Reverse | TAGCAGAGGAAGTTTATAgcGtACAAATGAAAAGACGCGT | |
| Ets-mt | Forward | TGTCCAATTTATAAACacagTCTGCTAATCTCAATTTTGT |
| Reverse | AAAATTGAGATTAGCAGActgtGTTTATAAATTGGACAAATG | |
| Oct-mt | Forward | CTGACGTTAACACCCAGGgccTGCATAGCCACCTCCAGTT |
| Reverse | AACTGGAGGTGGCTATGCAggcCCTGGGTGTTAACGTCAG |
Plasmid construction
A DNA fragment containing nt −2768 to +61 of the mouse resistin gene promoter was PCR-amplified using the forward primer, 5′-ACGCGTGCACATGTGCATGTATG-3′, and the reverse primer, 5′-AGATCTTAGCAGGACACAACTCAGTTCT-3′. The underlined sequences are MluI and BglII sites respectively, created to facilitate cloning. The DNA fragment was cloned into the MluI/BglII sites of the pGL3-basic luciferase reporter vector to obtain pResistin(−2768/+61)-Luc. The transcription start site (+1) was assigned according to Hartman et al. [25]. Various 5′ or internal deletion mutants of pResistin(−2768/+61)-Luc were created by PCR. Site-directed mutagenesis was performed by PCR using the QuikChange™ site-directed mutagenesis kit (Stratagene) and the sense and antisense oligonucleotides (Table 1). All constructs were verified by restriction mapping and sequencing.
Transfection and reporter gene activity assay
At 1 day before transfection, RAW264.7 cells were re-plated at a density of 3×105 cells/well in 1 ml of fresh culture medium in 12-well tissue-culture plates, then transiently transfected using the SuperFect Transfection Reagent (Qiagen). Briefly, 1.45 μg of resistin promoter–luciferase reporter plasmid and 0.05 μg of phRL-TK plasmid in 75 μl of serum-free DMEM were mixed with 7.5 μl of SuperFect reagent and incubated for 10 min at room temperature (25 °C), and then the DNA/SuperFect mixture was added to the cells. After 2 h at 37 °C in a humidified atmosphere of 5% CO2, the transfection mixture was removed, and the cells were washed with PBS and re-fed with 1 ml of fresh culture medium. At 20 h after transfection, the cells were either left untreated or were treated with LPS for 4 h, then were lysed by addition of 100 μl of passive lysis buffer (Dual-Luciferase Reporter Assay System, Promega) and incubation at room temperature for 30 min. Photinus (firefly) and Renilla (sea pansy) luciferase activities in the lysates were assayed using the Dual-Luciferase Reporter Assay System as described previously [26]. The light intensity produced by Photinus luciferase (test plasmid) was normalized to that produced by Renilla luciferase (control plasmid). To determine the role of Oct-1 or Oct-2 in resistin promoter activity, 0, 0.15, 0.3 or 0.6 μg of expression vector (pCG-Oct-1 or pCG-Oct-2) was mixed with 1.35 μg of pResistin(−2768/+61)-Luc and 0.05 μg of phRL-TK plasmid and the total amount of DNA was adjusted to 2 μg with pCG, then the DNA mixture was transfected into RAW264.7 cells as described above. At least three independent experiments in duplicate were performed using each construct.
EMSA (electrophoretic mobility-shift assay)
Nuclear extracts were prepared as described previously [26]. The double-stranded oligonucleotide from −927 to −895 bp was 32P-labelled. Binding reactions were performed by incubating 20 μg of nuclear extract from untreated or LPS-treated RAW264.7 cells with or without Ab against Oct-2 or Sp1 for 15 min, then adding 15000 c.p.m. of 32P-labelled probe for a further 20 min at 4 °C in a final volume of 20 μl of binding buffer [20 mM Hepes, pH 7.9, 60 mM KCl, 6 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.1 μg/μl poly(dI-dC)·(dI-dC), 160 μg/ml BSA, 0.008% Nonidet P40 and protease inhibitor]. Electrophoresis was carried out at 4 °C and the results were visualized by autoradiography.
DNA-affinity precipitation assay
A biotinylated oligonucleotide (5′-CTGGAGGTGGCTATGCAAATCCTGGGT-3′) containing the Octamer site (underlined) was annealed with its complementary oligonucleotide to form double-stranded DNA. The biotinylated double-stranded DNA was then mixed for 10 min at room temperature with Dynabeads® M-280 Streptavidin (Dynal Biotech ASA). The beads were pelleted using a magnet and washed three times with binding buffer. Nuclear protein extract (100 μg) from LPS-treated RAW264.7 cells was added, and the suspension was incubated in binding buffer for 20 min at 4 °C. After extensive washes, the proteins that remained bound to the beads were eluted by boiling for 5 min in 12 μl of binding buffer containing 10% SDS and were analysed by Western blotting.
ChIP (chromatin immunoprecipitation) assay
The ChIP assay was performed according to the manufacturer's instructions (Upstate Biotechnology). Briefly, after LPS exposure, the cells were fixed with 1% (w/v) formaldehyde for 10 min at 37 °C. After fixation, the cells were collected by scraping and were then sonicated on ice by pulsing ten times for 10 s at a power setting of 30% using a Microson Ultrasonic Cell Disrupter XL. Immunoprecipitation analysis was carried out using control rabbit IgG or anti-Oct-2 Ab. Cross-links were reversed at 65 °C for 4 h, and proteins were digested with proteinase K for 1 h at 45 °C. Immunoprecipitated DNA was recovered by phenol/chloroform extraction and ethanol precipitation, and was used as template for PCR with the following primers for the resistin promoter (−1193 to −895): forward, 5′-CCATGCACAGTGGTGCGTCC-3′, and reverse, 5′-CTGGAGGTGGCTATGCAAATCC-3′. A total of 10% of the chromatin DNA used for immunoprecipitation was similarly subjected to PCR analysis and indicated as ‘input’. The number of PCR cycles was as the follows: 36 for all of the ChIP experiments and 26 for the input samples.
Western blot analysis
Samples of cell lysates (20 μg of protein/lane) from RAW264.7 cells were separated by SDS/PAGE on 10% gels and transferred on to a PVDF membrane, which was blocked overnight at 4 °C with blocking buffer [10 mM Tris/HCl, pH 8.0, 0.15 M NaCl, 0.1% Tween 20 and 5% (w/v) non-fat dried milk]. The blots were then incubated for 1 h at room temperature with 0.5 μg/ml rabbit polyclonal anti-Oct-2 Ab and for 40 min at room temperature with peroxidase-conjugated anti-rabbit IgG Ab (Amersham Biosciences), then bound Ab was detected using an improved chemiluminescence detection system (NEN). Protein concentrations were determined by the Bradford method (DC Protein Assay, Bio-Rad).
