Abstract
Although Clostridium perfringens is recognized as an important cause of clostridial enteric diseases, only limited knowledge exists concerning the association of particular C. perfringens toxinotypes (type A to E) with gastrointestinal (GI) diseases in domestic animals. Some C. perfringens isolates also produce the newly discovered beta2-toxin (CPB2). Recent epidemiological studies suggested that C. perfringens isolates carrying the gene encoding CPB2 (cpb2) are strongly associated with clostridial GI diseases in domestic animals, including necrotic enteritis in piglets and typhlocolitis in horses. These putative relationships, obtained by PCR genotyping, were tested in the present study by further genotyping and phenotyping of 29 cpb2-positive C. perfringens isolates from pigs with GI disease (pig GI disease isolates). PCR and restriction fragment length polymorphism analysis reconfirmed the presence of cpb2 gene sequences in all the disease isolates included in the study. Furthermore, genotyping by pulsed-field gel electrophoresis analyses showed that the pig GI disease isolates included in this study all carry a plasmid cpb2 gene, yet no clonal relationships were detected between the cpb2-positive pig GI disease isolates surveyed. Finally, CPB2-specific Western blotting demonstrated CPB2 expression by all of the cpb2-positive isolates surveyed. The CPB2 proteins made by five of these pig GI disease isolates were shown to have the same deduced amino acid sequences as the biologically active CPB2 protein made by the original type C isolate, CWC245. Collectively, our present results support a significant association between CPB2-positive C. perfringens isolates and diarrhea in piglets.
Clostridium perfringens is a gram-positive, endospore-forming, anaerobic bacterium that has long been recognized as a significant cause of both histotoxic and gastrointestinal (GI) diseases in humans and domestic animals (20). The virulence of this bacterium largely results from its ability to produce at least 15 different C. perfringens toxins (21). A commonly used classification scheme (21) assigns C. perfringens isolates to one of five types, types A to E, depending upon the isolate's ability to produce the four major lethal toxins (i.e., the alpha, beta, epsilon, and iota toxins). C. perfringens type A strains are defined as producing alpha toxin, and type C strains are defined as producing alpha and beta toxins. The major lethal toxins, however, are not the only biomedically important toxins; some C. perfringens isolates (mostly belonging to type A) produce C. perfringens enterotoxin (CPE), and some type C isolates produce a newly discovered C. perfringens toxin, the beta2 toxin (CPB2) (10).
CPB2, a 28-kDa protein, was first purified from C. perfringens type C strain CWC245, which was isolated from a piglet that died of necrotizing enterocolitis (16). Purified CPB2 was reported to be cytotoxic for Chinese hamster ovary cells and to induce hemorrhagic necrosis of the intestinal mucosa in a guinea pig ligated intestinal loop (16). In 1997, the gene encoding this 28-kDa protein was cloned from C. perfringens type C strain CWC245, and its nucleotide sequence was determined (10). Since the deduced amino acid sequence showed no significant homology with beta toxin, the respective protein corresponded to a new C. perfringens toxin, referred to as CPB2.
C. perfringens type C is generally considered to be the primary cause of necrotic enteritis in piglets aged 0 to 2 weeks (29), but type A isolates have also been linked to enteric disease in suckling and feeding pigs with mild necrotic enterocolitis and villous atrophy (8, 15, 22, 24, 25). The role of alpha toxin (produced by all C. perfringens toxinotypes) as a virulence factor in type A enterotoxemia remains unclear (29). Type A enterotoxemia syndrome could be produced by oral inoculation of type A isolates into gnotobiotic colostrum-deprived pigs as well as conventional weaner pigs (14). However, this effect could not be explained by the contribution of alpha toxin alone because purified alpha toxin was unable to produce significant lesions and fluid loss in a pig ileal loop assay (25, 29). Although CPE has been implicated in porcine diarrheal disease (5, 29, 32), only nonenterotoxigenic C. perfringens type A and type C strains were isolated from some diarrheic piglets (9, 10, 17). Despite the probable central role of beta toxin, typical type C disease cannot be produced in the pig model by use of that toxin alone. Experimental reproduction of the disease in pigs requires viable bacteria as well as toxin (as crude culture supernatants) (23).
