Abstract
Group A human rotavirus strains belonging to the unusual serotype G9 were detected at high frequency in stool specimens from infected children with acute diarrhea in Bari, Italy, during a 15-month survey from March 2001 to June 2002. This may signify a local reemergence of the G9 rotaviruses detected in Italy in the early and mid-1990s or may be related to the global emergence of G9 rotaviruses in recent years.
Rotaviruses are the major etiologic agents of infantile diarrhea worldwide and are responsible for up to 3.3 million child deaths per year in developing countries (10, 13). Rotaviruses are nonenveloped and possess a trilaminar capsid containing 11 segments of double-stranded RNA (7). The two proteins constituting the outer capsid, VP7 and VP4, elicit a neutralizing antibody response and bear the G and P serotype specificities, respectively. At present, 10 G serotypes and 7 P serotypes have been identified in humans (10, 13). Epidemiological studies have demonstrated that rotavirus serotypes G1, G2, G3, and G4 are the most common types associated with disease globally (more than 90% of all the G serotypes detected), and therefore they are the targets of vaccine development (10, 13). Regarding P type specificity, P1A[8] is the most common P serotype detected worldwide, followed by P1B[4] and P2A[6] (10). G serotypes 1, 3, and 4 are most commonly associated with P type P1A[8], whereas G serotype 2 segregates preferentially with P type 1B[4] (10).
Recently, two unusual rotavirus serotypes, G5 and G8, have been described in association with diarrhea in various parts of the world (10). The detection of rotaviruses with the G9 specificity has been reported in the Asian (12, 19), American (2, 6, 17), African (6), and Australian (16) continents at either low or high frequency. In Europe, G9 rotaviruses have been detected sporadically in the United Kingdom, Ireland, France, and Italy. Moreover, a prolonged nosocomial outbreak caused by a P2A[6],G9 strain has been recently reported in Holland (1, 3, 4, 5, 15, 20). Because of the increasing number of reports, there is evidence for an apparently expanding distribution, on a global scale, of G9 rotaviruses, and currently the G9 serotype is considered the fifth most common type worldwide. In the present note we describe the detection at a high frequency in Bari, Italy, of rotaviruses belonging to the emerging G9 serotype. Thirty rotavirus-positive stool samples were collected between March 2001 and June 2002 from children under 5 years of age who presented with acute diarrhea to the Policlinico Hospital in Bari. The presence of rotavirus in those samples was determined by an immunochromatographic assay (Rotascreen dipstick; Microgen Bioproducts, Camberley, United Kingdom). Rotavirus double-stranded RNA was extracted from the stool samples by adsorption on cellulose CF11 (21) and analyzed by reverse transcription-PCR and sequence analysis for determination of the G (VP7) and P (VP4) specificity, as well as of the subgroup specificity (VP6) (8, 9, 11). The PCR products obtained from the VP7, VP4, and VP6 genes were purified through Ultrafree-Da columns (Millipore, Bedford, Mass.), and the sequences were determined using the BigDye sequencing kit (Applied Biosystems, Foster City, Calif.) and an ABI 377 automatic DNA sequencer (Applied Biosystems). Phylogenetic analysis was conducted using the MEGA software package, version 2.1 (14).
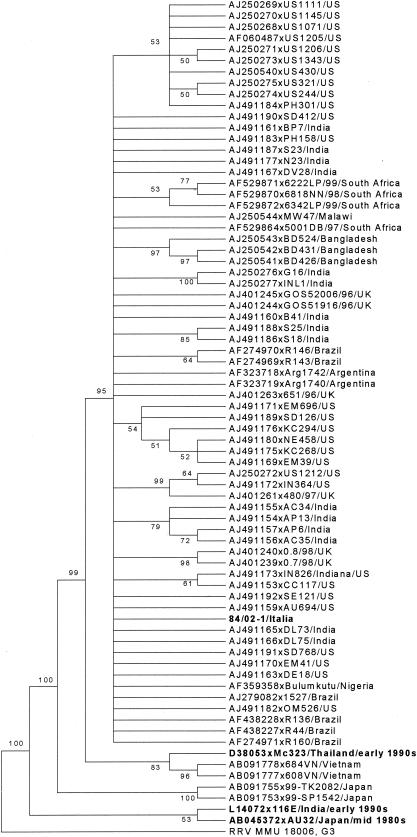
Sixteen of 30 samples (53.3%) were typed as rotavirus genotype G9, while 12 (40.0%) were G1 rotaviruses and one was a mixed infection (Table 1). The G9 strains were detected over the entire period of observation, even if there was apparently a higher frequency of detection of G9 rotaviruses at the beginning of 2001 and of G1 in late 2001 and early 2002 (Table 2). Sequence analysis of VP7 revealed high genetic similarity among the G9 strains detected (about 100%). Nucleotide variation from several G9 strains of recent identification in various parts of the world (Brazil, India, Japan, United States, Malawi, South Africa, and United Kingdom) ranged from 0.6 to 2.4%, with the highest identity (99.4% at the nucleotide and 98.4% at the amino acid level) being to the Brazilian G9 strain R136. Nucleotide variation from Vietnamese and Japanese G9 strains was more than 3 and 5%, respectively. Conversely, genetic relatedness was much lower to old G9 rotaviruses. For instance, there was a nucleotide and amino acid sequence similarity of about 89.6 and 91.4%, respectively, to the Indian strain 116E, isolated in 1985. In the phylogenetic tree (Fig. 1), the Italian strains segregated in a large group encompassing Asian, North and South American, African, and European G9 strains, which was clearly different (bootstrap value = 95%) from the Vietnamese and Japanese strains and from the “old” G9 strains 116E and AU32.