RNAi (RNA interference) experiments
Oligonucleotides were cloned into the pLL3.7 plasmid [27] and confirmed by sequencing. The Oct-2 RNAi target sequence is GGCACAGCAGAGTCAGCCA. The plasmid containing the Oct-2 RNAi target sequence was transfected into RAW264.7 cells using the SuperFect transfection reagent as described above. At 44 h after transfection, the cells were either left untreated or were treated with LPS for 4 h, RNA was isolated and levels of resistin, iNOS, Oct-2, Oct-1 and GAPDH were determined by RT–PCR as described above. A scrambled sequence was used as control.
Statistical analysis
Results are shown as means±S.D. Differences between mean values were evaluated using Student's t test and were considered significant at P<0.05.
RESULTS
LPS induces resistin mRNA expression in RAW264.7 cells
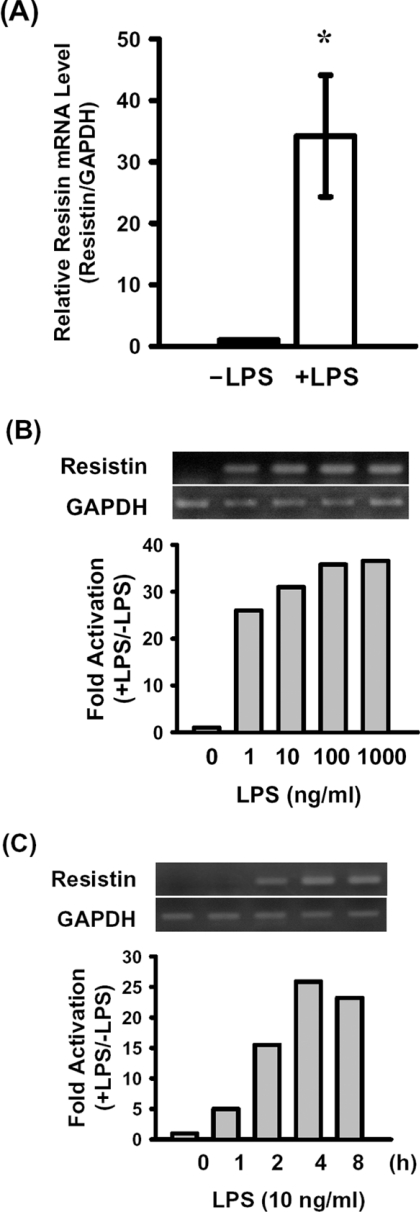
Resistin mRNA expression was identified in RAW264.7 cells. The mRNA level increased approx. 34-fold (34.2±9.9), as measured by real-time RT–PCR, after exposure to 100 ng/ml LPS for 8 h (Figure 1A). An approx. 25-fold increase in resistin mRNA was detected in cells treated with 1 ng/ml LPS for 4 h, and a maximal 35-fold increase was seen with 1000 ng/ml LPS (Figure 1B). When cells were exposed to 10 ng/ml LPS for 1–8 h, the levels of resistin mRNA increased in a time-dependent manner, and maximal levels were reached at 4 h (Figure 1C).
Figure 1. Resistin mRNA levels in RAW264.7 macrophages with or without LPS treatment.
(A) Total RNA was isolated from untreated RAW264.7 cells or cells treated with 100 ng/ml LPS for 8 h. Resistin and GAPDH mRNA were detected by real-time PCR as described in the Experimental section. Levels of resistin mRNA were normalized to those for GAPDH and the results were expressed relative to those in the untreated control (−LPS; relative value=1). Results are means±S.D.; n=3. *P<0.05 compared with the control. (B and C) RAW264.7 cells were treated with 0, 1, 10, 100 or 1000 ng/ml LPS for 4 h (B) or treated with 10 ng/ml LPS for 0, 1, 2, 4 or 8 h (C). Resistin and GAPDH mRNAs were amplified by RT–PCR and separated on a 1% agarose gel (upper panels). Resistin mRNA levels were normalized to those for GAPDH and expressed relative to those in the untreated cells (relative value=1) (lower panels). Similar results were obtained in three independent experiments.
Dissection of the 5′-flanking sequence of the mouse resistin gene
To determine whether the LPS-mediated induction of expression of the resistin gene was due to an increase in transcription, rather than to mRNA stabilization, RAW264.7 cells were treated with or without 5 μg/ml actinomycin D before addition of LPS to 10 ng/ml. In the presence of actinomycin D, LPS did not increase resistin mRNA levels (results not shown), showing that the LPS-induced increase in resistin mRNA was due to increased transcription.
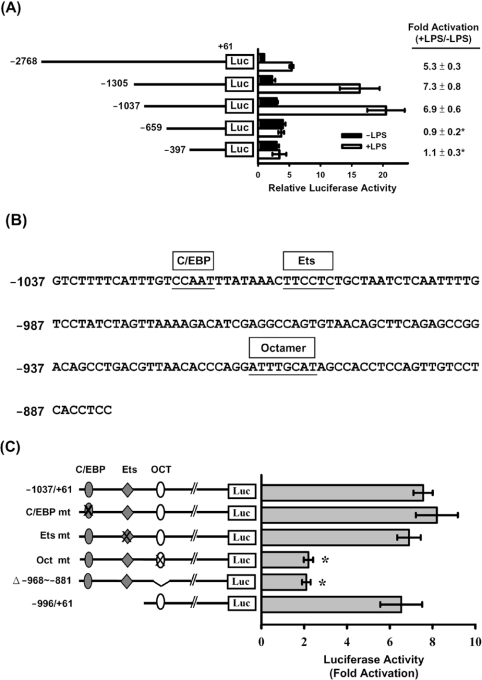
We then cloned the 5′-flanking regulatory sequence of the mouse resistin gene into a site upstream of the luciferase gene in the pGL3-basic reporter vector. A series of 5′-deleted resistin promoter–luciferase constructs and phRL-TK, as a control plasmid, were co-transfected into RAW264.7 macrophages. As shown in Figure 2(A), in non-LPS-treated cells, the gradual deletion of 5′ sequences from nt −2768 to −659 of the resistin promoter resulted in a gradual increase in luciferase activity, the highest activity (3-fold increase) being seen in pResistin(−659/+61)-Luc transfected cells. Upon further deletion to −397, luciferase activity dropped slightly. In LPS-treated cells, the luciferase activity in cells transfected with pResistin(−2768/+61)-Luc was 5-fold higher than in the untreated cells, while a maximal (approx. 7-fold) increase in luciferase activity compared with untreated cells was seen in cells transfected with pResistin(−1305/+61)-Luc. Near maximal LPS-mediated induction was seen using deletion to −1037. Further deletion to nt −659 resulted in no LPS-induced activation of luciferase activity. Sequence analysis of nucleotides −1037 to −881 using the MatInspector program [28] revealed potential cis-acting elements for C/EBP (CCAAT/enhancer-binding protein), Ets (E twenty-six) and Octamer starting at nt −1023, −1010 and −914 respectively, relative to the transcription start site (Figure 2B).