The availability of the cpb2 gene sequence has provided a powerful genetic tool for the detection of CPB2-positive C. perfringens isolates, which has greatly improved our understanding of the association between C. perfringens toxinotypes and animal GI diseases. For example, by using a multiplex PCR assay incorporating cpb2 gene-specific primers, cpb2-positive C. perfringens type A isolates could be detected from diarrheic piglets (9, 17), horses with typhlocolitis (13), diarrheic dogs (31), an African elephant with ulcerative enteritis (1), and calves with enterotoxemia (19).
In two studies performed in Switzerland and The Netherlands, all C. perfringens isolates obtained from diarrheic piglets were genotyped as nonenterotoxigenic types A and C (17). Interestingly, however, the cpb2 gene was found to be highly prevalent in isolates from diarrheic piglets in both studies. These data suggest a causal relationship between cpb2-positive strains and digestive tract diseases in piglets (17). In a recent PCR genotyping study (9), most of the C. perfringens isolates from pigs with GI disease (pig GI disease isolates) were found to be genotype A. Of the 33 isolates from piglets with diarrhea examined, 27 (82%) were positive for the cpb2 gene, whereas none of the isolates from the piglet controls were positive for the cpb2 gene, consistent with cpb2-positive type A isolates being significantly associated with diarrhea in piglets. However, these putative associations between cpb2-positive isolates and GI diseases in piglets remain tentative because this conclusion was drawn only on the basis of the results of PCR genotyping, in which only the presence of cpb2 sequences and not the expression of CPB2 were demonstrated.
To better appreciate the involvement of cpb2-positive C. perfringens isolates in GI diseases of piglets, the present study genotypically and phenotypically characterized 35 pig fecal C. perfringens isolates. Notably, this study includes the first in-depth genotypic analysis of cpb2-positive fecal isolates obtained from piglets with CPB2-associated GI diseases. Results from the present study indicate that all cpb2-positive C. perfringens fecal isolates carry the cpb2 gene on a plasmid and express CPB2, confirming their virulence potential. These new findings hold potential epidemiologic significance, as they are consistent with cpb2-positive C. perfringens isolates being responsible for GI diseases in piglets.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. perfringens isolates used in this study are listed and described in Table 1. Each strain was from a separate disease instance in piglets, as described earlier (9, 10, 17). A starter culture (6 ml) of each C. perfringens isolate was prepared by overnight growth at 37°C in fluid thioglycolate broth (FTG; Difco), as described previously (7). For DNA isolation or culture supernatant protein preparation, an aliquot (0.2 ml) of each FTG culture was inoculated into 10 ml of TGY broth (3% Trypticase, 2% glucose, 1% yeast extract, 0.1% cysteine) (7), which was then incubated at 37°C overnight.
TABLE 1.
