Abstract
A bovine rotavirus, NGRBg8, isolated from the feces of a calf with diarrhea in Nigeria was characterized by reverse transcription-PCR, nucleotide sequence analysis, and Northern blot hybridization. The nucleotide sequence of the VP7 gene of the strain was most closely related to that of a Nigerian human G8-serotype strain, HMG035 (99.9%). The NSP1 gene of strain NGRBg8 is highly related (99.4%) to that of a Thai G8 bovine strain, A5-10. Northern blot hybridization revealed a high overall genomic relatedness of bovine strain NGRBg8 with human strain HMG035; all 11 RNA segments hybridized to each other. Thus, the results show the close relationship between G8 bovine and human rotaviruses in Nigeria.
Rotaviruses are the most common cause of acute gastroenteritis in the young of a number of mammalian and avian species worldwide (17). Rotaviruses with genomes made up of 11 segments of double-stranded RNA (dsRNA) have two neutralization proteins, VP4 and VP7, on their outer capsids. These two proteins define the P and G types, respectively. Rotaviruses have been classified into 15 G and 22 P types so far (10).
Bovine rotaviruses are a major cause of enteric disease in calves of 1 to 3 weeks of age (18). Field surveys have demonstrated that G6 and G10 are the most prevalent serotypes among calf strains worldwide, whereas G8 is the least common serotype (7, 11, 27, 28, 32, 33). G8 strains have also been isolated from pigs (13) and a horse (15).
In humans, a G8 rotavirus (strain 69 M) was first isolated from a child with acute gastroenteritis in Indonesia (19), and its RNA electropherotype was characterized by a unique, supershort pattern. Subsequently, G8 strains with a long RNA electropherotype were detected among Finnish and Italian children (12) as well as children in Australia, South Africa, and the United Kingdom (22, 29).
The presence of rotaviruses in domestic livestock is an epidemiological problem whose significance is becoming increasingly evident with the detection in human subjects of viruses of animal origin (20). Recently, G8 viruses with unusual serotypic properties were identified in Africa (1, 4, 5, 8, 9). The relationship between the G serotype and P serotype (VP4 genotype) in representative human and animal rotavirus strains shows that reassortment of rotavirus genome segments has occurred as a result of intraspecies and interspecies mixed infections (31). It might be possible that there has been cross-species transmission of rotaviruses also in Nigeria.
Rotaviruses are active everywhere, and hence more data on the G and P types circulating in particular parts of the world will be valuable for those searching for essential constituents of a vaccine. In this study, we isolated a Nigerian bovine rotavirus strain, NGRBg8. The isolate was characterized by examining its genomic relatedness to other bovine and human rotavirus strains by reverse transcription-PCR (RT-PCR), nucleotide sequencing, and Northern blot hybridization.
One hundred eight fecal specimens from calves under 1 year of age with diarrhea were collected at various farms located in Borno (Maiduguri, Benisheik, Ngala, and Ngamdu), Yobe (Potiskum and Damaturu), and Gombe (Gombe and Ashaka) states in Nigeria between November and December 1998. Three rotavirus-positive specimens were stored at −20°C until being transported on ice to Japan, where they were analyzed.
Virus isolation with MA-104 cells in roller tube cultures was attempted from the three stool specimens for cell culture as previously described (35). One stool specimen (NGRBg8) from a 4-week-old male calf was successfully adapted to growth in MA-104 cells in roller tube cultures.
The following rotavirus strains with different G and P types were used as reference strains: KU (G1P[8]), S2 (G2P[4]), AU-1 (G3P[9]), SA11 (G3P[2]), B223 (G10P[11]), UK (G6P[5]), 61A (G10P[5]), NCDV (G6P[1]), A5-10 and A5-16 (G8P[1]), 69 M (G8P[10], and HMG035 (G8P[1]).
Viral RNA was extracted from a fecal suspension or culture fluid with a 1/5 volume of 6× disruption solution comprising 6% sodium dodecyl sulfate, 0.6% 2-mercaptoethanol, and 300 mM EDTA and then with phenol-chloroform. The RNA was electrophoresed in 10% acrylamide gels (2 mm thick) for 16 h at 20 mA at room temperature. RNA segments were visualized by silver staining.