Figure 2. Analysis of mouse resistin gene promoter activity in RAW264.7 cells.
Various 5′-deletion or mutant pResistin-Luc constructs were co-transfected with phRL-TK (internal control) into RAW264.7 cells using the SuperFect reagent. At 20 h after transfection, the cells were left untreated (■, −LPS) or were treated with 10 ng/ml LPS (□, +LPS) for 4 h, then luciferase activities were assayed as described in the Experimental section. (A) Schematic representation of the 5′-deletion promoter–luciferase constructs. The Photinus luciferase activity was normalized to the Renilla luciferase activity, and is shown on the x-axis as the relative activity compared with that for pResistin(−2768/+61)-Luc in cells with no LPS treatment (relative value=1). The fold activation (+LPS/−LPS) of luciferase activity is the ratio of the activity after LPS treatment to that in the absence of LPS and is shown on the right. Results are means±S.D. for at least three independent experiments. *P<0.05 compared with pResistin(−2768/+61)-Luc, pResistin(−1305/+61)-Luc or pResistin(−1037/+61)-Luc. (B) DNA sequence of the resistin promoter from nucleotide −1037 to −881. The C/EBP, Ets and Octamer elements are indicated by boxes above the sequence. (C) The different mutants (mt) are shown in the left-hand panel, the mutated elements being indicated by a cross. Fold activation (+LPS/−LPS) of luciferase activity by LPS treatment is shown in the right-hand panel. Results are means±S.D. for at least three independent experiments. *P<0.05 compared with the wild-type control.
Examination of potential regulatory elements by in vitro mutagenesis and transfection
To define the regulatory element(s) responsible for LPS-mediated induction of resistin expression, the C/EBP, Ets and Octamer sequences were mutated individually using pResistin(−1037/+61)-Luc as the template. The mutant constructs were then transfected into RAW264.7 cells, and the increase in luciferase activity induced by LPS was assayed as described above. As shown in Figure 2(C), mutation of the C/EBP or the Ets sequence did not decrease the LPS-mediated induction of luciferase activity, whereas mutation of the Octamer element resulted in a drastic decrease in luciferase activity. When deletion constructs of pResistin(−1037/+61)-Luc lacking internal nt −968 to −881 showed approx. 2-fold induction by LPS. A construct containing the −996/+61 promoter fragment (no C/EBP or Ets site) retained the ability to drive the LPS-mediated induction of luciferase expression. These results show that an Octamer element at −914 to −907 is required for LPS activation of the resistin promoter.
Identification of transcription factor(s) that interact with the Octamer element
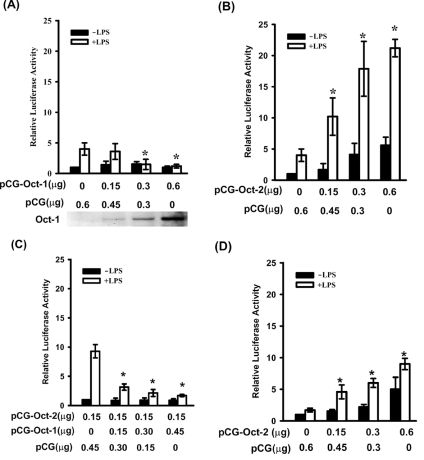
Two nuclear proteins, Oct-1 and Oct-2, have been reported to bind to the Octamer element. To determine which of the two was responsible for activation of the resistin promoter, RAW264.7 cells were co-transfected with the pResistin(−996/+61)-Luc construct and either an Oct-1 or an Oct-2 expression plasmid (pCG-Oct-1 or pCG-Oct-2 respectively). As shown in Figure 3(B), a dose-dependent increase in luciferase activity driven by pResistin(−996/+61)-Luc was seen in the presence of increasing amounts of pCG-Oct-2; in addition, another 4–6-fold increase in luciferase activity was detected when the cells were treated with LPS. However, co-transfection of cells with pResistin(−996/+61)-Luc and pCG-Oct-1 did not lead to an increase in luciferase activity in non-LPS treated cells and led to a decrease in the effect of LPS on luciferase activity with increasing amounts of pCG-Oct-1 plasmid (Figure 3A). A dose-dependent increase in Oct-1 protein in cells transfected with increasing amounts of pCG-Oct-1 is shown at the bottom of Figure 3(A). As shown in Figure 3(C), co-transfection of cells with a constant amount of pCG-Oct-2 and increasing amounts of pCG-Oct-1 resulted in a gradual decrease in LPS-mediated induction of luciferase activity. Surprisingly, co-transfection with Octamer-mutated pResistin(−1037/+61)-Luc and pCG-Oct-2 also resulted in a gradual increase in luciferase activity with increasing amounts of pCG-Oct-2 plasmid, but the effect was much lower than that in cells transfected with the wild-type promoter (Figure 3D).
Figure 3. Oct-2 transactivates the resistin promoter.
Wild-type (A, B and C) or Octamer-mutated (D) reporter plasmid pResistin(−996/+61)-Luc was transfected into RAW264.7 cells. Various amounts of expression vectors for Oct-1 (pCG-Oct-1), Oct-2 (pCG-Oct-2) or the empty vector (pCG) were co-transfected as indicated. At 20 h after transfection, cells were either left untreated (■) or treated with 10 ng/ml LPS (□) for 4 h. Oct-1 protein, detected by Western blot analysis, in cells transfected with various amounts of pCG-Oct-1 plasmid is shown in the bottom of (A). The relative luciferase activity is the activity relative to that of cells transfected with pCG alone (A, B and D) or pCG and pCG-Oct-2 only (C) in the absence of LPS. Results are means±S.D. for at least three independent experiments. *P<0.05 compared with cells transfected only with pCG in (A), (B) and (D). *P<0.05 compared with cells transfected with pCG-Oct-2 and pCG, but no pCG-Oct-1, in (C).