Summary of genotypic and phenotypic characterization results obtained in this study for cpb2-positive C. perfringens isolates
| Source and strain | Type | cpb2 PCR resulta | cpb2 RFLP pattern (kb) with HpaI | CPB2-specific Western blotting resultsb | cpb2 location determined by PFGE | Reference |
|---|---|---|---|---|---|---|
| cpb2-positive strain CWC245 | C | + | ∼5 | + | Plasmid | 10 |
| Healthy pigs | ||||||
| JGS1537 | A | − | − | − | NDc | 9 |
| JGS1540 | A | − | − | ND | ND | 9 |
| JGS1569 | A | − | − | ND | ND | 9 |
| JGS1580 | A | − | − | ND | ND | 9 |
| D9703881 | A | − | − | ND | ND | 9 |
| 106527 | C | − | − | − | − | 17 |
| Diarrheic pigs | ||||||
| 106526 | A | + | ∼5 | + | Plasmid | 17 |
| JGS1528 | A | + | ∼5 | + | ND | 9 |
| JGS1538 | A | + | ∼5 | + | ND | 9 |
| JGS1541 | A | + | ∼5 | + | Plasmid | 9 |
| JGS1548 | A | + | ∼6 | + | ND | 9 |
| JGS1552 | A | + | ∼5 | + | Plasmid | 9 |
| JGS1561 | A | + | ∼5 | + | Plasmid | 9 |
| JGS1570 | A | + | ∼5 | + | ND | 9 |
| JGS1580 | A | − | − | ND | ND | 9 |
| JGS1807 | A | + | ∼5 | + | Plasmid | 9 |
| JGS1817 | A | + | ∼5 | + | Plasmid | 9 |
| JGS1818 | A | ± | >20 | + | Plasmid | 9 |
| 120-98 | A | + | ∼5 | + | Plasmid | 9 |
| 170-98 | A | + | ∼5 | + | Plasmid | 9 |
| 855-97#3 | A | + | ∼5 | + | ND | 9 |
| 855-97#4 | A | + | ∼5 | + | Plasmid | 9 |
| 2011 | A | + | ∼8 | + | Plasmid | 9 |
| 2142 | A | + | ∼5 | + | ND | 9 |
| 9461-97 | A | + | ∼5 | + | Plasmid | 9 |
| 9777colon | A | + | ∼5 | + | Plasmid | 9 |
| 10618 J | A | + | ∼5 | + | Plasmid | 9 |
| 10728 | C | + | ∼5 | + | ND | 9 |
| 97-4029 | C | + | ∼6 | + | ND | 9 |
| JGS1071 | C | + | ∼5 | + | Plasmid | 9 |
| JGS1090 | C | + | ∼5 | + | Plasmid | 9 |
| JGS1495 | C | + | ∼5 | + | ND | 9 |
| JGS1504 | C | + | ∼5 | + | ND | 9 |
| JGS1508 | C | + | ∼5 | + | ND | 9 |
| 106640 | C | + | ∼5 | + | Plasmid | 17 |
+, presence of both 1,373-bp and 318-bp products; −, absence of both 1,373-bp and 318-bp PCR products; ±, presence of 318-bp product but absence of 1,373-bp product.
+, presence of ∼28-kDa immunoreactive band; −, absence of ∼28-kDa immunoreactive band.
ND, not determined.
cpb2-specific PCR analysis.
Total C. perfringens DNA was isolated from the overnight TGY cultures by a previously described protocol (7). The isolated DNA was then subjected to screening by a cpb2-specific PCR with two sets of primers. The primer set 5′-AAACTGAATTTTTAAATGGTGC-3′ (primer 1F) and 5′-TCCACATCCAATGATCTACAA-3′ (primer 2R) was used to amplify a cpb2 gene-specific internal 318-bp PCR product. The primer set 5′-GCTCTAGAGGATATCTTAAATTTAGCACAG-3′ (primer 3F) and 5′-CCGGAATTCTTTTTTAAGCTCAATTTTTACTGG-3′ (primer 4R) was used to amplify a 1,373-bp DNA fragment carrying the entire cpb2 open reading frame (ORF) and 263-bp upstream and 312-bp downstream sequences. These PCRs used 100 ng of template DNA, 25 pM each primer, 200 μM deoxynucleoside triphosphates (Roche), 2.5 mM MgCl2, and 1 U of TaqDNA polymerase (Fermentas) in a total volume of 50 μl. The reaction mixture was placed in a thermal cycler (Techne) for an initial period of 5 min at 94°C (denaturation) and was then subjected to 28 cycles, each consisting of 1 min at 94°C, 1 min at 58°C (annealing), and 1 min at 72°C (extension), followed by an additional period of extension for 10 min at 72°C. After PCR, the presence of an amplified product was analyzed by subjecting an aliquot of each PCR sample to electrophoresis at 100 V in 1.5% agarose gels, followed by ethidium bromide staining and visualization under UV illumination.
Preparation of DIG-labeled cpb2 probe for Southern blotting experiments.
