Abstract
The leading cause of morbidity and mortality in cystic fibrosis (CF) patients stems from repeated bacterial respiratory infections. Many bacterial species have been cultured from CF specimens and so are associated with lung disease. Despite this, much remains to be determined. In the present study, we characterized without prior cultivation the total bacterial community present in specimens taken from adult CF patients, extracting DNA directly from 14 bronchoscopy or sputum samples. Bacterial 16S ribosomal DNA (rRNA) gene PCR products were amplified from extracted nucleic acids, with analyses by terminal restriction fragment length polymorphism (T-RFLP), length heterogeneity PCR (LH-PCR), and sequencing of individual cloned PCR products to characterize these communities. Using the same loading of PCR products, 12 distinct T-RFLP profiles were identified that had between 3 and 32 T-RFLP bands. Nine distinct LH-PCR profiles were identified containing between one and four bands. T-RFLP bands were detected in certain samples at positions that corresponded to pathogens cultured from CF samples, e.g., Burkholderia cepacia and Haemophilus influenzae. In every sample studied, one T-RFLP band was identified that corresponded to that produced by Pseudomonas aeruginosa. A total of 103 16S rRNA gene clones were examined from five patients. P. aeruginosa was the most commonly identified species (59% of clones). Stenotrophomonas species were also common, with eight other (typically anaerobic) bacterial species identified within the remaining 17 clones. In conclusion, T-RFLP analysis coupled with 16S rRNA gene sequencing is a powerful means of analyzing the composition and diversity of the bacterial community in specimens sampled from CF patients.
Cystic fibrosis (CF) is the most common severe monogenic disease of autosomal recessive inheritance in Caucasian populations (12, 40) and is carried by 1 in 25 European Caucasians (40). CF patients suffer from repeated bacterial infections. Traditionally, the primary bacterial pathogens associated with CF pulmonary infections have been identified as Burkholderia cepacia, Stenotrophomonas maltophilia, Haemophilus influenzae, Staphylococcus aureus, and Pseudomonas aeruginosa (18, 20, 25, 26), with P. aeruginosa considered to elicit the greatest inflammatory response (16, 40). These inflammatory responses cause an irreversible loss of lung function that determines morbidity and mortality for CF patients (37). CF patients, however, are screened typically only for the presence of pathogens, such as those described above, that are perceived to be of most clinical significance. It is, however, important to determine whether other bacterial species are present since they may also be involved in the progressive loss of lung function. Such information would have profound clinical significance in terms of ameliorating existing and developing novel therapeutic approaches.
Currently, the bacterial pathogens in CF airways are characterized through cultivation of expectorated sputum samples on selective media. Such culture-based analysis can, however, be problematic since the process is time-consuming (41), potentially inaccurate (32), and requires species-specific selective media. Cultivation also only allows at best a semiquantitative assessment of the load of the targeted pathogen. More fundamentally, it excludes the detection of unculturable bacteria that may predominate in many environments (2).
By assessing nucleic acids extracted from clinical samples directly, molecular biology-based assays obviate the requirement for cultivation. As such, these approaches are becoming increasingly important and are finding more applications in clinical microbiology. Some PCR-based assays have already been developed to detect specific CF bacterial pathogen species (23, 28, 41, 42). Although valuable, these assays by definition, however, do not characterize the total bacterial community. As such, potentially clinically important pathogens can remain unidentified (11, 23, 27, 41). Methodologies, however, have been developed that allow the total bacterial community in a given environment to be characterized (24). Such studies typically exploit phylogenetically informative 16S rRNA sequences. Previous studies have enabled a range of specific pathogens to be detected in clonal libraries comprised of 16S ribosomal DNA (rDNA) PCR products amplified from DNA extracted directly from sputa (38). However, the same study also indicated that many clones did not hybridize to the pathogen-specific probes used, thus indicating that there may be many other bacterial species present in CF sputa.
It is therefore important to develop a means of rapidly characterizing the total CF bacterial community. A number of approaches have been effective in characterizing complex microbial communities, including length heterogeneity PCR (LH-PCR) analysis (33, 36) and terminal restriction fragment length polymorphism (T-RFLP) analysis (4, 7, 13, 24). The first step in such community analyses, common to both LH-PCR and T-RFLP, is the amplification of ribosomal sequences from nucleic acids extracted directly from clinical samples. LH-PCR resolves amplicons generated from different bacterial species on the basis of length (36). T-RFLP generates fragments that differ in length due to the variation in the position of the first specific restriction endonuclease site in ribosomal sequences amplified from individual bacterial species. These informative fragments are typically fluorescently labeled and so allow their detection on automated DNA sequencing machines (7). Therefore, both LH-PCR and T-RFLP allow complex bacterial communities to be profiled rapidly in a single electrophoretic track.
The aim of the present study was to examine the composition and diversity of bacterial communities within sputa and bronchoscopies sampled from CF patients. Profiling data generated by using LH-PCR and T-RFLP were also compared to information derived from 16S rRNA clone libraries prepared from the same samples.
MATERIALS AND METHODS
In silico sequence analysis.
Published bacterial 16S rRNA gene sequence data, stored at GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide), were retrieved. Computer programs were then used to predict the band sizes for both LH-PCR and T-RFLP analysis. Specifically, LH-PCR analysis was carried out by using GAP (Wisconsin Package version 10.3; Accelrys, Inc., San Diego, Calif.), a program that can be used to locate the relative positions of, and hence the distance between, pairs of PCR primers. In this manner, GAP was used to determine the length (in bases) from the 5′ end of primer 8f-700IR to the 5′ end of primer 338r (see below). T-RFLP sequence analysis was carried out using MapSort (Wisconsin Package version 10.3; Accelrys), a program that locates the position of restriction endonuclease recognition motifs in a given sequence. In this manner, MapSort was used to determine the length (in bases) from the 5′ end of primer 8f-700IR (see below) to the first cleavage position of the restriction endonuclease CfoI in each 16S rRNA gene sequence.