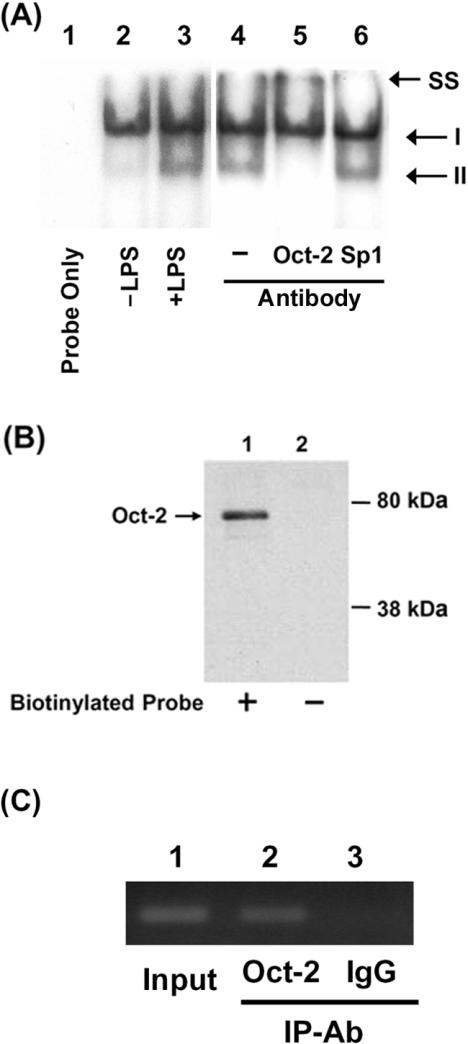
In order to verify binding of Oct-2 to the Octamer element, we performed EMSAs using a 32P-labelled DNA fragment (−927 to −895) and nuclear extracts from untreated or LPS-treated RAW264.7 cells. When the labelled probe was incubated with nuclear extract from control RAW264.7 cells, a major DNA–protein complex (I) and a minor complex (complex II) were detected (Figure 4A, lane 2). Increase in complex II was seen when the probe was incubated with nuclear extract from LPS-treated cells (Figure 4A, lanes 3 and 4). The formation of complex II disappears when nuclear extracts were pre-incubated with Oct-2 Ab (Figure 4A, lane 5); however, a control Ab against Sp1 did not affect the formation of complex II (Figure 4A, lane 6). These results suggest that Oct-2 is involved in the formation of complex II. Binding of Oct-2 to the Octamer element was demonstrated further using a DNA-affinity precipitation assay using a biotinylated probe containing the Octamer site and detected the precipitated protein with anti-Oct-2 Ab. Figure 4(B) shows that a protein with a molecular mass of approx. 75 kDa was detected by the assay (Figure 4B, lane 1), whereas no protein was detected in control experiment without the biotinylated DNA probe (Figure 4B, lane 2). A ChIP assay was carried out to examine binding of Oct-2 to the Octamer element in vivo. Figure 4(C) shows that a resistin promoter region encompassing the Octamer element could be pulled down by anti-Oct-2 Ab, while a control Ab could not. These results show that Oct-2 exists in RAW264.7 macrophages and binds to the Octamer element of the resistin promoter.
Figure 4. Binding of Oct-2 to the Octamer element in the resistin promoter.
Binding of Oct-2 to the Octamer element was demonstrated by EMSA (A), DNA-affinity precipitation assay (B) and ChIP (C). (A) EMSAs were carried out as described in the Experimental section using a probe covering nucleotides −927 to −895 of the resistin promoter. Lane 1, probe alone; lane 2, 20 μg of nuclear extract from untreated cells; lanes 3–6, 20 μg of nuclear extract from LPS-treated cells; lane 4, no Ab; lane 5, Oct-2 Ab; lanes 6, Sp1 Ab. SS, supershift. (B) Nuclear protein (100 μg) from LPS-treated cells (lane 1) was incubated with a biotinylated oligonucleotide from the Octamer region (−927 to −895 bp) of the resistin promoter, and the complex captured using streptavidin-coated magnetic beads. The captured proteins were separated by SDS/8% PAGE and immunoblotted for Oct-2. Nuclear extracts incubated without the biotinylated oligonucleotide were used as negative controls (lanes 2). (C) RAW264.7 cells were treated with LPS (10 ng/ml) for 12 h, then ChIP assays were performed as described in the Experimental section. IgG was used as a negative control. A 299 bp resistin promoter (−1193 to −895 bp) was amplified by PCR and separated on an agarose gel. A total of 10% of the chromatin DNA used for immunoprecipitation (IP) was subjected to PCR and is indicated as ‘Input’.
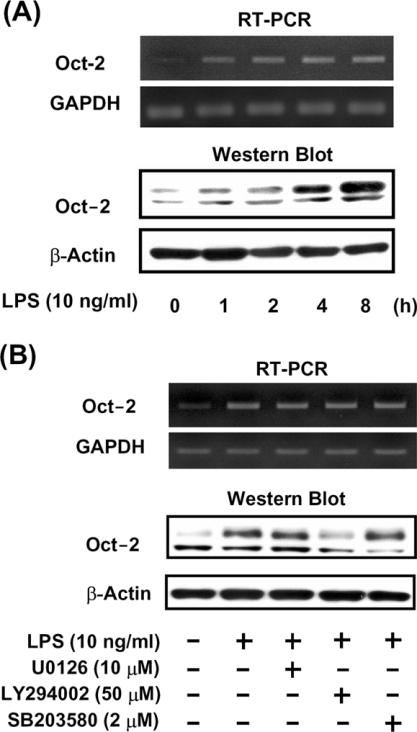
LPS treatment increases both Oct-2 mRNA and protein levels in RAW264.7 cells
To examine the effect of LPS stimulation on Oct-2 mRNA and protein levels in macrophages, RAW264.7 cells were exposed to LPS for 1–8 h, then Oct-2 mRNA and protein were measured by RT–PCR and Western blot analysis respectively. Figure 5(A) shows that Oct-2 mRNA and the 75 kDa Oct-2 protein were barely detectable in untreated cells, but their levels increased considerably after 1 h of LPS treatment and increased in a time-dependent manner (Figure 5A). A protein with a molecular mass of approx. 60 kDa was also detected when total cell lysate was used in the Western blot analyses. However, this protein signal was not detected in the DNA-affinity precipitation assay (Figure 4A) or by Western blot analysis when nuclear protein was used (results not shown) suggesting that the 60 kDa protein existed only in the cytoplasm but not in the nuclear extract. To understand the signalling cascade triggering the increase in Oct-2 gene expression induced by LPS, specific pharmacological antagonists (LY294002, a PI3K inhibitor, SB203580, a p38 MAPK inhibitor, and U0126, a MEK inhibitor) were used to pre-treat RAW264.7 cells for 30 min before the addition of LPS. Figure 5(B) shows that the LPS-induced increase in Oct-2 mRNA levels was unaffected by any of the pre-treatments, but the LPS-induced increase in 75 kDa Oct-2 protein was blocked by LY294002.