Rotavirus dsRNA was extracted by the guanidine-silica method with an RNaid kit (Bio 101, Inc., La Jolla, Calif.), and the extract was used as a template for RT-PCR.
For G and P typing of the strain, the method of Ishizaki et al. (16) was used with some modifications. Briefly, a full-length VP7 gene (1,062 bp) was amplified with a pair of primers, T31 and T32, corresponding to the common 5′ and 3′ ends of the gene, respectively. The second and multiplex seminested PCR comprised a mixture of G6-, G8-, and G10-specific primers and a common primer, T32, as previously described (34). For VP4 typing, a pair of primers, PC1 and PC2, were used to amplify partial-length copies (1,085 bp) of the rotavirus VP4 gene. For the second and multiplex seminested PCR, a mixture of P[1]-, P[5]-, and P[11]-specific primers and a conserved primer, PC1, was used to identify P types (16). The NSP1 gene was amplified using the T182 and T183 primers, which have common 3′- and 5′-end sequences, respectively, as described previously (30). PCR products were electrophoresed in 1% agarose gels, stained with ethidium bromide, and then visualized with a UV transilluminator.
The nucleotide sequence was determined with an automated sequencer (ABI PRISM 310 Genetic Analyzer) and a PRISM Ready Reaction dye terminator cycle sequencing kit (ABI Inc., Foster City, Calif.). Sequence data were analyzed with the Genetyx-Mac software package for sequence alignment and for the construction of a phylogenetic tree by the unweighted pair group method with arithmetic means.
Northern blot hybridization was carried out as previously described (24). Briefly, following polyacrylamide gel electrophoresis (PAGE) analysis, dsRNA was denatured by soaking the gel in 0.1 N NaOH and 0.25 M NaCl for 20 min and then neutralized in 4× Tris-acetate-EDTA for 20 min twice and in 1× Tris-acetate-EDTA for 20 min. Electrotransfer of rotavirus RNA to Hybond N+ (Amersham) was conducted at 0.2 A overnight at 4°C. Hybridization was performed with an enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham) according to the instructions of the manufacturer. Stringency was regulated by changing the concentration of the SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution in primary and secondary wash buffers.
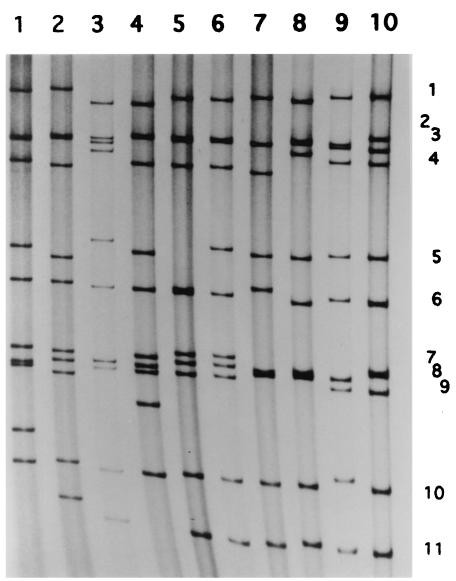
RT-PCR for G and P typing of a field fecal sample, NGRBg8, showed it to have G8P[1] type specificity (data not shown). Strain NGRBg8 was adapted to growth in MA-104 cell culture by inoculating fecal specimen NGRBg8. RNA extracted from the culture fluid of strain NGRBg8 was also analyzed for G and P type specificities. The results confirmed that the strain was of the G8P[1] type. The RNA profile of strain NGRBg8 on PAGE is shown in Fig. 1, together with those of Nigerian human G8 HMG035 and representative bovine rotaviruses. The RNA pattern of bovine G8 strain NGRBg8 was very similar to that of human G8 strain HMG035 detected in a previous study (1), except that the mobility of gene 5 of strain HMG035 was very high and was almost the same as that of gene 6.
FIG. 1.
RNA profile of bovine strain NGRBg8 in comparison with those of other representative human and bovine rotavirus strains. Lanes: 1, S2 (G2P[4], human); 2, KU (G1P[8], human); 3, A5-10 (G8P[1], bovine); 4, 69 M (G8P[10], human); 5, HMG035 (G8P[1], human); 6, NGRBg8 (G8P[1], bovine); 7, NCDV (G6P[1], bovine); 8, UK (G6P[5], bovine); 9, B223 (G10P[11], bovine); and 10, 61A (G10P[5], bovine). The numbers on the right refer to the corresponding gene segment number. The migration is from top to bottom.
