Abstract
Type III secretion genes in Aeromonas salmonicida subsp. salmonicida are located on a large plasmid of approximately 140 kb. Cultivation of this organism at elevated temperatures such as 25°C can, however, result in loss of this plasmid. This is accompanied by a loss of virulence for cultured fish cells.
Aeromonas salmonicida subsp. salmonicida (referred to here as A. salmonicida) is an important fish pathogen causing furunculosis, a systemic disease of the salmonids. Despite the economic significance of this bacterium, very little is known about the factors that it uses to cause disease.
We have recently identified a number of genes in A. salmonicida that comprise part of a functional type III secretion system (TTSS) (4). (For a comprehensive review of TTSSs, see reference 5.) In A. salmonicida, the TTSS is responsible for secretion of the ADP-ribosylating toxin AexT (4). However, we have observed that the bacterium often loses its ability to secrete AexT, particularly when grown at the recommended temperature of 25°C. We therefore examined the location of the TTSS genes in A. salmonicida and the response of these genes to growth under laboratory conditions. Here, we provide evidence that the TTSS genes in A. salmonicida are located on a large plasmid of approximately 140 kb but that this plasmid is lost when the cells are passaged at 25°C. Loss of the plasmid is accompanied by an inability to secrete AexT and by a loss of virulence for cultured fish cells. These findings have broad implications regarding the growth conditions used when studying A. salmonicida pathogenicity.
Secretion of AexT.
We examined secretion of the AexT toxin by three A. salmonicida strains: ATCC 33658T (American Type Culture Collection), strain JF2267 (a virulent strain isolated from an Arctic char [Savelinus alpinus] [3]), and strain JF2397 (a strain derived directly from strain JF2267 by nine consecutive passages at 25°C).
All strains were grown overnight at 18°C in Trypticase soy broth. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (6) was then carried out on the culture supernatants, and immunoblotting was performed using polyclonal anti-AexT antibodies (3). AexT was detected in the culture supernatant of strain JF2267 but not JF2397 or ATCC 33658T. However, the aexT gene was present in all three A. salmonicida strains as examined by Southern blot hybridization with a digoxigenin-labeled probe against the aexT gene (Fig. 1A).
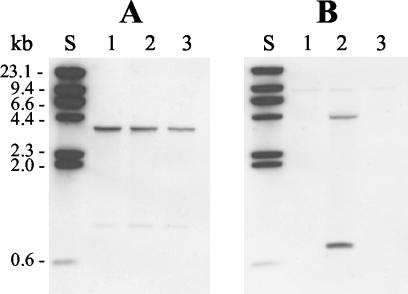
FIG. 1.
Presence of aexT and ascV genes. Shown are the results of Southern blot analysis of total DNA extracted from A. salmonicida strains ATCC 33658T (lanes 1), JF2267 (lanes 2), and JF2397 (lanes 3) and digested with the restriction endonuclease EcoRI. (A) Hybridization with aexT probe. (B) Hybridization with ascV probe. Lanes S contain lambda phage DNA digested with HindIII; fragment sizes are indicated.
We then utilized Southern blotting to examine all strains for the presence of the TTSS gene ascV (a gene encoding an inner membrane component of the TTSS). As expected, hybridization with DNA from strain JF2267, which is capable of AexT secretion, was positive for ascV (Fig. 1B). The presence of two bands which hybridize with the ascV probe in the sample of DNA from this strain can be explained by the fact that there is an EcoRI cut site between the binding sites of the primers used to generate the ascV probe. In contrast, strains ATCC 33658T and JF2397 did not react with the ascV probe (Fig. 1B), indicating that these strains lack the ascV gene. This finding explains the inability of these strains to secrete AexT even though both strains contain the aexT gene. The absence of ascV in JF2397, which was derived from the ascV-positive strain JF2267, suggests that the TTSS genes in A. salmonicida are located on a mobile genetic element that is easily lost during laboratory cultivation.
Plasmid location of type III secretion genes.