Figure 5. LPS increases both levels of Oct-2 mRNA and protein in RAW264.7 macrophages.
(A) RAW264.7 cells were treated with 100 ng/ml LPS for 0, 1, 2, 4 or 8 h, or (B) pre-treated for 30 min with U0126 (10 μM), LY294002 (50 μM) or SB203580 (2 μM), then LPS was added to a final concentration of 10 ng/ml and incubation continued for a further 4 h. Total RNA and protein were isolated for Oct-2 mRNA and protein detection by RT–PCR and Western blot analysis respectively.
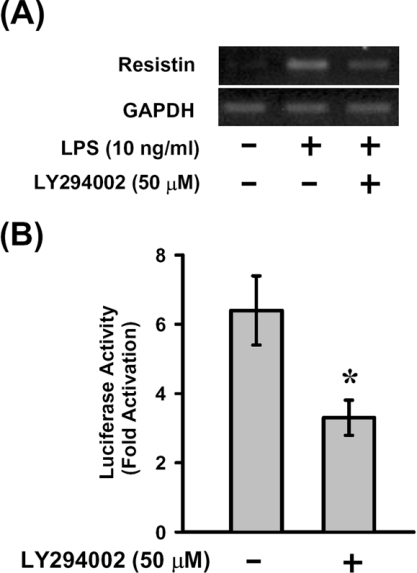
LPS-induced increases of resistin mRNA and promoter activity are inhibited by LY294002
The role of the PI3K pathway in LPS-induced up-regulation of resistin mRNA and promoter activity was evaluated in RAW264.7 cells and cells transfected with pResistin(−996/+61)-Luc. Exposure of the cells to LY294002 resulted in inhibition of LPS-induced increase in resistin mRNA (Figure 6A) and inhibition of the LPS-induced luciferase activity (Figure 6B). These results also support that Oct-2 is required for the up-regulation of resistin gene expression in response to LPS treatment.
Figure 6. LPS-induced increase in resistin mRNA and promoter activity were inhibited by LY294002.
Untransfected (A) or pResistin(−996/+61)-Luc-transfected (B) RAW264.7 cells were pre-treated for 30 min with LY294002 (50 μM), then treated with 10 ng/ml LPS for 4 h. Resistin mRNA and luciferase activities were assayed as described. Fold activation (+LPS/−LPS) of luciferase activity by LPS treatment is shown. Results are means±S.D. for three independent experiments. *P<0.05 compared with that without pre-treatment with LY294002.
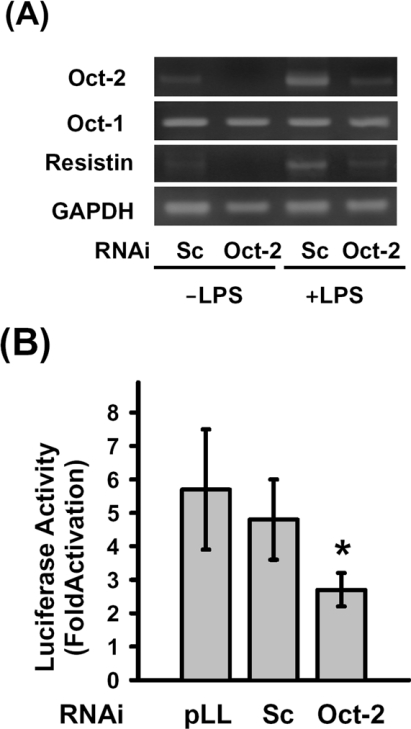
Knockdown of Oct-2 expression by RNAi prevented LPS-induced resistin and iNOS expression
In order to confirm the role of Oct-2 in resistin gene expression regulated by LPS treatment, we constructed an Oct-2 RNAi vector (pLL3.7-Oct-2) that could specifically knockdown the expression of Oct-2 mRNA. Figure 7(A) shows that RNAi-mediated knockdown of Oct-2 resulted in decrease of LPS-induced resistin mRNA level, but the levels of Oct-1 and GAPDH mRNA were not affected. Co-transfection of cells with pLL3.7-Oct-2 and pResistin(−996/+61)-Luc plasmids led to an decrease of the LPS-mediated induction of luciferase activity; however, the LPS-induced luciferase activity was not affected when the pResistin(−996/+61)-Luc plasmid was co-transfected with a scrambled control RNAi plasmid (Figure 7B). These results support the above findings that Oct-2 is involved in the LPS-mediated induction of resistin expression in macrophages.
Figure 7. Knockdown of Oct-2 inhibits the expression of resistin in response to LPS treatment.
(A) The pLL3.7-Oct-2 plasmid was transfected into RAW264.7 cells using the SuperFect reagent. At 44 h after transfection, the cells were left untreated or were treated with 100 ng/ml LPS for 4 h, then Oct-2, Oct-1, resistin, iNOS and GAPDH mRNA were detected by RT–PCR as described in the Experimental section. (B) RAW264.7 cells were co-transfected with pResistin(−996/+61)-Luc and pLL3.7-Oct-2 plasmids for 24 h, then were either left untreated or were treated with LPS for 4 h. Fold activation (+LPS/−LPS) of luciferase activity by LPS treatment is shown. Results are means±S.D. for three independent experiments. pLL, pLL3.7 plasmid; Sc, pLL3.7-scrambled plasmid. *P<0.05 compared with the pLL3.7 or scramble control.
DISCUSSION
Although expression of resistin was reported mainly in the adipose tissues in rodents, our previous study showed that small amount of resistin mRNA is expressed in white blood cells of rats and that the expression was up-regulated by LPS [16]. Although the level of resistin mRNA is relatively low in RAW264.7 cells, it is readily detectable under certain conditions. Thommesen et al. [29] showed that the levels of resistin mRNA in RAW264.7 cells can be induced 6-fold and 300-fold after exposure to 200 ng/ml PMA or 100 μM chlorophenylthio-cAMP for 12 h respectively. In the present study, we have shown a 34-fold increase in resistin mRNA in RAW264.7 cells treated with 100 ng/ml LPS for 8 h (Figure 1). However, we failed to detect resistin protein by Western blot which might be due to a very low level of resistin expressed in RAW264.7 cells. Recently, Lehrke et al. [21] detected resistin mRNA and protein in primary human macrophages, and showed that the expression of resistin was up-regulated by LPS in vivo and in vitro. These results suggest that resistin is expressed in macrophages and may play a role in the inflammatory process.