16S rRNA genes from 15 different strains of P. aeruginosa (accession numbers AJ536419, AF502423, AB073312, AB062598, AF094720, AB037563, AB037562, AB037558, AB037545, AJ249451, AF157689, M34133, AB037546, AB037560, and AB037549) were used, in addition to the genome sequence strain PAO1 (accession numbers AE004091 [rrs, 722096 to 723631], AE004091 [rrs, 4792196 to 4793731], AE004091 [rrs, 5267724 to 52692590], and AE004091 [rrs, 6043208 to 6044743]).
DNA extraction. (i) DNA extraction from bacterial cultures.
Actinobacillus pleuropneumoniae, B. cepacia (genomovar III), Escherichia coli, Klebsiella pneumoniae, P. aeruginosa, and S. maltophilia were cultured from clinical specimens by the routine Diagnostic Laboratories at the Royal Liverpool Hospital. The remaining strains, H. influenzae NCTC12699 and S. aureus NCTC12232 used were obtained from the National Collection of Type Cultures (London, United Kingdom). Bacterial strains were cultured at 37°C on 2.8% (wt/vol) nutrient agar (Oxoid, Basingstoke, United Kingdom).
DNA was extracted from bacterial cells by a previously described method (30). The extracted DNA was verified by Tris-acetate-EDTA (TAE)-agarose gel electrophoresis on 0.8% (wt/vol) TAE-agarose gels stained in ethidium bromide (0.5 mg/liter) and images captured by using a Herolab image analyzer with E.A.S.Y STOP win 32 software (Herolab, Wiesloch, Germany).
(ii) DNA extraction from clinical specimens.
Sputa and bronchoscopy samples were obtained from adult CF patients at the Royal Liverpool University Hospital under full ethical approval. Of these, 10 were sputum samples (patients 1, 2, 5, 7, 8, 10, 11, 12, 13, and 14), and 4 were bronchoscopy samples (patients 3, 4, 6, and 9). One additional sputum sample was obtained to develop the approaches used here. All samples were washed three times in phosphate-buffered saline (Oxoid) prior to DNA extraction. Sputum samples were treated with Sputasol (Oxoid) in accordance with the manufacturer's instructions, followed by centrifugation for 5 min at 12,000 × g. Pellets were resuspended in 1.5 ml of phosphate-buffered saline and then centrifuged for 5 min at 12,000 × g. This step was repeated three times.
DNA was isolated from clinical specimens by a modification of a procedure described previously (7). Approximately 0.2 ml of each clinical sample was resuspended in 800 μl of 200 mM sodium phosphate buffer (pH 8.0) and 100 μl of guanidinium thiocyanate-EDTA-Sarkosyl (as described above). Then, 0.2 g of 0.18-mm-diameter glass beads (B. Braun Biotech International GmbH, Melsungen, Germany) was added, and homogenization performed for 30 s at 30 Hz in a Qiagen Mixer Mill 300 (Qiagen, Crawley, United Kingdom). Samples were heated to 70°C for 20 min and then placed on ice for 20 min, and beads and other debris were pelleted by centrifugation at 12,000 × g for 5 min at room temperature. The supernatant was transferred to a fresh microfuge tube, and NaCl (to a final concentration of 0.5 M) and polyethylene glycol (to a final concentration of 15%) were added. Samples were left to precipitate at 4°C for 1 h. DNA was pelleted by centrifugation at 12,000 × g for 10 min at room temperature, and the pellet was resuspended in 300 μl of sterile distilled water. Next, 0.3 ml of Tris-buffered phenol (pH 8.0) was added to each sample before the tubes were vortexed vigorously. After centrifugation at 12,000 × g for 5 min, supernatants were transferred to fresh microfuge tubes. A further 0.3 ml of Tris-buffered phenol (pH 8.0)-chloroform-isoamyl alcohol (25:24:1) was added, and the mixture was vortexed vigorously. After centrifugation at 12,000 × g for 10 min, supernatants were precipitated by using an equal volume of isopropanol and a 1/10 volume of 10 M ammonium acetate for 1 h at −20°C. The pellets formed by centrifugation at 12,000 × g for 10 min at room temperature were washed three times in 70% ethanol. After being dried, pellets were resuspended in 100 μl of sterile distilled water and stored at −20°C. The extracted DNA was verified by TAE-agarose gel electrophoresis as described above.
DNA quantification.
Extracted DNA was quantified on a CytoFluor series 4000 multiwell plate reader (PerSeptive Biosystems, Foster City, Calif.) by using the PicoGreen DS DNA quantitation kit (Molecular Probes, Leiden, The Netherlands) according to the manufacturer's instructions.
LH-PCR amplification and profiling. (i) LH-PCR amplification.
PCR products for LH-PCR analysis were amplified with the universal bacterial primers 8f-700IR (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 338r (5′-GCT GCC TCC CGT AGG AGT-3′), obtained from MWG-Biotech (Milton Keynes, United Kingdom), which are specific for 16S rDNA from 20 ng of extracted DNA (24, 33). Using these primers, an LH-PCR product size of 330 bases would be generated from E. coli (accession no. NC_000913). Primer 8f-700IR was labeled at the 5′ end with IRD700; primer 338r was unlabeled. PCR mixtures were composed of 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.2 μM concentrations of each primer, and 1 U of REDTaq DNA polymerase (Sigma-Aldrich, Dorset, United Kingdom) in a final volume of 50 μl. After an initial denaturation step of 94°C for 2 min, samples were subjected to 32 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min, followed by a final extension step at 72°C for 10 min. Amplification was carried out by using a GeneAmp PCR System 2400 (Perkin-Elmer, Beaconsfield, United Kingdom) with LH-PCR products stored at −20°C. Prior to LH-PCR analysis, the amplified PCR products were verified by TAE-agarose gel electrophoresis as described above.