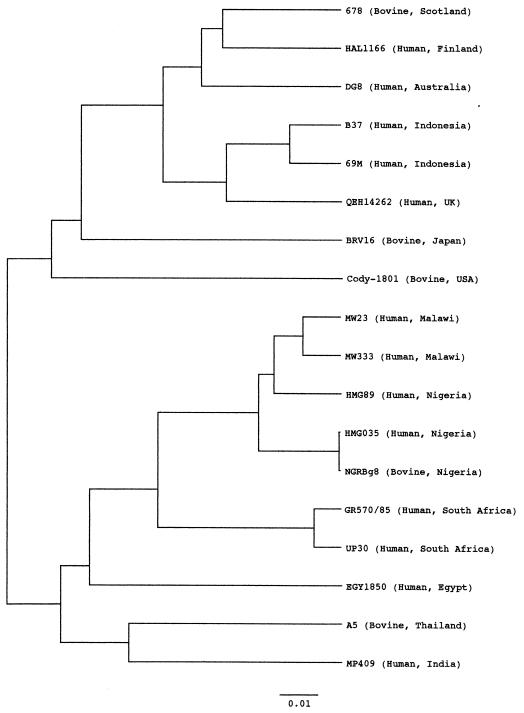
The complete nucleotide sequence of the VP7 gene of Nigerian bovine G8 strain NGRBg8 was determined using the RT-PCR product. The primary structure of the VP7 gene of this strain is similar to those of other strains representing G1 to G15 from animal and human species. It was observed that the VP7 gene of the Nigerian bovine G8 strain was most closely related (99.9% at the nucleotide level) to that of a Nigerian G8 human strain, HMG035. It also exhibited close relatedness to African G8 human strains from Malawi, South Africa, and Egypt (89.3 to 96.0% at the nucleotide level). In contrast, the VP7 gene of NGRBg8 showed low identities (from 64.4% for G7 strain Ty-1 to 77.0% for G11 strain YM) to non-G8 rotaviruses. On the phylogenetic tree, Nigerian strain NGRBg8 forms a separate cluster with an African human G8 strain (Fig. 2), showing geographical segregation.
FIG. 2.
Phylogenetic tree for the nucleotide sequences of the VP7 genes of G8 human and bovine rotaviruses. The bar indicates the variation scale. UK, United Kingdom; USA, United States.
Since migration in PAGE of gene 5 (NSP1 gene) of HMG035 was unusually fast, we analyzed the NSP1 gene of the two strains NGRBg8 and HMG035 by sequence determination. The NSP1 gene of strain NGRBg8 is 1,590 nucleotides in length and is very similar (99.4% identity at the nucleotide level) to that of a Thai bovine strain, A5-10, which is 1,587 nucleotides long. As found for the A5-10 NSP1 gene, the NSP1 gene of NGRBg8 codes for only 40 amino acids since a nonsense codon is present at nucleotides 153 to 155. In contrast, the HMG035 NSP1 gene was found to have a long deletion of nucleotides (222 nucleotides) compared to the NGRBg8 NSP1 gene (Fig. 3) and codes for 418 amino acids, although other NSP1 genes code for 486 to 571 amino acids. On comparison of the NSP1 gene of HMG035 with other representative published NSP1 sequences of rotavirus strains of different species and G/P types, the former exhibited only low identities, from 48.3% (strain EW) to 77.7% (strain HMG035) at the nucleotide level (data not shown) in the calculation including the deletion sequence.
FIG. 3.
Alignment of parts of the NSP1 genes of strains NGRBg8 and HMG035. Upper sequence, bovine strain NGRBg8; lower sequence, human strain HMG035, with a deletion of 222 nucleotides in total. Asterisks represent nucleotides identical between the two strains. Dashes show sequence deletions.