TABLE 1.
G and P type distribution of the rotavirus strains detected in Bari in 2001-2002a
| No. of sample | VP7 G type | VP4 P type and lineage | VP6 genogroup |
|---|---|---|---|
| 13 | G9 | P[8] Malawian-like | II |
| 1 | G9b | P[8]b | II |
| 2 | G9b | ND | ND |
| 5 | G1 | P[8] Japanese-like | II |
| 7 | G1 | P[8] American-like | II |
| 1 | G1 + G9b | P[8]b | ND |
| 1 | ND | P[8]b | ND |
Thirty samples positive by a quick diagnostic test were subjected to molecular analysis. ND, not determined. Of the 30 samples, 29 were typed for VP7 6 type, 28 were typed for VP4 p type and lineage, and 26 were typed for VP6 genogroup.
The VP7 and VP4 specificities were determined by a typing assay based on a heminested PCR with type-specific primers, since poor amplification of the first-round PCR did not allow sequence analysis.
TABLE 2.
Temporal distribution of the G9 and G1 strains detected in Bari during 2001-2002
| Yr, mo | No. of strains
|
|
|---|---|---|
| G9 | G1 | |
| 2001, March | 2 | |
| 2001, April | 2 | |
| 2001, May | 1 | |
| 2001, June | 1 | 1 |
| 2001, July | 2 | |
| 2001, August | 1 | |
| 2001, September | 2 | |
| 2001, October | 1 | |
| 2001, November | 1 | |
| 2001, December | ||
| 2002, January | 1a | 3 + 1a |
| 2002, February | ||
| 2002, March | 1 | 3 |
| 2002, April | ||
| 2002, May | 3 | 1 |
| 2002, June | 1 | 2 |
Mixed infection, G9 + G1.
FIG. 1.
Consensus tree based on the VP7 genes of a wide selection of G9 rotaviruses. The tree was elaborated by the neighbor-joining method and by calculating the distances with the Kimura two-parameter algorithm and supplying a statistical support by bootstrapping over 1,000 replicates. Bootstrap values lower than 50% are not shown. The branches of the tree are not drawn to scale. The tree is rooted using the VP7 gene of simian strain RRV MMU 18006, P5B[3],G3.
Analysis of the VP4 gene revealed that both the G1 and G9 strains belonged to the P type P1A[8]. However, by sequence and phylogenetic analysis it was possible to distinguish three different P[8] lineages, resembling the VP4 of a Malawian rotavirus for G9 strains and of either a Japanese or an American strain for G1 rotaviruses (data not shown). The G9 strains displayed the highest degree of genetic similarity to the P[8],G1 Malawian strain OP351 (more than 99%). The VP6 of all the strains analyzed was characterized as genogroup II.
Thus far, rotavirus serotype G9 has been associated with VP4 genotype P[4], P[6], P[8], P[11], or P[19] (3, 15, 18). Analysis of a global collection of G9 viruses showed that strains from different geographic locations may share the same constellation of genes and be virtually identical, while strains from the same country may have a highly conserved VP7 gene but a diverse assortment of other genes (18). Here we found G9 rotaviruses bearing the P[8] genotype and subgroup II specificity. In Italy, rotavirus strains bearing P1A[8],G9 and subgroup II specificity have been previously described but at very low frequency (from 0.7 to 1.7%) in two surveys conducted in the early and mid-1990s in Palermo, Sicily (1, 4). Of note, no G2, G3, or G4 rotavirus strains, the most common worldwide after G1 rotaviruses, were detected during the 15-month period of observation in the hospital in Bari. This was particularly surprising since those G types accounted for about 26 to 50% of the strains typed in Palermo in the 1990s, with G1 always being highly frequent (40.9 to 50.6%) (1, 4). This might be explained as a geographical variation of the serotype distribution or as a temporal variation occurring in recent years in Italy. However, it may be possible that a correlation exists between the spread of the G9 serotype strains and the relevant influxes of immigrants from countries with poor sanitation who have traveled through the region of Puglia and the town of Bari since the early 1990s. The Malawian-like P[8] lineage identified in VP4 and the high genetic relatedness in VP7 with G9 rotaviruses recently identified throughout the world seem to give support to the latter hypothesis.