A 318-bp digoxigenin (DIG)-labeled, double-stranded cpb2-specific DNA gene probe was prepared by a two-step PCR amplification method. Briefly, a 318-bp cpb2 internal fragment (nucleotides 581 to 899 of the cpb2 ORF) (10) was first amplified from the total DNA of C. perfringens type C strain CWC245 by PCR, as described above.
The 318-bp PCR product obtained from this amplification was then gel purified by using the Prep-A-Gene DNA Purification system (Bio-Rad) and was used as the template for a second PCR in order to produce a 318-bp PCR product containing DIG-labeled nucleotides. This second PCR involved the incubation of 50 ng of the 318-bp purified product obtained from the previous PCR; 2.5 mM MgCl2; 100 μM dATP, dCTP, and dGTP; 65 μM dTTP; and 35 μM DIG-dUTP (Roche) under the same amplification conditions described above.
Restriction fragment length polymorphism (RFLP) and Southern blotting analyses.
Isolated C. perfringens DNA samples, prepared as described above, were digested to completion with BamHI, BglII, EcoRI, EcoRV, HpaI, or XbaI (New England Biolabs); separated by electrophoresis on 1% agarose gels; transferred to positively charged nylon membranes (Roche); and fixed to the membranes with UV light, as described previously (26). The blots were hybridized with the DIG-labeled cpb2 probe, and the hybridized cpb2 probe was then detected with a DIG chemiluminescence detection system with CSPD [disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3,3.1.13.7]decan}-4-yl) phenyl phosphate] ready-to-use substrate (Roche), as described earlier (26).
Pulsed-field gel electrophoresis (PFGE) and Southern blotting analyses.
Cells from overnight TGY cultures were collected by centrifugation and used to prepare agarose plugs containing genomic C. perfringens DNA, as described previously (2, 3, 6, 27, 30). The undigested total unsheared genomic DNA in 100 μl of an agarose plug was electrophoresed at 200 V with a Bio-Rad CHEF-DRII apparatus, with pulse times ramped from 50 to 90 s over 20 h (27, 30). These pulsed-field gels were then subjected to cpb2 Southern blotting analysis by the same procedure described above for RFLP and Southern blotting analyses.
PFGE studies for clonal relationships.
Total DNA in 100 μl of an agarose plug was digested overnight with SmaI at 30°C or with MluI at 37°C in 200 μl of the buffer solution recommended by the enzyme manufacturer. Both SmaI- and MluI-digested DNA samples were then analyzed by PFGE with 1% agarose gels prepared with PFGE-grade agarose (Bio-Rad), with pulse times ramped from 5 to 120 s over 46 h at 150 V (4). After PFGE, these gels were subjected to ethidium bromide staining and photographed under a UV transilluminator (Bio-Rad).
Isolation of culture supernatant proteins.
An aliquot (0.2 ml) of each C. perfringens FTG starter culture was inoculated into two tubes containing 10 ml of TGY broth and grown overnight at 37°C. The culture supernatant (∼20 ml) was separated from the bacterial pellet by centrifugation. To precipitate the supernatant proteins, ammonium sulfate [4.76 g of (NH4)2SO4/10 ml of supernatant] was added, and the mixture was incubated overnight at 4°C. The precipitated proteins were then collected by centrifugation and resuspended in 25 μl of sterile water and 25 μl of sample buffer. The prepared proteins were immediately used for Western blotting or were stored at −80°C.
Isolation of total cell proteins.
Total cell protein was prepared from the bacterial pellet of the 20-ml TGY culture that was used to prepare the culture supernatant proteins. After aspiration of the culture supernatant, the bacterial pellet was resuspended in 500 μl of sterile water, transferred to an Eppendorf tube, and then washed twice with 500 μl of sterile water. The resultant pellet was resuspended in 500 μl of sterile water and 500 μl of sample buffer, and the mixture was boiled for 5 min. Five microliters of this total cell protein was immediately electrophoresed and Western blotted or stored at −80°C.
CPB2 Western blotting analysis.