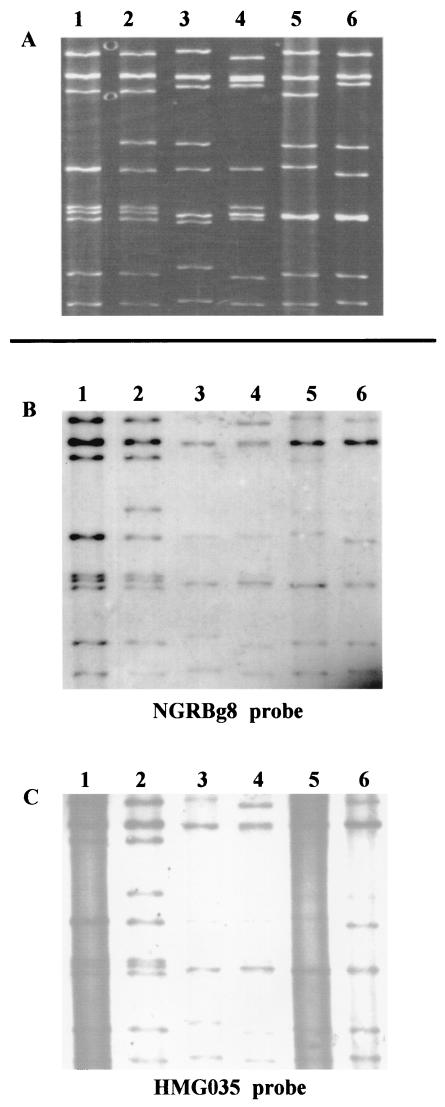
On Northern blot analysis, the NGRBg8 probe reacted with eight or nine RNA segments of reference bovine strains. Interestingly, the probe of Nigerian bovine strain NGRBg8 hybridized to all the RNA segments of a Nigerian human strain, HMG035, isolated from a diarrheic child (Fig. 4). In the reciprocal hybridization assay involving the HMG035 probe, this relationship was confirmed: the probe reacted with eight or nine segments of representative bovine rotaviruses and with all the segments of bovine NGRBg8 (Fig. 4). In contrast, a human rotavirus KU probe did not react with any of the RNA segments from the Nigerian human and bovine strains (data not shown). Thus, the two Nigerian rotaviruses isolated from a cow and a human were found to be highly related.
FIG. 4.
Northern blot analysis of bovine G8 rotavirus strain NGRBg8 detected in Nigeria. (A) RNA profiles in PAGE. Lanes: 1, HMG035 (G8P[1], human); 2, NGRBg8 (G8P[1], bovine); 3, B223 (G10P[11], bovine); 4, A5-16 (G8P[1], bovine); 5, NCDV (G6P[1], bovine); and 6, UK (G6P[5], bovine). (B and C) Northern blot analysis using the NGRBg8 (B) and HMG035 (C) probes. Northern blot analysis in panels B and C was performed using the same blot, which was transferred from the polyacrylamide gel shown in panel A.
For group A rotaviruses, great genome diversity has been described. The diversity results from the accumulated effects of three distinct mechanisms: point mutation, reassortment, and rearrangement (31). Group A rotaviruses are classified into G and P types according to the specificities of two neutralization antigens, VP4 and VP7. Among the 15 G types and 22 P types, G1P[8] is the most prevalent worldwide. Recently, however, rotavirus strains with unusual G or P type specificity or ones which may be derived from a different species have been detected. Since G8 strains were first detected in diarrheic children in Indonesia (19), they have been detected across the globe among humans in Finland, Italy, Nigeria, Brazil, Malawi, South Africa, Egypt, Australia, the United States, and the United Kingdom (1, 4, 8, 9, 12, 14, 22, 23, 25, 26, 29). G8 strains have also been found in calves in many countries of the world such as Scotland, Thailand, and Japan (28, 31, 32). The detection of a bovine G8 strain in Nigeria in this study extends the global distribution of bovine rotavirus G8 strains. As judged on phylogenetic analysis, the G8 strains in Africa (Malawi, South Africa, Egypt, and Nigeria) are similar to one another and form a distinct cluster, showing geographical segregation.