(ii) LH-PCR analysis.
Approximately 0.7 μg of the LH-PCR products was separated by length with a 25-cm SequagelXR denaturing acrylamide gel (National Diagnostics, Ashby de la Zouch, United Kingdom) prepared in accordance with the manufacturer's instructions with the addition of 8.3 M urea and 10% (final concentration) formamide and a LI-COR IR2 automated DNA sequencer (LI-COR Biosciences, Cambridge, United Kingdom) at 55°C and 1,200 V. The gels were analyzed by using GeneimageIR v.3.56 (Scanalytics, Fairfax, Va.). When profile data were assessed, only peaks of >0.5% of the total lane signal were classified as bands for further analysis. The positions of these individual bands were calculated in relation to microSTEP 15a (700-nm) size marker (Microzone, Lewes, United Kingdom).
Cloning and sequence analysis of LH-PCR products.
Samples of the LH-PCR products generated for LH-PCR analysis, as described above, were cloned with a pGEM-T Easy Vector system (Promega, Southampton, United Kingdom) according to the manufacturer's instructions. DNA was extracted from clones as follows: a single white colony was resuspended in 200 μl of sterile distilled water and then boiled for 10 min, with the cell debris pelleted by centrifugation at 12,000 × g for 5 min. Then, 15 μl of the supernatant was added to a 50-μl (final volume) PCR mixture, comprising 1× PCR buffer, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 μM, and 1 U of REDTaq DNA polymerase (Sigma-Aldrich) in a final volume of 50 μl. Amplification was carried out with the primers gemsp6 (5′-GCT GCG ACT TCA CTA GTG AT-3′) and gemt7-700IR (5′-GTG GCA GCG GGA ATT CGA T-3′) (designed for the present study). Primer gemt7-700IR was labeled at the 5′ end with IRD700; primer gemsp6 was unlabeled. An initial denaturation step of 94°C for 2 min was followed by 32 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. Amplification was carried out by using a GeneAmp PCR System 2400 (Perkin-Elmer) with LH-PCR products stored at −20°C. Sequencing of individual clones was carried out by the Genetics Core Facility, Hammersmith Hospital, London, England. Sequences were analyzed by BLAST (1) (www.ncbi.nlm.nih.gov/blast/) and by using the Ribosomal Database Project II (http://www.rdp.cme.msu.edu/html/). Sequences obtained in the present study are stored under accession numbers AJ491776 to AJ491780 and accession numbers AJ496326 to AJ496330.
T-RFLP amplification and profiling. (i) T-RFLP PCR product amplification.
PCR products for T-RFLP analysis were amplified with the universal bacterial primers 8f-700IR and 926r (5′-CCG TCA ATT CCA TTT RAG TTT-3′) specific for 16S rDNA from 20 ng of extracted DNA (24). Primer 8f700 was labeled at the 5′end with IRD700; primer 926r was unlabeled. PCR mixtures comprised 1× PCR buffer, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 μM, and 1 U of REDTaq DNA polymerase (Sigma-Aldrich) in a final volume of 50 μl. An initial denaturation step of 94°C for 2 min was followed by 32 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. Amplification was carried out by using a GeneAmp PCR System 2400 (Perkin-Elmer) with PCR products for T-RFLP analysis stored at −20°C after verification on TAE-agarose gels as described above.
(ii) T-RFLP analysis.
PCR products (ca. 20 ng) were digested by using the restriction endonuclease CfoI (Roche, Lewes, United Kingdom) for 3 h at 37°C with the reaction buffer supplied by the manufacturer. All restriction endonuclease digestions were carried out to complete digestion as shown by comparing PCR products after various digestion incubation times (data not shown). The restriction endonuclease was inactivated by heating at 90°C for 20 min. An approximately 0.7-μg portion of T-RFLP PCR products was separated by length by using a 25-cm SequagelXR denaturing acrylamide gel (National Diagnostics) prepared in accordance with the manufacturer's instructions, with the addition of 8.3 M urea and 10% (final concentration) formamide, and with a LI-COR IR2 automated DNA sequencer (LI-COR Biosciences) at 55°C and 1,200 V. The T-RFLP product size that would be generated under these conditions from E. coli (accession no. NC_000913) is 373 bases. The gels were analyzed with GeneimageIR v.3.56 (Scanalytics). When we assessed the profile data, only peaks of >0.5% of the total lane signal were classified as bands for further analysis. The positions of these individual bands were calculated in relation to the microSTEP 15a (700-nm) size marker (Microzone).
RESULTS
LH-PCR and T-RFLP analysis of bacterial species.
Experiments were carried out to determine LH-PCR and T-RFLP band sizes for each 16S rRNA gene operon within the fully sequenced P. aeruginosa strain PAO1 and the corresponding 16S rRNA gene GenBank database entry for 15 distinct strains of P. aeruginosa identified in Materials and Methods. These experiments identified that the LH-PCR and T-RFLP band sizes for the 16S rRNA sequences from all four PAO1 operons, and the 15 other P. aeruginosa strains tested were identical. Similarly, analyses also confirmed that Burkholderia multivorans and B. cepacia genomovar III had the same LH-PCR and T-RFLP profile. Moreover, this initial analysis of T-RFLP band sizes for 165 bacterial species showed that 133 (81%) gave distinct T-RFLP band sizes (data not shown [http://www.kcl.ac.uk/kis/schools/life_sciences/life_sci/TRFLPandLHdata.html]).