We compared the DNA profile of the wild-type (wt) strain JF2267 with that of its passaged derivative, strain JF2397, by pulsed-field gel electrophoresis (PFGE). The pattern of NotI restriction fragments of DNA from these strains appears to be identical except for a large fragment of approximately 110 kb that is found only in strain JF2267 (Fig. 2A). Southern blot analysis of NotI-digested DNA from these strains with a probe against ascV indicated that the ascV gene and presumably the remainder of the TTSS genes are located on this 110-kb fragment (Fig. 2B).
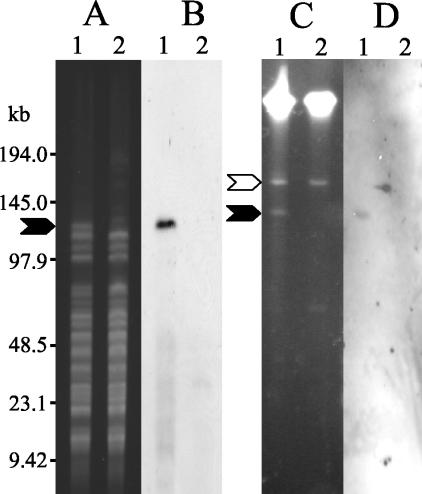
FIG. 2.
Location of the ascV gene by PFGE analysis. A. salmonicida strain JF2267 (lanes 1) and the passaged derivative strain JF2397 (lanes 2) were analyzed by PFGE. (A) Total DNA digested with NotI, separated by PFGE, and stained with ethidium bromide. (B) Southern blot of the gel shown in panel A, hybridized with a probe directed against ascV. (C) Samples prepared by S1 treatment, separated by PFGE, and stained with ethidium bromide. (D) Southern blot of the gel shown in panel C, hybridized with a probe directed against ascV. The positions of the standards are indicated by horizontal bars. The locations of the plasmid pASvirA (140 kb) in panels C and D and the 110-kb fragment of plasmid pASvirA in panels A and B are indicated by black arrows. The location of the cryptic 160-kb plasmid is indicated by an open arrow.
As TTSS genes are often located on plasmids, we wished to examine whether the 110-kb NotI fragment observed on PFGE of strain JF2267 represented plasmid DNA. To accomplish this, we utilized S1 nuclease treatment and PFGE, which is a reliable method of detecting and sizing large plasmids (1). The results of this experiment are shown in Fig. 2C. Two distinct plasmid bands of 140 and 160 kb can be seen in DNA from the wt isolate JF2267, whereas strain JF2397 contains only the 160-kb plasmid. Southern blot analysis revealed that the 140-kb plasmid in strain JF2267 hybridized with the ascV probe (Fig. 2D). No hybridization signal was found with the passaged strain JF2397. (ATCC 33658T does not contain the 140-kb plasmid and does not hybridize with the ascV probe [results not shown].) We conclude that the TTSS genes in A. salmonicida strain JF2267 are found on a large plasmid of approximately 140 kb, which we have named pASvirA. The difference in size between the 140-kb fragment, seen in the S1-treated DNA (Fig. 2D), and the 110-kb NotI fragment that hybridizes with ascV (Fig. 2B) indicates that the 140-kb plasmid contains more than one NotI restriction site. However, because of the relatively small size of the other NotI fragment(s), 30 kb or less, it cannot be distinguished under the PFGE conditions used. The finding that strain JF2397 does not possess plasmid pASvirA indicates that this strain has lost the plasmid during growth at 25°C. In a control experiment with the aexT probe for hybridization, we could localize the aexT gene to the chromosome of the three strains, JF2267, JF2397, and ATCC 33658T (results not shown).
Instability of plasmid pASvirA.
We examined strain JF2267 for the presence of ascV during growth at different temperatures to learn how quickly plasmid pASvirA is lost during laboratory cultivation of A. salmonicida. Strain JF2267 was inoculated into Luria-Bertani broth to an optical density at 600 nm of 0.08 and incubated for 2 h at 18°C. After this time, the culture was split into two equal parts; one part was incubated at 18°C, while the other was incubated at 25°C. Samples were taken from each culture at regular intervals and plated onto Luria-Bertani agar plates which were then grown at 18°C for 24 h. After this time, colony blot hybridizations were performed on all plates according to the method described by Braun et al. (2). The percentage of ascV-positive colonies relative to the total number of colonies on each plate was then determined. At 25°C, all cells contained the ascV gene for the first 5 h following the temperature shift. However, after this time, the gene was rapidly lost, and 1 h later it could not be detected within the population. In contrast, all colonies from the culture incubated at 18°C retained ascV even after 24 h of incubation.