Ohshima et al. (21) and Browning et al. (6) suggested that G8 strains are reassortant viruses between human and bovine rotaviruses. In RNA-RNA hybridization assays, human strains 69 M and HAL1166 of the G8 type exhibited overall genomic relatedness with a bovine genogroup and the human DS-1 genogroup (21). Between Nigerian bovine strain NGRBg8 and representative bovine rotavirus strains, eight or nine RNA segments hybridized well, but the origin of the remaining two or three segments was not clear. Furthermore, the strains NGRBg8 and HMG035 are highly related: all 11 RNA segments hybridized to each other. However, the NSP1 gene of HMG035 has a long deletion (222 bases) and the sequence of the NSP1 gene of strain NGRBg8 showed 90.3% identity to that of strain HMG035 even when calculated excluding the deletion sequence. These results substantiate the potential for human or bovine rotaviruses in Nigeria to have been made through reassortment after transmission between humans and cows, but this does not appear to be a direct event. Reassortant viruses have been much more frequently detected in developing countries than in developed ones. The incidence of mixed infections is high in developing countries. In Bangladesh, after a devastating flood, mixed infection cases drastically increased. More intimate contact of humans with livestock is conceivable in developing countries. Such an environmental situation may facilitate mixed infection, leading to the occurrence of reassortant viruses.
Two G8 strains detected in Nigeria (bovine NGRBg8 and human HMG035) have almost the same RNA electropherotype except for gene 5. The VP7 genes of these two Nigerian G8 rotavirus strains exhibit the closest identity at both the nucleotide and amino acid levels, in contrast to that exhibited by two South African G8 rotavirus strains (29). Furthermore, in Northern blot analysis, it was surprising that all the segments from bovine NGRBg8 hybridized with those of human HMG035, suggesting the cross-species transmission. The places where the two strains were obtained are geographically close. In the present study area, northeast Nigeria, there is close contact between herdsmen and their livestock, including the sharing of common sources of drinking water. It will be of value to search for more rotavirus strains that might have resulted from cross-species transmission and/or reassortment in this area.
The NSP1 gene of NGRBg8 showed high identity to that of a Thai G8 bovine strain, A5-10. It is also related to that of a Nigerian human strain, HMG035, which is unique; the NSP1 gene has a long deletion of 222 nucleotides and the NSP1 amino acid sequence exhibits low identity to those of representative rotaviruses. Although the identity at nucleotide level between the two Nigerian strains, NGRBg8 and HMG035, was relatively low (77.7%), the identity excluding the deletion region was 90.2%. It is, therefore, reasonable that the NSP1 genes of the two strains hybridized in Northern blot analysis, since the stringency in the present assays appears to allow the hybridization of segments exhibiting more than 85% homology.
It is interesting that the NSP1 gene of strain NGRBg8 has a nonsense codon at nucleotides 153 to 155 and codes for only 40 amino acids, as found for the A5-10 clone isolated in Thailand (30). NSP1 carries a zinc finger motif and RNA binding capacity for the assembly process and is associated with the cytoskeleton. Furthermore, the protein is recognized to be related to species specificity. However, it is unknown whether the large deletion of NSP1 has an advantage for the possible interspecies transmission of the strain. Characterization of the NSP1 synthesis in vitro and in vivo of strains NGRBg8 and HMG035 having unusual NSP1 genes would be useful for analyzing the function of NSP1 in rotavirus infection. It is also of interest that G8 strains 69 M, B37, HMG035, NGRBg8, and A5-16 all have rearranged RNA segments. There is a possibility that unusual strains (reassortment strains) tend to undergo rearrangement for some unknown reason.
The epidemiological data on rotavirus infections in Africa are not yet sufficient (1-5, 8, 9, 14, 29), although recent studies have revealed the high prevalence of strains with an unusual G serotype: G8 was detected in 27.7% of samples in our previous study (1). The protective efficacy of rotavirus vaccine was not satisfactory in vaccine trials in this area. The different prevalence of G and/or P types might be one reason for this finding. The inclusion of G8 and other serotypes for future vaccines should be considered. It would be valuable to obtain more epidemiological data including serotype distribution through long-term and large-scale surveys.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the DDBJ and EMBL/GenBank data libraries under accession numbers AF361439 (NGRBg8 VP7 gene), AB096861 (NGRBg8 NSP1 gene), and AB096862 (HMG035 NSP1 gene).
Acknowledgments
The work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. M. I. Adah received an award under the “Long Term FY2000 JSPS Invitation Fellowship Program for Research in Japan” from the JSPS (Japan Society for the Promotion of Science).