The DNA extracted from eight bacterial species was used to verify both LH-PCR and T-RFLP assays. The results of the LH-PCR analyses and the T-RFLP analyses for these eight species are shown in Fig. 1 and 2, respectively. Only one LH-PCR peak was produced by these bacterial strains (Fig. 1). The size of each bacterial species LH-PCR peak corresponded to the value for that species determined by in silico analysis (Table 1). Similarly, only one T-RFLP peak was produced by these bacterial strains (Fig. 2). The size of each bacterial T-RFLP species peak corresponded to the value for that species determined by in silico analysis (Table 1). These analyses were in addition performed for five clinical strains of both S. aureus and P. aeruginosa. In each case, the LH-PCR and T-RFLP band sizes generated matched that obtained for each species by in silico analysis (data not shown). The same LH-PCR and T-RFLP protocols were then applied to the study of bronchoscopy and sputum samples taken from CF patients.
FIG. 1.
LH-PCR (700IR-labeled) profiles of 16S rRNA gene products amplified from single bacterial strains and separated on a LI-COR IR2 automated sequencer. (a) S. maltophilia; (b) P. aeruginosa, (c) E. coli; (d) H. influenzae; (e) A. pleuropneumoniae; (f) B. cepacia; (g) K. pneumoniae; (h) S. aureus. The x axis on each graph corresponds to the signal intensity, and the y axis on each graph corresponds to the retardation factor.
FIG. 2.
CfoI-generated T-RFLP profiles formed from 16S rRNA gene PCR products (700IR labeled) amplified from single bacterial strains and separated on a LI-COR IR2 automated sequencer. (a) S. maltophilia; (b) P. aeruginosa; (c) E. coli; (d) K. pneumoniae; (e) A. pleuropneumoniae; (f) H. influenzae; (g) S. aureus; (h) B. cepacia. The x axis on each graph corresponds to the signal intensity, and the y axis on each graph corresponds to the retardation factor.
TABLE 1.
Fragment sizes predicted by in silico analysis for eight bacterial species used in this study for LH-PCR and T-RFLP profiling
| Bacterial strain | Fragment size predicted (bases)
|
|
|---|---|---|
| LH-PCR | T-RFLP | |
| A. pleuropneumoniae | 350 | 920a |
| B. cepacia | 343 | 209 |
| E. coli | 348 | 373 |
| H. influenzae | 348 | 364 |
| K. pneumoniae | 346 | 371 |
| P. aeruginosa | 342 | 155 |
| S. aureus | 355 | 238 |
| S. maltophilia | 349 | 214 |
Uncut over this region by CfoI.
LH-PCR profiling of clinical sample DNA.
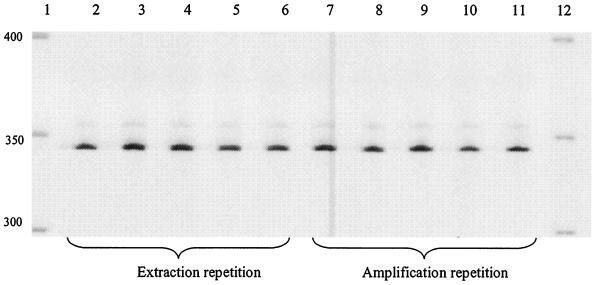
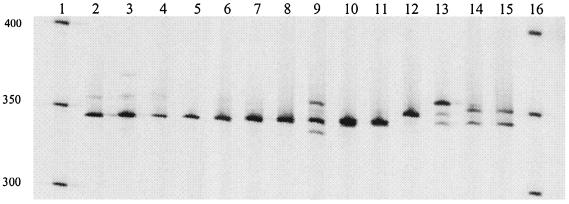
LH-PCR profiles generated from PCR products amplified from DNA extracted directly from clinical samples were first examined with respect to their reproducibility. LH-PCR profiles generated from DNA extracted from a sputum sample were indistinguishable both in terms of repeated extractions from different subsamples of the same clinical material (Fig. 3, lanes 2 to 6) and also in terms of PCR amplification from an individual DNA extraction (Fig. 3, lanes 7 to 11). Figure 4 shows the LH-PCR profiles generated from PCR products amplified from DNA extracted directly from 10 sputa and 4 bronchoscopies sampled from 14 CF patients. Table 2 shows the sizes of the LH-PCR bands detected for samples taken from these patients. The number of bands visualized ranged from 1 (Fig. 4, lane 5) to 4 (Fig. 4, lane 9). On average, 2.7 bands were detected for sputum samples and 1.25 bands were detected for the bronchoscopy samples. The size range of bands detected was 330 to 360 bases, with the dominant band being detected at 342 bases, corresponding to that which would be produced by P. aeruginosa. The LH-PCR band sizes predicted by in silico analysis for B. cepacia (344 bases), H. influenzae (348 bases), S. maltophilia and Fusobacterium gonidoformans (both 349 bases), and S. aureus (355 bases) were compared to empirically derived data. Of these selected sizes, only a band of 355 bases was detected by LH-PCR analysis (Fig. 4, lanes 3, 4, 9, and 13). Despite the limited range in sizes of bands generated, variation was observed when the LH-PCR profiles generated from this group of individuals were compared. Ten distinct LH-PCR profiles were detected for these 14 individuals (Fig. 4).
FIG. 3.
LH-PCR product (700IR-labeled) profiles generated from DNA extracted directly from a sputum sample separated on a LI-COR IR2 automated sequencer. Lanes 1 and 12, microSTEP 15a ladder (sizes in bases). Lanes 2 to 6 were loaded with equal quantities of PCR product amplified from five separate DNA extractions from a single sputum sample; lanes 7 to 11 were loaded with equal quantities of PCR product amplified in five separate reactions with a template from a single DNA extraction.
FIG. 4.