Resistin promoter activity, assayed by transfection with a resistin promoter–luciferase reporter gene, was increased in parallel with the increase in resistin mRNA levels in response to LPS, suggesting that LPS increases resistin mRNA levels mainly by increased transcription. Dissection of the resistin promoter by deletion and mutation studies clearly demonstrated that the Octamer element plays a crucial role in LPS-mediated induction of resistin promoter activity (Figure 2C). However, the Octamer may not be the only cis-element that responsible for the LPS-mediated induction of resistin promoter activity, since the luciferase activity in cells transfected with Octamer deleted or mutated pResistin(−1037/+61)-Luc still can be increased 2-fold after stimulation with LPS (Figure 2C). An Octamer element is reported to be required for LPS-mediated induction of iNOS promoter activity in macrophages, but an NF-κB (nuclear factor κB)-binding element, located 20 bp upstream of the Octamer element, is also required, and mutation of either results in loss of LPS-mediated induction of promoter activity to a level 2–3% of that of the wild-type [30]. These results suggest that the Octamer element alone is not sufficient for maximal LPS activation of the iNOS promoter and that this depends on interaction of the Octamer element with the NF-κB-binding element. It is not clear which of the two Octamer-binding proteins, Oct-1 or Oct-2, is responsible for iNOS activation in response to LPS. In the present study, we showed that co-transfection with a resistin promoter–luciferase reporter construct and an Oct-2 expression plasmid led to a robust increase in resistin promoter activity, particularly in the presence of LPS stimulation (Figure 3B). In contrast, co-transfection with a resistin promoter–luciferase reporter construct and an Oct-1 expression plasmid had no effect on basal resistin promoter activity, but co-transfection with an increasing amount of the Oct-1 expression plasmid resulted in a gradual decrease in LPS-mediated induction of resistin promoter activity (Figure 3A). Moreover, RNAi-mediated knockdown of endogenous Oct-2 resulted in inhibition of LPS-induced increase in resistin mRNA and promoter activity (Figure 7). These results strongly suggest that Oct-2, but not Oct-1, plays a crucial role in mediating LPS-induced activation of the resistin promoter. As both Oct-1 and Oct-2 bind equally well to the Octamer sequence, it remains unclear why Oct-2 specifically directs the expression of resistin in macrophages. Tanaka et al. [31] showed that Oct-1 and Oct-2 can differentially activate transcription through the use of promoter-selective activation domains, but not through DNA-binding specificity. It is also possible that interactions with another transcription factor and/or cofactors are essential for Oct-2-specific transcriptional activation. The decrease in LPS-mediated induction of promoter activity in Oct-1-overexpressing cells is presumably due to Oct-1 competing with endogenous Oct-2 for binding to the Octamer. The results shown in Figure 3(C) in which cells were transfected with a constant amount of Oct-2 expression plasmid and increasing amounts of Oct-1 expression plasmid support this idea. Unexpectedly, a resistin promoter bearing a mutant Octamer was also activated when co-transfected with the Oct-2 expression plasmid (Figure 3D), although the magnitude of activation was much lower than that using a wild-type promoter. One possible explanation is that Oct-2 might bind to the mutant sequence with a lower affinity and activate transcription less efficiently. However, binding of Oct-2 to sequences unrelated to the Octamer element cannot be excluded, since it has been reported to bind to a TAATGARAT-like heptamer motif in the tyrosine hydroxylase promoter [32].
Oct-1 and Oct-2 both belong to the POU-domain activator proteins; Oct-1 is ubiquitously expressed, whereas Oct-2 is expressed predominantly in B-cells [33], but is also present in neuronal cells [34], T-cells [35] and macrophages [36]. Oct-2 is known to be a critical mediator of immunoglobulin gene transcription in B-cells [37], but little is known of its functions in macrophages. Lopez-Rodriguez et al. [36] showed that Oct-2 is induced during the monocytic differentiation of U937 cells and is involved in transcription of CD11c integrin during the process. In the present study, we detected Oct-2 mRNA and Oct-2 protein in RAW264.7 cells and found that levels of both were increased by LPS treatment (Figure 5). An LPS-induced increase in Oct-2 mRNA and protein levels is also seen in primary mouse peritoneal macrophages (T.-Y. Wu and L.-C. Lu, unpublished work). Several Oct-2 isoforms have been identified in mouse B-cell lines [38,39]. Miller et al. [39] have shown that expression of both Oct-2A (60 kDa) and Oct-2B (75 kDa) proteins were induced by LPS in mouse B-cell lines. However, only a 75 kDa Oct-2 protein, which may represent Oct-2B protein, appeared to be LPS-inducible in macrophages (Figure 5). It is not clear what mechanism is responsible for macrophages not producing the 60 kDa Oct-2A protein. Wirth et al. [40] have isolated multiple cDNAs encoding multiple Oct-2 isoforms in mouse B-cells and have demonstrated that the multiple cDNAs were generated from a single gene by an alternative splicing mechanism.
While investigating the signalling molecules that involve in Oct-2 activation, we found that LPS-induced increase in Oct-2B protein was inhibited by a PI3K inhibitor, LY294002 (Figure 5B), but that the Oct-2 mRNA was unaffected. These results suggest that LPS may up-regulate Oct-2B protein at the post-transcriptional level through the PI3K pathway. In addition, the LPS-induced increase in resistin mRNA and promoter activity were also inhibited by LY294002 in cells transfected with pResistin(−996/+61)-Luc (Figure 6). These results also support the hypothesis that activation of Oct-2 is essential for up-regulation of resistin gene expression. Increased protein production induced by LPS has been reported to be regulated via an increased translation rate or reduced proteasome degradation. Raabe et al. [41] have shown that translation rate of TNF-α mRNA increased approx. 2–3-fold after stimulation with LPS for 2 h. On the other hand, LPS-induced nuclear accumulation and transcriptional activation of β-catenin through the PI3K-dependent pathway that reduces the ubiquitination and proteasome degradation of β-catenin in alveolar macrophages [42]. The signals downstream of PI3K that involve in regulation of Oct-2B protein level in LPS-treated macrophages are currently under investigation.