We are grateful to the staff of the Veterinary Division of the Ministry of Animal and Forest Resources in Borno, Yobe, and Gombe states, Nigeria, for their cooperation in the fecal sample collection.
REFERENCES
- 1.Adah, M. I., A. Wade, and K. Taniguchi. 2001. Molecular epidemiology of rotavirus in Nigeria: detection of unusual strains with G2P[6] and G8P[1] specificities. J. Clin. Microbiol. 39:3969-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adah, M. I., A. Rohwedder, O. D. Olaleye, O. A. Durojaiye, and H. Werchau. 1997. Serotype of Nigerian rotavirus strains. Trop. Med. Int. Health 2:363-370. [DOI] [PubMed] [Google Scholar]
- 3.Adah, M. I., A. Rohwedder, O. D. Olaleye, O. A. Durojaiye, and H. Werchau. 1997. Further characterization of field strains of rotavirus from Nigeria VP4 genotype P6 most frequently identified among symptomatically infected children. J. Trop. Pediatr. 43:267-274. [DOI] [PubMed] [Google Scholar]
- 4.Adah, M. I., A. Rohwedder, O. D. Olaleye, and H. Werchau. 1997. Nigerian rotavirus serotype G8 could not be typed by PCR due to nucleotide mutation at the 3′ end of the primer binding site. Arch. Virol. 142:1881-1887. [DOI] [PubMed] [Google Scholar]
- 5.Armah, G. E., C. T. Pager, R. H. Asma, F. R. Anto, A. B. Oduro, F. Binka, and D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 6.Browning, G. F., D. R. Snodgrass, O. Nakagomi, E. Kaga, A. Sarasini, and G. Gerna. 1992. Human and bovine serotype G8 strains may be derived by reassortment. Arch. Virol. 125:121-128. [DOI] [PubMed] [Google Scholar]
- 7.Chang, K. O., A. V. Parwani, and L. J. Saif. 1996. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotaviruses from field samples using RT-PCR and RFLP analysis. Arch. Virol. 141:1727-1739. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe, N. A., J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and C. A. Hart. 1999. Rotavirus G and P types in children with acute diarrhoea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J. Med. Virol. 57:308-312. [PubMed] [Google Scholar]
- 9.Cunliffe, N. A., J. R. Gentsch, C. D. Kirkwood, J. S. Gondwe, W. Dove, O. Nakagomi, T. Nakagomi, Y. Hoshino, J. S. Bresee, R. I. Glass, M. E. Molyneux, and C. A. Hart. 2000. Molecular and serologic characterization of novel serotype G8 human rotavirus strains detected in Blantyre, Malawi. Virology 274:309-320. [DOI] [PubMed] [Google Scholar]
- 10.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Falcone, E., M. Tarantino, L. Di Trani, P. Cordioli, A. Lavazza, and M. Tollis. 1999. Determination of bovine rotavirus G and P serotypes in Italy by PCR. J. Clin. Microbiol. 37:3879-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerna, G. A., A. Scraisisi, L. Zenhlin, A. DiMatteo, P. Mirando, A. Parea, M. Battaglia, and G. Milanese. 1990. Isolation in Europe of 69M-like (serotype 8) human rotavirus strains with either subgroup I or II specificity and a long RNA electropherotype. Arch. Virol. 112:27-40. [DOI] [PubMed] [Google Scholar]
- 13.Gouvea, V. N., N. Santos, and M. D. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes, J. L., C. D. Kirkwood, G. Gerna, J. D. Clemens, M. R. Rao, A. B. Naficy, R. Abu-Elyazeed, S. J. Savarino, R. I. Glass, and J. R. Gentsch. 1999. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch. Virol. 144:1381-1396. [DOI] [PubMed] [Google Scholar]
- 15.Isa, P., A. R. Wood, T. Netherwood, M. Ciarlet, H. Imagawa, and D. R. Snodgrass. 1996. Survey of equine rotaviruses shows conservation of one P genotype in background of two G serotypes. Arch. Virol. 141:1601-1612. [DOI] [PubMed] [Google Scholar]
- 16.Ishizaki, H., T. Sakai, T. Shirahata, K. Taniguchi, T. Urasawa, S. Urasawa, and H. Goto. 1996. The distribution of G and P types within isolates of bovine rotavirus in Japan. Vet. Microbiol. 48:367-372. [DOI] [PubMed] [Google Scholar]