LH-PCR product (700IR-labeled) profiles generated from DNA extracted directly from bronchoscopy and sputum samples separated on a LI-COR IR2 automated sequencer. Lanes 1 and 16, microSTEP 15a ladder (sizes in bases); lane 2, individual 1, sputum sample; lane 3, individual 2, sputum sample; lane 4, individual 3, bronchoscopy sample; lane 5, individual 4, bronchoscopy sample; lane 6, individual 5, sputum sample; lane 7, individual 6, bronchoscopy sample; lane 8, individual 7, sputum sample; lane 9, individual 8, sputum sample; lane 10, individual 9, bronchoscopy sample; lane 11, individual 10, sputum sample; lane 12, individual 11, sputum sample; lane 13, individual 12, sputum sample; lane 14, individual 13, sputum sample; lane 15, individual 14, sputum sample.
TABLE 2.
Band positions for LH-PCR and T-RFLP profiles generated from samples from CF patients
| Analysis and CF patient no. | Band position(s) (bases) |
|---|---|
| LH-PCR | |
| 1 | 357, 354, 342 |
| 2 | 360, 355, 342 |
| 3 | 355, 342 |
| 4 | 342 |
| 5 | 356, 342 |
| 6 | 342 |
| 7 | 342 |
| 8 | 355, 350, 342, 330 |
| 9 | 342 |
| 10 | 342 |
| 11 | 350, 342 |
| 12 | 359, 355, 350, 342 |
| 13 | 357, 353, 352, 342 |
| 14 | 358, 352, 342 |
| T-RFLP | |
| 1 | 917, 902, 859, 572, 568, 213, 175, 155, 104, 102 |
| 2 | 596, 584, 569, 172, 155, 130, 118, 107 |
| 3 | 567, 173, 155 |
| 4 | 567, 212, 172, 155 |
| 5 | 595, 566, 549, 155, 136, 128 |
| 6 | 902, 870, 569, 565, 172, 155, 102 |
| 7 | 567, 212, 155, 102 |
| 8 | 632, 565, 535, 365, 335, 270, 215, 207, 171, 155, 103 |
| 9 | 567, 212, 172, 155 |
| 10 | 913, 568, 566, 380, 209, 171, 155, 113, 102 |
| 11 | 566, 563, 538, 372, 369, 214, 183, 172, 155, 131, 113, 102 |
| 12 | 942, 927, 916, 630, 601, 591, 586, 579, 577, 567, 473, 419, 405, 392, 380, 364, 306, 275, 269, 264, 257, 250, 238, 209, 206, 201, 181, 172, 155, 132, 106, 104 |
| 13 | 564, 429, 374, 288, 275, 214, 186, 173, 155, 133, 102 |
| 14 | 564, 375, 288, 214, 204, 173, 155, 132, 119, 106, 102 |
T-RFLP profiling of clinical sample DNA.
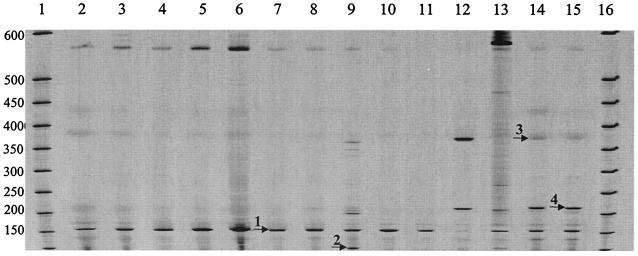
As found for the LH-PCR profiling data, the T-RFLP profiles generated from DNA extracted from a sputum sample were indistinguishable, both in terms of repeated extractions from different subsamples of the same clinical material (Fig. 5, lanes 2 to 6) and in terms of PCR amplification, from an individual DNA extraction (Fig. 5, lanes 7 to 11). Figure 6 shows the T-RFLP profiles obtained after CfoI restriction endonuclease digestion of PCR products amplified from DNA extracted directly from ten sputum samples and four bronchoscopies sampled from the same CF patients used in the LH-PCR study above. The size range of bands detected was from 100 to 942 bases. The data are shown only for the region between 100 and 600 bases since the remaining ca. 340 bases contributed comparatively few extra T-RFLP bands to the analyses. Table 2 also shows the sizes of the T-RFLP bands detected for samples taken from these patients. Over the entire T-RFLP size region, the number of bands detected ranged from 3 (Fig. 6, lane 4) to 32 (Fig. 6, lane 13) with, on average, 11 bands detected for sputum samples and 5 bands detected for the bronchoscopy samples.
FIG. 5.
CfoI-generated T-RFLP profiles formed from 16S rRNA gene PCR products (700IR labeled) amplified from DNA extracted directly from a sputum sample separated on a LI-COR IR2 automated sequencer. Lane 1 and 12, microSTEP 15a ladder (sizes in bases). Lanes 2 to 6 were loaded with equal quantities of CfoI-digested T-RFLP PCR products amplified from five separate DNA extractions from a single sputum sample; lanes 7 to 11 were loaded with equal quantities of CfoI-digested T-RFLP PCR products amplified in five separate reactions with a template from a single DNA extraction.
FIG. 6.
CfoI-generated T-RFLP profiles formed from 16S rRNA gene PCR products (700IR labeled) amplified from DNA extracted directly from bronchoscopy and sputum samples separated on a LI-COR IR2 automated sequencer. Lanes 1 and 16, microSTEP 15a ladder (sizes in bases); lane 2, individual 1, sputum sample; lane 3, individual 2, sputum sample; lane 4, individual 3, bronchoscopy sample; lane 5, individual 4, bronchoscopy sample; lane 6, individual 5, sputum sample; lane 7, individual 6, bronchoscopy sample; lane 8, individual 7, sputum sample; lane 9, individual 8, sputum sample; lane 10, individual 9, bronchoscopy sample; lane 11, individual 10, sputum sample; lane 12, individual 11, sputum sample; lane 13, individual 12, sputum sample; lane 14, individual 13, sputum sample; lane 15, individual 14, sputum sample. Labeled locations within these profiles correspond to position 1, band size 155 bases (P. aeruginosa); position 2, band size 102 bases (P. oris); position 3, band size 373 (C. murliniae); and position 4, band size 215 (A. defectiva).