The prepared proteins were electrophoresed on a 0.1% sodium dodecyl sulfate (SDS)-12% polyacrylamide gel and transferred to Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad) as described before (18, 26). The membrane was then washed once in 10 ml of Tris-buffered saline (TBS) for 5 min and blocked by incubation in 10 ml of 5% bovine serum albumin (BSA) for 2 h at room temperature, followed by three washes with Tween plus TBS (TTBS; 10 ml for each wash). The membrane was then incubated overnight in a rabbit polyclonal CPB2 antibody solution (1:500 dilution in 5% BSA) (10). After four more washes in TTBS, the membrane was incubated in anti-rabbit-horseradish peroxidase antibody (Pierce) solution (1:20,000 dilution in 5% BSA) for 1 h. Finally, the membrane was washed three times in TTBS and twice in TBS, incubated for 5 min in SuperSignal West Pico Chemiluminescent Substrate (Pierce) according to the protocol of the manufacturer, and exposed to autoradiography film (Kodak).
Cloning and sequencing of the cpb2-containing fragments from various cpb2-positive isolates.
To facilitate nucleotide sequencing, the 1,373-bp DNA fragment carrying the cpb2 ORF and the ∼262-bp upstream and ∼312-bp downstream sequences from each of several representative pig GI disease isolates were first amplified by PCR and cloned into Escherichia coli. To reduce the PCR-induced mutation(s) introduced during the amplification process, the strategy described by Collie et al. (4) was adopted. Briefly, the 1,373-bp cpb2-containing DNA fragment from each cpb2-positive isolate was amplified in five identical, but independent, reaction tubes, as described above. After confirmation of the presence of the expected amplification products in all reaction tubes by agarose gel electrophoresis, the five individual PCR mixtures with one isolate's DNA were combined. These mixed PCR products were then ligated into the pCR-XL-TOPO vector by using the TOPO XL cloning kit (Invitrogen), and TOP10 E. coli was transformed with the ligated mixture according to the instructions of the manufacturer. Plasmid-containing transformants were then selected on tryptic soy agar plates containing kanamycin (50 μg/ml). The presence and the orientation of the expected cpb2-containing insert in plasmids isolated from these transformants were confirmed by restriction analysis with EcoRI or HindIII. Five clones carrying the cpb2 insert of one C. perfringens isolate, placed in the same orientation in pCR-XL-TOPO, were then grown as separate overnight cultures in 3.0 ml of tryptic soy broth with kanamycin (50 μg/ml). These overnight cultures were combined, and plasmid DNA was isolated by using a Qiagen mini plasmid prep kit, according to the instructions of the manufacturer. The insert present in the plasmid DNA was then sequenced by the fluorescence-based automated DNA sequencing facilities available in the Center for Gene Research and Biotechnology at Oregon State University by using the M13 forward and reverse primers.
cpe-specific PCR.
The cpe-specific PCR analysis was performed as described before (7, 26) with the primer set 5′-GGTACCTTTAGCCAATCA-3′ (primer 2F) and 5′-TCCATCACCTAAGGACTG-3′ (primer 5R).
Alpha toxin expression by cpb2-positive C. perfringens pig GI disease isolates.
Alpha toxin expression by pig GI disease isolates was tested by the reverse CAMP test (12).
RESULTS
cpb2-specific PCR screening for presence of entire cpb2 ORF sequences in C. perfringens isolates associated with GI diseases in piglets.
The isolates in our collection of pig GI disease isolates have previously been identified (9, 17) as cpb2 positive by a multiplex PCR assay by monitoring the isolates for the presence of a 516-bp PCR product from the internal cpb2 sequence. In our present study, we determined (Fig. 1) whether these isolates carry the entire cpb2 ORF sequence by PCR analysis with two sets of primers designed from the CWC245 cpb2 gene sequence published earlier (10).
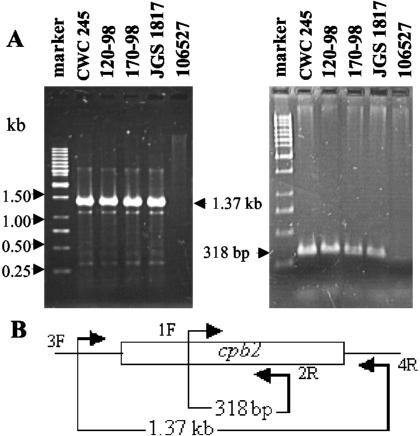
FIG. 1.