- 17.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 18.Lucchelli, A., S. E. Lance, P. B. Bartlett, G. Y. Miller, and L. J. Saif. 1992. Prevalence of bovine group A rotavirus shedding among dairy calves in Ohio. Am. J. Vet. Res. 53:169-174. [PubMed] [Google Scholar]
- 19.Matsuno, S., A. Hasegawa, A. Mukoyama, and S. Inouye. 1985. A candidate for a new serotype of human rotavirus. J. Virol. 54:623-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagomi, O., and T. Nakagomi. 1991. Genetic diversity and similarity among mammalian rotaviruses in relation to interspecies transmission of rotavirus. Arch. Virol. 120:43-55. [DOI] [PubMed] [Google Scholar]
- 21.Ohshima, A., T. Takagi, T. Nakagomi, S. Matsuno, and O. Nakagomi. 1990. Molecular characterization by RNA-RNA hybridization of a serotype 8 human rotavirus with a super-short RNA electropherotype. J. Med. Virol. 30:107-112. [DOI] [PubMed] [Google Scholar]
- 22.Palombo, E. A., R. Clark, and R. F. Bishop. 2000. Characterization of a “European-like” serotype G8 human rotavirus isolated in Australia. J. Med. Virol. 60:56-62. [PubMed] [Google Scholar]
- 23.Parwani, A. V., H. A. Hussein, B. I. Rosen, A. Lucchelli, L. Navarro, and L. J. Saif. 1993. Characterization of field strains of group A bovine rotaviruses by using polymerase chain reaction-generated G and P type-specific cDNA probes. J. Clin. Microbiol. 31:2010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pongsuwanna, Y., K. Taniguchi, M. Chiwakul, T. Urasawa, F. Wakasugi, C. Jayavasu, and S. Urasawa. 1996. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J. Clin. Microbiol. 34:1050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 26.Santos, N., R. C. C. Lima, C. F. A. Pereira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato, M., T. Nakagomi, K. Tajima, K. Ezuru, H. Akashi, and O. Nakagomi. 1997. Isolation of serotype G8, P6[1] bovine rotavirus from adult cattle with diarrhea. J. Clin. Microbiol. 35:1266-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snodgrass, D. R., T. Fitzgerald, I. Campbell, F. M. M. Scott, G. F. Browning, D. L. Miller, A. J. Herring, and H. B. Greenberg. 1990. Rotavirus serotypes 6 and 10 predominate in cattle. J. Clin. Microbiol. 28:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steele, A. D., S. P. Parker, I. Peenze, C. T. Pager, M. B. Taylor, and W. D. Cubitt. 1999. Comparative studies of human rotavirus serotype G8 strains recovered in South Africa and the United Kingdom. J. Gen. Virol. 80:3029-3034. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi, K., K. Kojima, and S. Urasawa. 1996. Nondefective rotavirus mutants with an NSP1 gene which has a deletion of 500 nucleotides, including a cysteine-rich zinc finger motif-encoding region (nucleotides 156 to 248), or which has a nonsense codon at nucleotides 153 to 155. J. Virol. 70:4125-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi, K., and S. Urasawa. 1995. Diversity in rotavirus genomes. Semin. Virol. 6:123-131. [Google Scholar]
- 32.Taniguchi, K., T. Urasawa, and S. Urasawa. 1993. Independent segregation of the VP4 and the VP7 genes in bovine rotaviruses as confirmed by VP4 sequence analysis of G8 and G10 bovine rotavirus strains. J. Gen. Virol. 74:1215-1221. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi, K., T. Urasawa, Y. Pongsuwanna, M. Choonthanom, C. Jayavasu, and S. Urasawa. 1991. Molecular and antigenic analyses of serotypes 8 and 10 of bovine rotaviruses in Thailand. J. Gen. Virol. 72:2929-2937. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi, K., F. Wakasugi, Y. Pongsuwanna, T. Urasawa, S. Ukae, S. Chiba, and S. Urasawa. 1992. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol. Infect. 109:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urasawa, T., S. Urasawa, and K. Taniguchi. 1981. Sequential passages of human rotavirus in MA-104 cells. Microbiol. Immunol. 25:1025-1035. [DOI] [PubMed] [Google Scholar]