In summary, we have shown that LPS induces an increase in resistin gene expression in RAW264.7 macrophages and that this requires an Octamer element, located at −914 to −907, in the resistin promoter. Our results demonstrate that up-regulation of Oct-2B protein is required for LPS-mediated induction of resistin gene expression in macrophages. Although previous studies have suggested that Oct-2 expression was restricted primarily to B-lymphocytes [33,37], our results show, for the first time, that LPS treatment increases Oct-2 mRNA and protein levels in macrophages. Inhibition of Oct-2 by a PI3K inhibitor (LY294002) or by Oct-2-specific RNAi impaired the LPS-induced up-regulation of resistin gene expression. Taken together, our results show that activation of Oct-2 is required for up-regulation of resistin gene expression in macrophages and suggest that Oct-2 may play an important role in the regulation of LPS-responsive genes, such as those coding for iNOS and G-CSF (granulocyte colony-stimulating factor), which contain Octamer elements in their promoters.
Acknowledgments
This work was supported by research grant NSC92-2320-B-002-124 from the National Science Council of Taiwan. We are highly indebted to Dr Winship Herr (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.) for providing the pCG-Oct-1, pCG-Oct-2 and pCG plasmids. We thank Dr Lih Kuo (Texas A&M University, College Station, TX, U.S.A.) for pre-submission critical reviewing of the manuscript and Dr Chia-Chu Tsai for helping in the set-up of real-time PCR.
References
- 1.Steppan C. M., Bailey S. T., Bhat S., Brown E. J., Banerjee R. R., Wright C. M., Patel H. R., Ahima R. S., Lazar M. A. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 2.Degawa-Yamauchi M., Bovenkerk J. E., Juliar B. E., Watson W., Kerr K., Jones R., Zhu Q., Considine R. V. Serum resistin (FIZZ3) protein is increased in obese humans. J. Clin. Endocrinol. Metab. 2003;88:5452–5455. doi: 10.1210/jc.2002-021808. [DOI] [PubMed] [Google Scholar]
- 3.Azuma K., Katsukawa F., Oguchi S., Murata M., Yamazaki H., Shimada A., Saruta T. Correlation between serum resistin level and adiposity in obese individuals. Obes. Res. 2003;11:997–1001. doi: 10.1038/oby.2003.137. [DOI] [PubMed] [Google Scholar]
- 4.Rajala M. W., Obici S., Scherer P. E., Rossetti L. Adipose-derived resistin and gut-derived resistin-like molecule-β selectively impair insulin action on glucose production. J. Clin. Invest. 2003;111:225–230. doi: 10.1172/JCI16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee R. R., Rangwala S. M., Shapiro J. S., Rich A. S., Rhoades B., Qi Y., Wang J., Rajala M. W., Pocai A., Scherer P. E., et al. Regulation of fasted blood glucose by resistin. Science. 2004;303:1195–1198. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- 6.Rangwala S. M., Rich A. S., Rhoades B., Shapiro J. S., Obici S., Rossetti L., Lazar M. A. Abnormal glucose homeostasis due to chronic hyperresistinemia. Diabetes. 2004;53:1937–1941. doi: 10.2337/diabetes.53.8.1937. [DOI] [PubMed] [Google Scholar]
- 7.Pravenec M., Kazdova L., Landa V., Zidek V., Mlejnek P., Jansa P., Wang J., Qi N., Kurtz T. W. Transgenic and recombinant resistin impair skeletal muscle glucose metabolism in the spontaneously hypertensive rat. J. Biol. Chem. 2003;278:45209–45215. doi: 10.1074/jbc.M304869200. [DOI] [PubMed] [Google Scholar]
- 8.Satoh H., Nguyen M. T., Miles P. D., Imamura T., Usui I., Olefsky J. M. Adenovirus-mediated chronic “hyper-resistinemia” leads to in vivo insulin resistance in normal rats. J. Clin. Invest. 2004;114:224–231. doi: 10.1172/JCI20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Way J. M., Gorgun C. Z., Tong Q., Uysal K. T., Brown K. K., Harrington W. W., Oliver W. R., Jr, Willson T. M., Kliewer S. A., Hotamisligil G. S. Adipose tissue resistin expression is severely suppressed in obesity and stimulated by peroxisome proliferator-activated receptor γ agonists. J. Biol. Chem. 2001;276:25651–25653. doi: 10.1074/jbc.C100189200. [DOI] [PubMed] [Google Scholar]
- 10.Nagaev I., Smith U. Insulin resistance and Type 2 diabetes are not related to resistin expression in human fat cells or skeletal muscle. Biochem. Biophys. Res. Commun. 2001;285:561–564. doi: 10.1006/bbrc.2001.5173. [DOI] [PubMed] [Google Scholar]
- 11.Savage D. B., Sewter C. P., Klenk E. S., Segal D. G., Vidal-Puig A., Considine R. V., O'Rahilly S. Resistin/Fizz3 expression in relation to obesity and peroxisome proliferator-activated receptor-γ action in humans. Diabetes. 2001;50:2199–2202. doi: 10.2337/diabetes.50.10.2199. [DOI] [PubMed] [Google Scholar]
- 12.Janke J., Engeli S., Gorzelniak K., Luft F. C., Sharma A. M. Resistin gene expression in human adipocytes is not related to insulin resistance. Obes. Res. 2002;10:1–5. doi: 10.1038/oby.2002.1. [DOI] [PubMed] [Google Scholar]
- 13.Lee J. H., Chan J. L., Yiannakouris N., Kontogianni M., Estrada E., Seip R., Orlova C., Mantzoros C. S. Circulating resistin levels are not associated with obesity or insulin resistance in humans and are not regulated by fasting or leptin administration: cross-sectional and interventional studies in normal, insulin-resistant, and diabetic subjects. J. Clin. Endocrinol. Metab. 2003;88:4848–4856. doi: 10.1210/jc.2003-030519. [DOI] [PubMed] [Google Scholar]
- 14.Holcomb I. N., Kabakoff R. C., Chan B., Baker T. W., Gurney A., Henzel W., Nelson C., Lowman H. B., Wright B. D., Skelton N. J., et al. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 2002;19:4046–4055. doi: 10.1093/emboj/19.15.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Ambrosi J., Fruhbeck G. Do resistin and resistin-like molecules also link obesity to inflammatory diseases? Ann. Intern. Med. 2001;135:306–307. doi: 10.7326/0003-4819-135-4-200108210-00030. [DOI] [PubMed] [Google Scholar]