Of the T-RFLP banding patterns generated, two were found to be indistinguishable (Fig. 6, lanes 5 and 10), in which each contained four bands over this size range. Profiles generated by LH-PCR analysis for these samples were also found to be indistinguishable (Fig. 4, lanes 5 and 10). The dominant T-RFLP band in these samples was detected at 155 bases, corresponding to that produced by P. aeruginosa (Fig. 6, position 1). This band of 155 bases was generated by all of the samples studied, although it is extremely faint in lane 12 (Fig. 6). Other T-RFLP bands in these samples were detected at 102 bases (Fig. 6, position 2) corresponding to that produced by Prevotella oris, at 373 bases (Fig. 6, position 3) corresponding to that produced by Citrobacter murliniae, and at 215 bases (Fig. 6, position 4) corresponding to that produced by Abiotrophia defectiva. In this group, a band at base position 209 corresponding to that produced by B. cepacia was observed in two patients (Fig. 6, lanes 11 and 13); a band at base position 214 corresponding to that produced by either S. maltophilia or F. gonidoformans was observed in three patients (Fig. 6, lanes 12, 14 and 15); a band at base position 238 corresponding to that produced by S. aureus was observed in one patient (Fig. 6, lane 13), and a band at base position 364 corresponding to that produced by H. influenzae was observed in one patient (Fig. 6, lane 13). The common “band” at ca. 580 bases (Fig. 6) is an artifact of many separate bands appearing as one due to the image being compressed for display.
16S rDNA clone data.
Table 3 shows the bacterial species that were identified through sequencing 16S rDNA PCR products amplified from DNA extracted directly from sputum samples and bronchoscopies sampled from five CF patients within the group of 14 studied above. Of these five samples, four were sputa and one was a bronchoscopy specimen. In all, 103 clones were examined. P. aeruginosa was the species most commonly identified; it was found in 61 of the 103 clones (59%) and also in every individual studied (Table 3). This finding correlates well with T-RFLP data that indicated that P. aeruginosa was present in each patient. S. maltophilia was also commonly detected by cloning (patients 8, 13, and 14 [Table 3]). Patients 13 and 14 were also found to have a band corresponding to S. maltophilia based on T-RFLP data generated from the same samples. Eight distinct bacterial species were detected in the 17 non-P. aeruginosa or non-S. maltophilia clones. Patients 8, 13, and 14, who had more than two bacterial species identified by 16S rDNA clone sequencing, were found to have more T-RFLP bands than patients 6 and 10.
TABLE 3.
Incidence of bacterial species present in 16S rRNA genes cloned from PCR products amplified from DNA extracted directly from five clinical samples
| Bacterial speciesa | No. of clones in CF patient no.:
|
||||
|---|---|---|---|---|---|
| 6b | 8c | 10c | 13c | 14c | |
| P. aeruginosa | 32 | 6 | 11 | 6 | 6 |
| S. maltophilia | 2 | 12 | 11 | ||
| P. oris | 1 | 1 | |||
| F. gonidoformans | 4 | ||||
| B. fragilis | 3 | ||||
| Leptotrichia-like sp. | 1 | 1 | |||
| A. defectiva | 2 | ||||
| C. murliniae | 2 | ||||
| L. mirabilis | 1 | ||||
| S. ventriculi | 1 | ||||
| Total | 32 | 15 | 11 | 21 | 24 |
Indicates a match of >98% nucleotide similarity.
Bronchoscopy sample.
Sputum sample.
DISCUSSION
Fresh insight into the structure and diversity of the bacterial communities present within a wide range of different environments has been gained by introducing molecular biological profiling methodologies. In the present study, we sought to determine the composition and diversity of the bacterial community present in sputa and bronchoscopies sampled from CF patients. Two profiling methodologies were employed, LH-PCR and T-RFLP, with the data derived compared to phylogenetic information derived from sequences of individual 16S rRNA gene clones amplified from DNA extracted from the same patient sample.
T-RFLP profiling has been used to examine the total bacterial community in ecosystems as diverse as sludge, sand, termite guts (24), soil (13), a phenol bioreactor (17), the rhizosphere (35), and feces (21). Although T-RFLP profiling has been used in many different contexts, as far as we are aware the present study is the first application of this technique to a clinically important scenario.
Before these approaches were used to study the bacteria present in clinical samples, both T-RFLP and LH-PCR methodologies were verified by analyzing pure cultures of a range of bacterial species derived from clinical specimens. These preliminary studies showed that for both approaches, empirically derived data matched those derived from in silico analyses; for each species and approach, single discrete bands were generated that matched the size predicted by computer program (MapSort)-driven analyses. Other preliminary studies demonstrated that T-RFLP profiling could detect species when they were present at a level of 1% of the original template DNA (data not shown). This finding is in line with the detection limits observed in other molecular biological profiling studies; for example, a member of the total community present at ∼1% would be detected by denaturing gradient gel electrophoresis (29).
Of the two profiling approaches, T-RFLP provided much greater resolution of the bacterial community than did LH-PCR. Although LH-PCR has been used to examine the bacterial community in soil samples (33), this approach was of limited efficacy in determining either the bacterial species present or the overall diversity of bacteria in CF clinical samples, primarily due to the relatively small differences in amplicon length generated from different bacterial species. This finding also indicates that the diversity of the CF lung bacterial community is likely to be significantly lower than that found in soil bacterial communities. In addition to greater resolution of the bacterial community, T-RFLP analysis was able to assign a tentative bacterial species identity based on the sizes of the individual T-RFLP bands generated. These tentative bacterial species assignments could be strengthened either by performing T-RFLP with restriction endonucleases of distinct recognition sequences other than the CfoI used here or by comparison with 16S rDNA data from the same samples. The latter approach, sequence analysis of 16S rDNA clones generated from the same clinical samples, provided the main means of comparison with CfoI T-RFLP data. It is important that although analyzing individual bacterial 16S rDNA sequences cloned from mixed pools of PCR products has been extremely informative regarding the composition of a wide range of clinical and nonclinical environments, the clones that are sequenced can only represent a fraction of the total bacterial community in each sample. Profiling approaches are therefore valuable since they allow the range of sequence types to be displayed in a single electrophoretic run.