PCR analysis of pig GI disease isolates. (A) Representative results of a PCR assay with primers designed to amplify 1,373- and 318-bp PCR products. The migration of the PCR products derived from each primer set is indicated between the two gels. Results are shown for control strains CWC245 (a known cpb2-positive C. perfringens type C strain) and 106527 (a cpb2-negative type C strain) and representative pig GI disease isolates 120-98, 170-98, and JGS1817. DNA size markers (GeneRuler 1-kb ladder; Fermentas) are shown in the left lane of each gel. (B) Schematic diagram showing the location of each primer in CWC245 DNA (10).
To ensure the reliability of the PCR results obtained with the template DNAs isolated from our pig GI disease strains, control PCRs were run by using template DNAs isolated from CWC245 (a known cpb2-positive C. perfringens type C isolate) and 106527 (previously determined to be a cpb2-negative C. perfringens type C strain). As shown in Fig. 1, the expected 318-bp PCR product (which is internal to the cpb2 ORF) and the expected 1,373-bp PCR product (which contains the whole cpb2 ORF) were amplified from CWC245 DNA with primer pairs 1F-2R and 3F-4R, respectively. However, no such PCR-amplified products were obtained when DNA from cpb2-negative strain 106527 was used as the template. These results indicate that our PCR results are reliable and specific for the detection of isolates carrying the cpb2 ORF sequence.
When template DNA isolated from each pig GI disease isolate was subjected to these same PCR analyses, both 318- and 1,373-bp products were obtained from the DNA of 28 of 29 isolates tested with primer pairs 1F-2R and 3F-4R, respectively (Table 1; see Fig. 1 for representative results). The 318-bp cpb2 internal fragment but not the 1,373-bp fragment containing the whole cpb2 ORF was amplified from the DNA of the remaining isolate, JGS1818. These results confirm that our pig GI disease isolates carry the cpb2 ORF sequence.
Genotyping of cpb2-positive C. perfringens isolates by RFLP analysis.
Next, our collection of cpb2-positive isolates was subjected to RFLP analysis. We first performed RFLP and Southern blotting analyses with a representative cpb2-positive C. perfringens isolate (isolate CWC245) and a representative cpb2-negative C. perfringens isolate (isolate 106527) using a series of restriction enzymes which have no cleavage site within the cpb2 ORF. When RFLP analysis with XbaI, EcoRI, BamHI, or BglII and Southern blotting assays were performed, a DIG-labeled cpb2 probe always hybridized to a >20-kb fragment of DNA from cpb2-positive isolate CWC245 (data not shown). However, when RFLP analysis with HpaI or EcoRV and Southern blotting assays were performed with that isolate (Fig. 2), the cpb2 probe hybridized to an ∼5-kb HpaI DNA fragment and an ∼7.5-kb EcoRV DNA fragment of CWC245, respectively. No hybridizing band was observed with DNA from cpb2-negative healthy pig isolate 106527, indicating that our cpb2-specific RFLP and Southern blotting analyses were reliable and specific.
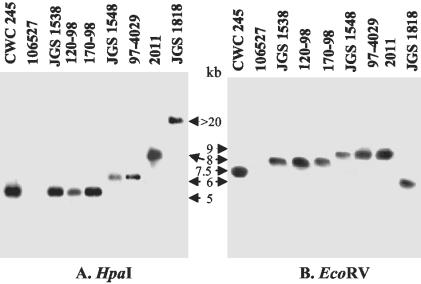
FIG. 2.
RFLP and Southern blotting analyses of HpaI- or EcoRV-digested DNA from pig GI disease isolates. Total DNA isolated from each of the specified C. perfringens strains was digested with HpaI (A) or EcoRV (B) and then Southern transferred. The Southern blots were probed with a 318-bp, DIG-labeled cpb2-specific probe. Results are shown for control strains CWC245 (a known cpb2-positive type C isolate) and 106527 (a cpb2-negative type C strain) and representative cpb2-positive pig GI disease isolates JGS1538, 120-98, 170-98, JGS1548, 97-4029, 2011, and JGS1818. The migration of the hybridizing band derived from each strain is indicated between the two blots.