- 16.Lu S. C., Shieh W. Y., Chen C. Y., Hsu S. C., Chen H. L. Lipopolysaccharide increases resistin gene expression in vivo and in vitro. FEBS Lett. 2002;530:158–162. doi: 10.1016/s0014-5793(02)03450-6. [DOI] [PubMed] [Google Scholar]
- 17.Kaser S., Kaser A., Sandhofer A., Ebenbichler C. F., Tilg H., Patsch J. R. Resistin messenger-RNA expression is increased by proinflammatory cytokines in vitro. Biochem. Biophys. Res. Commun. 2003;309:286–290. doi: 10.1016/j.bbrc.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Harsch I. A., Koebnick C., Wallaschofski H., Schahin S. P., Hahn E. G., Ficker J. H., Lohmann T., Konturek P. C. Resistin levels in patients with obstructive sleep apnoea syndrome: the link to subclinical inflammation? Med. Sci. Monit. 2004;10:CR510–CR515. [PubMed] [Google Scholar]
- 19.McTernan P. G., Fisher F. M., Valsamakis G., Chetty R., Harte A., McTernan C. L., Clark P. M., Smith S. A., Barnett A. H., Kumar S. Resistin and Type 2 diabetes: regulation of resistin expression by insulin and rosiglitazone and the effects of recombinant resistin on lipid and glucose metabolism in human differentiated adipocytes. J. Clin. Endocrinol. Metab. 2003;88:6098–6106. doi: 10.1210/jc.2003-030898. [DOI] [PubMed] [Google Scholar]
- 20.Stejskal D., Adamovska S., Bartek J., Jurakova R., Proskova J. Resistin concentrations in persons with Type 2 diabetes mellitus and in individuals with acute inflammatory disease. Acta Univ. Palacki. Olomouc. Fac. Med. 2003;147:63–69. [PubMed] [Google Scholar]
- 21.Lehrke M., Reilly M. P., Millington S. C., Iqbal N., Rader D. J., Lazar M. A. An inflammatory cascade leading to hyperresistinemia in humans. PloS Med. 2004;1:161–168. doi: 10.1371/journal.pmed.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Hartman H. B., Hu X., Tyler K. X., Dalal C. K., Lazar M. A. Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem. 2003;277:19754–19761. doi: 10.1074/jbc.M201451200. [DOI] [PubMed] [Google Scholar]
- 26.Chou S. F., Chen H. L., Lu S. C. Sp1 and Sp3 are involved in up-regulation of human deoxyribonuclease II transcription during differentiation of HL-60 cells. Eur. J. Biochem. 2003;270:1855–1862. doi: 10.1046/j.1432-1033.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- 27.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Zhang M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 28.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thommesen L., Stunes A. K., Monjo M., Grosvik K., Tamburstuen M. V., Kjobli E., Lyngstadaas S. P., Reseland J. E., Syversen U. Expression and regulation of resistin in osteoblasts and osteoclasts indicate a role in bone metabolism. J. Cell Biochem. 2006;99:824–834. doi: 10.1002/jcb.20915. [DOI] [PubMed] [Google Scholar]
- 30.Xie Q. A novel lipopolysaccharide-response element contributes to induction of nitric oxide synthase. J. Biol. Chem. 1997;272:14867–14872. doi: 10.1074/jbc.272.23.14867. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka M., Lai J. S., Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 32.Dawson S. J., Yoon S. O., Chikaraishi D. M., Lillycrop K. A., Latchman D. S. The Oct-2 transcription factor represses tyrosine hydroxylase expression via a heptamer TAATGARAT-like motif in the gene promoter. Nucleic Acids Res. 1994;22:1023–1028. doi: 10.1093/nar/22.6.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staudt L. M., Clerc R. G., Singh H., LeBowitz J. H., Sharp P. A., Baltimore D. Cloning of a lymphoid-specific cDNA encoding a protein binding the regulatory Octamer DNA motif. Science. 1988;241:577–580. doi: 10.1126/science.3399892. [DOI] [PubMed] [Google Scholar]
- 34.He X., Treacy M. N., Simmons D. M., Ingraham H. A., Swanson L. W., Rosenfeld M. G. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 35.Kang S. M., Tsang W., Doll S., Scherle P., Ko H. S., Tran A. C., Lenardo M. J., Staudt L. M. Induction of the POU domain transcription factor Oct-2 during T-cell activation by cognate antigen. Mol. Cell. Biol. 1992;12:3149–3154. doi: 10.1128/mcb.12.7.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Rodriguez C., Zubiaur M., Sancho J., Concha A., Corbi A. L. An Octamer element functions as a regulatory element in the differentiation-responsive CD11c integrin gene promoter: Oct-2 inducibility during myelomonocytic differentiation. J. Immunol. 1997;158:5833–5840. [PubMed] [Google Scholar]
- 37.Clerc R. G., Corcoran L. M., LeBowitz J. H., Baltimore D., Sharp P. A. The B-cell-specific Oct-2 protein contains POU box- and homeobox-type domains. Genes Dev. 1988;2:1570–1581. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber E., Matthias P., Müller M. M., Schaffner W. Identification of a novel lymphoid specific Octamer binding protein (OTF-2B) by proteolytic clipping bandshift assay (PCBA) EMBO J. 1988;7:4221–4229. doi: 10.1002/j.1460-2075.1988.tb03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller C. L., Feldhaus A. L., Rooney J. W., Rhodes L. D., Sibley C. H., Singh H. Regulation and a possible stage-specific function of Oct-2 during pre-B-cell differentiation. Mol. Cell. Biol. 1991;11:4885–4894. doi: 10.1128/mcb.11.10.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirth T., Priess A., Annweiler A., Zwilling S., Oeler B. Multiple Oct2 isoforms are generated by alternative splicing. Nucleic Acids Res. 1991;19:43–51. doi: 10.1093/nar/19.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raabe T., Bukrinsky M., Currie R. A. Relative contribution of transcription and translation to the induction of tumor necrosis factor-α by lipopolysaccharide. J. Biol. Chem. 1998;273:974–980. doi: 10.1074/jbc.273.2.974. [DOI] [PubMed] [Google Scholar]
- 42.Monick M. M., Carter A. B., Robeff P. K., Flaherty D. M., Peterson M. W., Hunninghake G. W. Lipopolysaccharide activates Akt in human alveolar macrophages resulting in nuclear accumulation and transcriptional activity of β-catenin. J. Immunol. 2001;166:4713–20. doi: 10.4049/jimmunol.166.7.4713. [DOI] [PubMed] [Google Scholar]