To our knowledge, this is the first report describing the detection of rotavirus with the unusual G9 specificity with such remarkable frequency (about 53%) among diarrheic patients in the European continent. The outbreak of G9 serotype infection in Bari suggests that such a rotavirus serotype is an emerging pathogen in Italy and that the presence of G9 rotaviruses in Europe, as in other regions (2, 12, 17), does not always follow a sporadic pattern. From this perspective, it will be important to continue and extend rotavirus strain surveillance to determine whether a similar phenomenon occurs in the other parts of Italy and Europe. The results described here emphasize the role of rotavirus G9 as an epidemiologically important serotype and the need to include G9 specificity in candidate rotavirus vaccines.
Nucleotide sequence accession number.
The sequence of the VP7 gene of the G9 Italian rotavirus strains is available in GenBank under accession number AY184813.
Acknowledgments
We thank Donato Narcisi for his expert technical assistance and L. E. Carmichael for encouragement throughout the study.
REFERENCES
- 1.Arista, S., E. Vizzi, D. Ferraro, A. Cascio, and R. Di Stefano. 1997. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch. Virol. 142:2065-2071. [DOI] [PubMed] [Google Scholar]
- 2.Bok, K., G. Palacios, K. Sijvarger, D. Matson, and J. Gomez. 2001. Emergence of G9 P[6] human rotaviruses in Argentina: phylogenetic relationships among G9 strains. J. Clin. Microbiol. 39:4020-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bon, F., C. Fromantin, S. Aho, P. Pothier, E. Kohli, and The Azay Group. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascio, A., E. Vizzi, C. Alaimo, and S. Arista. 2001. Rotavirus gastroenteritis in Italian children: can severity of symptoms be related to the infecting virus? Clin. Infect. Dis. 32:1126-1132. [DOI] [PubMed] [Google Scholar]
- 5.Cubitt, W. D., A. D. Steele, and M. Iturriza-Gomara. 2000. Characterization of rotavirus from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 6.Cunliffe, N. A., W. Dove, J. E. G. Bunn, M. B. Ramadam, J. W. O. Nyangao, R. L. Riveron, L. E. Cuevas, and C. A. Hart. 2001. Expanding global distribution of rotavirus serotype G9: detection in Libya, Kenya and Cuba. Emerg. Infect. Dis. 5:890-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z.-Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino, Y., and A. Z. Kapikian. 2000. Rotavirus serotypes: classification and importance in rotavirus epidemiology, immunity and vaccine development. J. Health Popul. Nutr. 18:5-14. [PubMed] [Google Scholar]
- 11.Iturriza Gómara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence for independent segregation. J. Virol. 76:6596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang, G., J. Green, C. I. Gallimore, and D. W. G. Brown. 2002. Molecular epidemiology of rotaviral infection in South Indian children with acute diarrhea from 1995-1996 to 1998-1999. J. Med. Virol. 67:101-105. [DOI] [PubMed] [Google Scholar]
- 13.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott William & Wilkins, Philadelphia, Pa.
- 14.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 15.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palombo, E., P. J. Masendycz, H. C. Bugg, N. Bogdanovic-Sakran, G. L. Barnes, and R. F. Bishop, 2000. Emergence of serotype G9 human rotavirus in Australia. J. Clin. Microbiol. 38:1305-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. Woods, L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresse, and R. I. Glass. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran, M., C. D. Kirkwood, L. Unicomb, N. A. Cunliffe, R. L. Ward, M. K. Bhan, H. F. Clark, R. I. Glass, and J. R. Gentsch. 2000. Molecular characterization of serotype G9 rotavirus from strains from a global collection. Virology 278:436-444. [DOI] [PubMed] [Google Scholar]
- 19.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widdowson, M.-A., G. J. J. van Doornum, W. H. M. van der Poel, A. S. de Boer, U. Mahdi, and M. Koopmans. 2000. Emerging group-A rotavirus and a nosocomial outbreak of diarrhoea. Lancet 356:1161-1162. [DOI] [PubMed] [Google Scholar]
- 21.Wilde, J., J. Eiden, and R. Yolken. 1990. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J. Clin. Microbiol. 28:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]