When the same assay consisting of RFLP analysis with HpaI and Southern blotting was applied to our collection of cpb2-positive pig GI disease isolates, the cpb2-specific probe hybridized to an ∼5-kb HpaI DNA fragment for 25 of 29 pig GI disease isolates (Table 1; see Fig. 2 for representative results). Four pig GI disease isolates (isolates JGS1548, JGS1818, 2011, and 97-4029) produced different hybridizing bands of >5 kb (Table 1 and Fig. 2). In order to confirm that these different results by RFLP analysis with HpaI were not simply due to the loss of an HpaI restriction site, further RFLP analyses were performed with EcoRV-digested DNA (Fig. 2). The results of RFLP analysis with EcoRV confirmed our results obtained by RFLP analysis with HpaI, demonstrating that the patterns of these four isolates obtained by RFLP analysis with EcoRV are different from those of the other pig GI disease isolates surveyed (Fig. 2 and data not shown).
Finally, the specificity of the cpb2-specific probe used in the present RFLP studies has been confirmed by further control studies demonstrating (Table 1; see Fig. 2 for representative results) that this probe does not hybridize to HpaI-digested DNA from C. perfringens healthy pig isolates (isolates 106527, JGS1537, JGS1540, JGS1569, JGS1580 and D9703881) that are genotyped as cpb2-specific PCR negative (Table 1).
PFGE evidence supporting the plasmid localization of cpb2 in isolates from pigs with GI diseases.
To determine whether the cpb2 gene is located on the chromosome or plasmid of cpb2-positive pig GI disease isolates, representative cpb2-positive C. perfringens isolates were subjected to PFGE and Southern blotting analyses, which have successfully been used (2, 3, 6, 27, 30) to formally establish the chromosomal or plasmid localization of the cpe gene. Briefly, the principle of this method is that, without any restriction enzyme digestion, unsheared C. perfringens chromosomal DNA is too large to enter a pulsed-field gel. However, because of its smaller size, at least some plasmid DNA should enter a pulsed-field gel, even without any restriction enzyme digestion. Therefore, when total DNA from cpb2-positive isolates is subjected to this PFGE analysis and then Southern blotted for cpb2, either of two specific profiles should be observed: (i) if the cpb2 gene is located on the chromosome, all cpb2-containing DNA should remain in the gel wells and no hybridizing band should be observed in the gel; or (ii) if the cpb2 gene is located on the plasmid, some cpb2-containing plasmid DNA should enter the pulsed-field gels and should produce a hybridizing band in the blot.
When PFGE genotyping experiments were performed with the DNA of control cpb2-positive type C isolate CWC245, cpb2-containing DNA entered the pulsed-field gel in the absence of restriction enzyme digestion and produced a hybridizing band of ∼50 kb (Fig. 3). These PFGE results confirmed previous observations that the cpb2 gene is located on the plasmid of cpb2-positive isolate CWC245 (10). The apparent size of the CWC245 plasmid carrying cpb2 matches the calculated size of the plasmid from type A strain 13 whose cpb2 gene was sequenced (28).
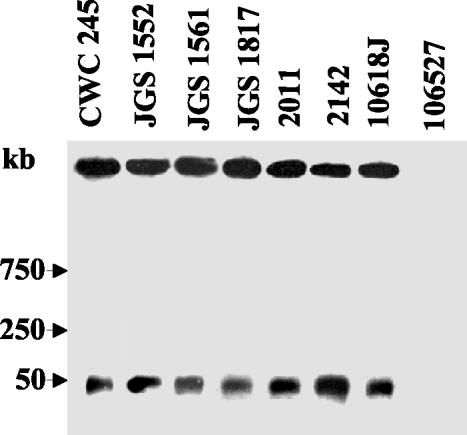
FIG. 3.