Species that were detected in these samples included those previously identified as CF pathogens, including P. aeruginosa. This species was present in every set of 16S rDNA clones studied. Moreover, a band corresponding to that produced by P. aeruginosa was present in every T-RFLP track. Bands corresponding to those produced by a range of other CF pathogens, including B. cepacia, S. aureus, H. influenzae, and S. maltophilia, were also detected by T-RFLP in a number of the CF patients sampled. In the case of S. maltophilia, it is possible that these bands may all or in part be due to F. gonidoformans, a species for which no pathogenic role in CF lung disease has yet been described. No evidence, however, except for S. maltophilia or F. gonidoformans, was found in the corresponding CF patient 16S rDNA cloning data for these species. This may indicate that, as has been shown in earlier studies, cloning can introduce biases when mixed pools of PCR products are used (39). It has, for example, been demonstrated that discrepancies between the numerical distribution of subgroups of methanogen sequences identified by either cloning or by T-RFLP analysis can occur (19).
Analysis of 16S rDNA clones did, however, identify a number of other bacterial species, namely, P. oris, F. gonidoformans, Bacteroides fragilis, Leptotrichia-like sp., A. defectiva, C. murliniae, Lautropia mirabilis, and Sarcina ventriculi. Certain of these organisms are considered to be oral-associated microbes (e.g., P. oris, Leptotrichia-like sp., etc.), others (e.g., L. mirabilis) have been reported as CF pathogens previously (3), and one species (A. defectiva) is associated with many different body sites (8). However, certain species, including C. murliniae, F. gonidoformans, B. fragilis (6), and S. ventriculi are considered to be of gut origin typically and are, as such, not associated with the lung. C. murliniae, in particular, has not been associated with CF patients with previously human-associated strains being isolated from stool, wound, blood, and urine samples (5). These species may therefore be “novel” potential CF pathogens. Whereas these species were found comparatively infrequently compared to Pseudomonas and Stenotrophomonas spp., these data support the conclusion drawn from T-RFLP analyses that diverse communities of bacteria are not uncommon in these environments. This may be particularly important given that mixed populations may be able to regulate gene expression in a coordinated manner within the bacterial biofilm in the CF lung (9, 22).
Many of the observed T-RFLP band positions could not be assigned to a bacterial species. This result may reflect the general lack of characterization of this ecosystem. Ongoing studies are currently characterizing these unassigned bands. The numbers of T-RFLP bands observed varied markedly between different patients, indicating that the bacterial communities present in these individuals were typically distinct. Further, it implies that there is no evidence for identically composed bacterial communities to be shared within this patient group. This raises interesting questions that will require further study to determine whether the variation in bacterial community diversity observed among these patients can be explained in terms of clinical parameters and in relation to the development of lung disease (34).
Two types of clinical samples, bronchoscopy and sputa, from CF patients were evaluated here. As with any process of recovering clinical samples from the lung, it is possible that sample contamination occurred with microbes from other body sites either during expectoration or, potentially less so, through introduction and removal of the bronchoscope. Efforts were taken to minimize this potential error by, for example, washing samples prior to DNA extraction. Moreover, it has been shown that the contribution of oropharyngeal bacteria to the analysis by cultivation of sputa sampled from CF patients is not significant (15). More diversity was observed in T-RFLP profiles from sputum samples compared to bronchoscopy samples. This cannot be interpreted as meaning that sputa were inherently more diverse however because of the overall low number of samples and the particular lack of paired sputum and bronchoscopy samples from the same patient.
As with all PCR-based methods, biases can be introduced in terms primarily of the initial DNA extraction step and the PCR process itself. To minimize these biases, we used protocols here that have already been used successfully to extract DNA and amplify sequences from complex microbial communities (7). It is also important to note that data from any analytical technique that examines 16S rDNA can be influenced by the different number of copies of ribosomal operons in different species in terms of the number of clones or relative intensities of bands profiled (14).
Compared to traditional culture-based analyses, however, T-RFLP is much more rapid in providing information on the composition of the bacterial community: DNA extraction, PCR, restriction endonuclease digestion, and electrophoretic separation and analysis can be completed within a 12-h period. There are many cases in which this rapidity could be of critical importance in terms of clinical diagnosis. Moreover, the speed and accuracy of T-RFLP will allow the impact of therapeutic interventions to be assessed and altered as required. The additional strength of this approach over other molecular biology-based diagnostic tools, such as gene microarrays, is that it allows any novel or “unexpected” species to be detected and as such is not limited by preconceptions as to what bacteria are important in a given system. Recent studies have shown the importance of this in analyzing CF-associated bacteria (10, 31).
Culturing bacteria from CF specimens remains important both diagnostically and as a resource for subsequent genotypic analyses. It is likely, however, that combined culture and molecular biology-based methodologies will become increasingly important. Of these molecular methodologies, T-RFLP analysis has been shown here to be valuable in determining bacterial community diversity in samples from CF patients. Moreover, it is a flexible technique that will have many applications to the study of other important infections.
Acknowledgments
This work was supported by MWG and the School of Health and Life Sciences, King's College London.