Toxicity of A. salmonicida.
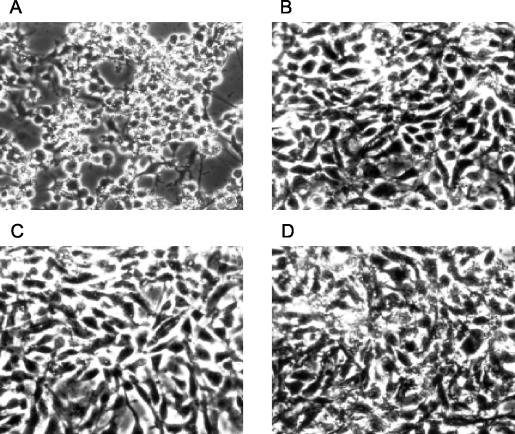
The impact of plasmid pASvirA on the virulence of A. salmonicida was analyzed. Rainbow trout (Oncorhynchus mykiss) gonad cells (RTG-2, ATCC CCL-55) were grown as described previously (3). RTG-2 cells were inoculated with either strain JF2267, JF2397, or ATCC 33658T at a multiplicity of infection of 20:1 (bacteria/fish cells). Two and six hours following infection at 18°C, the cells were examined by phase-contrast microscopy (Axiovert 100; Zeiss). Two hours following infection, RTG-2 cells inoculated with the wt strain JF2267 had undergone cell rounding and retraction (Fig. 3A) while cells inoculated with strain JF2397 or with ATCC 33658T underwent no morphological changes after 2 or 6 h of incubation (Fig. 3B and C).
FIG. 3.
Virulence of A. salmonicida strains for RTG-2 cells. Shown are RTG-2 cells inoculated with strains JF2267 (A), ATCC 33658T (B), and JF2397 (C) and with phosphate-buffered saline only (D). Pictures were taken 2 h after infection.
Conclusion.
The TTSS genes in A. salmonicida are found on a 140-kDa plasmid, which we have termed pASvirA. This plasmid is thermolabile and is rapidly lost during growth at 25°C. Loss of plasmid pASvirA is accompanied by an inability of A. salmonicida to secrete AexT and by a loss of virulence for RTG-2 cells. A. salmonicida ATCC 33658T does not contain plasmid pASvirA, does not secrete AexT, and is avirulent for RTG-2 cells. These findings have marked implications for the study of A. salmonicida pathogenicity, as (i) such studies often refer to the type strain and (ii) the recommended growth temperature for A. salmonicida is 22 to 28°C as reported by Bergey's Manual of Systematic Bacteriology and other guidelines. However, cultures grown at these temperatures are likely to have lost the TTSS genes, an event that undoubtedly has a significant impact on the virulence of the organism. We recommend that A. salmonicida be grown at temperatures below 20°C in order to ensure that all virulence genes are retained during cultivation. Studies of molecular mechanisms of virulence in A. salmonicida should refer to wt strains that are known to harbor plasmid pASvirA. Furthermore we suggest using the presence or absence of plasmid pASvirA to assess virulence of A. salmonicida subsp. salmonicida isolates.
Acknowledgments
We are grateful to Yvonne Schlatter and Lea Lagcher for valuable technical assistance.
This work was supported by the Priority Program Biotechnology of the Swiss National Science foundation (grant 5002-045027) and by the Institute of Veterinary Bacteriology Research Fund.
REFERENCES
- 1.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Braun, M., P. Kuhnert, J. Nicolet, A. P. Burnens, and J. Frey. 1999. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J. Bacteriol. 181:2501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, M., K. Stuber, Y. Schlatter, T. Wahli, P. Kuhnert, and J. Frey. 2002. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burr, S. E., K. Stuber, T. Wahli, and J. Frey. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:5966-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 6.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]