We also acknowledge the generous support of the Molecular Psychiatry Unit of University College London.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Dekhil, S. M., M. M. Peel, V. A. Lennox, E. Stackebrandt, and L. I. Sly. 1997. Isolation of Lautropia mirabilis from sputa of a cystic fibrosis patient. J. Clin. Microbiol. 35:1024-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., H. L. Ayala-Del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner, D. J., C. M. O'Hara, P. A. D. Grimont, J. M. Janda, E. Falsen, E. Aldova, E. Ageron, J. Schindler, S. L. Abbott, and A. G. Steigerwalt. 1999. Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11). J. Clin. Microbiol. 37:2619-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressane, M. A., L. E. Durigon, and M. J. Avila-Campos. 2001. Prevalence of the Bacteroides fragilis group and enterotoxigenic Bacteroides fragilis in immunodeficient children. Anaerobe 7:277-281. [Google Scholar]
- 7.Bruce, K. D., and M. R. Hughes. 2000. Terminal restriction fragment length polymorphism monitoring of genes amplified directly from bacterial communities in soils and sediments. Mol. Biotechnol. 16:261-269. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, J. J., and R. R. Facklam. 2001. Granulicatella and Abiotrophia species from human clinical specimens. J. Clin. Microbiol. 39:3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Coenye, T., J. Goris, T. Spilker, P. Vandamme, and J. J. LiPuma. 2002. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J. Clin. Microbiol. 40:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva Filho, L. V., J. E. Levi, C. N. Oda Bento, S. R. da Silva Ramos, and T. Rozov. 1999. PCR identification of Pseudomonas aeruginosa and direct detection in clinical samples from cystic fibrosis patients. J. Med. Microbiol. 48:357-361. [DOI] [PubMed] [Google Scholar]
- 12.Davis, P. B., M. Drumm, and M. W. Konstan. 1996. Cystic fibrosis. Am. J. Respir. Crit. Care Med. 154:1229-1256. [DOI] [PubMed] [Google Scholar]
- 13.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilljam, H., A. S. Malmborg, and B. Strandvik. 1986. Conformity of bacterial growth in sputum and contamination free endobronchial samples in patients with cystic fibrosis. Thorax 41:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guieysse, B., P. Wikstrom, M. Forsman, and B. Mattiasson. 2001. Biomonitoring of continuous microbial community adaptation toward more efficient phenol-degradation in a fed-batch bioreactor. Appl. Microbiol. Biotechnol. 56:780-787. [DOI] [PubMed] [Google Scholar]
- 18.Hart, C. A., and C. Winstanley. 2002. Persistent and aggressive bacteria in the lungs of cystic fibrosis children. Br. Med. Bull. 61:81-96. [DOI] [PubMed] [Google Scholar]
- 19.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchison, M. L., and J. R. W. Govan. 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 1:1005-1014. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, C. W., J. C. Astaire, M. E. Sanders, B. S. Reddy, and C. L. Kitts. 2001. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 67:1935-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewenza, S., M. B. Visser, and P. A. Sokol. 2002. Interspecies communication between Burkholderia cepacia and Pseudomonas aeruginosa. Can. J. Microbiol. 48:707-716. [DOI] [PubMed] [Google Scholar]
- 23.LiPuma, J. J., B. J. Dulaney, J. D. Mmenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 27.Martin, C., M. A. Ichou, P. Massicot, A. Goudeau, and R. Quentin. 1995. Genetic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis revealed by restriction fragment length polymorphism of the rRNA gene region. J. Clin. Microbiol. 33:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell, A., E. Mahenthiralingam, J. E. Moore, K. E. A. Dunbar, A. K. Webb, M. E. Dodd, S. L. Martin, B. C. Millar, C. J. Scott, M. Crowe, and J. S. Elborn. 2001. PCR-based detection and identification of Burkholderia cepacia complex pathogens in sputum from cystic fibrosis patients. J. Clin. Microbiol. 39:4247-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 31.Pitulle, C., D. M. Citron, B. Bochner, R. Barbers, and M. D. Appleman. 1999. Novel bacterium isolated from a lung transplant patient with cystic fibrosis. J. Clin. Microbiol. 37:3851-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeld. M., R. L. Gibson, S. McNamara, J. Emerson, J. L. Burns, R. Castile, P. Hiatt, K. McCoy, C. B. Wilson, A. Inglis, A. Smith, T. R. Martin, and B. W. Ramsey. 2001. Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr. Pulm. 32:356-366. [DOI] [PubMed] [Google Scholar]
- 35.Sakai, M., N. Matsunaga, A. Matsuka, and S. Kanazawa. 2001. Application of T-RFLP analysis to the study of bacterial community structure in the rhizosphere. Soil Sci. Plant Nutr. 47:773-778. [Google Scholar]
- 36.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tümmler, B., and B. C. Kiewitz. 1999. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol. Med. Today 5:351-357. [DOI] [PubMed] [Google Scholar]
- 38.van Belkum, A., N. H. Renders, S. Smith, S. E. Overbeek, and H. A. Verbrugh. 2000. Comparison of conventional and molecular methods for the detection of bacterial pathogens in sputum samples from cystic fibrosis patients. FEMS Immunol. Med. Microbiol. 27:51-57. [DOI] [PubMed] [Google Scholar]
- 39.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 40.Welsh, M. J., L. C. Tsui, T. F. Boat, and A. L. Beaudet. 1995. Cystic fibrosis, p. 3799-3876. In C. R. Sciver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease. McGraw-Hill Professional, New York, N.Y.
- 41.Whitby, P. W., H. L. N. Dick, P. W. Preston III, D. E. Tullis, A. Matlow, and T. L. Stull. 1998. Comparison of culture and PCR for detection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J. Clin. Microbiol. 36:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitby, P. W., L. C. Pope, K. B. Carter, J. J. LiPuma, and T. L. Stull. 2000. Species-specific PCR as a tool for the identification of Burkholderia gladioli. J. Clin. Microbiol. 38:282-285. [DOI] [PMC free article] [PubMed] [Google Scholar]