PFGE evidence supporting the plasmid localization of cpb2 in pig GI disease isolates. PFGE and Southern hybridization were used to analyze undigested DNA, prepared in agarose plugs, from each of the C. perfringens isolates specified. The blots were probed with a 318-bp cpb2-specific probe. Results are shown for control strains CWC245 (a cpb2-positive type C isolate carrying the cpb2 gene on a large plasmid) and 106527 (a cpb2-negative type C isolate) and representative cpb2 pig isolates JGS1552, JGS1561, JGS1817, 2011, 2142, and 10618J. The pulsed-field gel was calibrated with bacteriophage lambda DNA markers, whose migration is shown at the left of the blot.
When similar PFGE genotyping analyses were performed with the DNA of representative cpb2-positive isolates, their cpb2-containing plasmid DNA entered the pulsed-field gels and produced hybridizing bands that comigrated with the cpb2-containing plasmid DNA from CWC245 (Table 1; see Fig. 3 for representative results). These results indicate that both type A and C cpb2-positive C. perfringens pig GI disease isolates carry the cpb2 gene on a plasmid.
PFGE analysis for clonal relationship between cpb2-positive isolates.
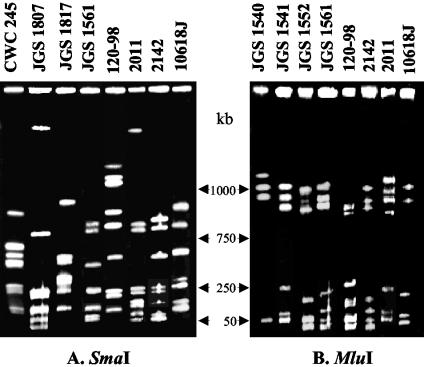
The final series of experiments conducted for the genotyping of cpb2-positive isolates involved the use of PFGE analyses to examine the possibility that cpb2-positive pig isolates share a common clonal relationship with each other. PFGE analyses of SmaI-digested DNA identified no clonal relationship between the 17 pig GI disease isolates tested (see Fig. 4 for representative results). In order to confirm that the results of the SmaI digestions were not due to uneven migration throughout the gel, further PFGE analyses were performed with MluI-digested gel blocks. The results of PFGE with MluI-digested DNA confirmed the lack of a clonal relationship between 17 pig GI disease isolates (see Fig. 4 for representative results).
FIG. 4.
Analysis of clonal relationships among C. perfringens pig GI disease isolates. DNA from each of the specified C. perfringens isolates in agarose plugs was digested with SmaI (A) or MluI (B) and subjected to PFGE and ethidium bromide straining. The gel was calibrated with bacteriophage lambda ladder DNA. The molecular sizes of the DNA markers are between the two gels.
CPB2 expression by cpb2-positive isolates.
The phenotypic characteristic of the cpb2-positive C. perfringens pig GI disease isolates examined in this study was whether these isolates can, in fact, express CPB2. Because cpb2 sequencing information (10) supports the fact that CPB2 is a secreted protein, we first analyzed culture supernatant proteins from cpb2-positive C. perfringens isolates. C. perfringens culture supernatants (Fig. 5) contained more than 15 protein bands in SDS-polyacrylamide gels stained with Coomassie blue. When CPB2-specific Western blotting was performed with supernatant proteins from C. perfringens CWC245 (Fig. 5), an ∼28-kDa immunoreactive band was obtained, which is consistent with the results presented previously (10). However, no CPB2 expression could be detected in the culture supernatant of cpb2 PCR-negative isolate 106527. These results indicate that our CPB2-specific Western blotting analyses are specific and reliable.
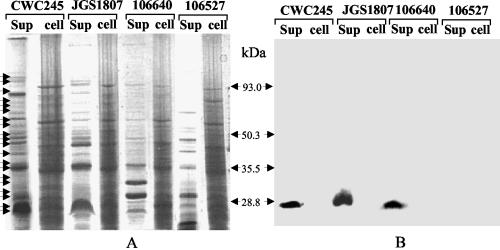
FIG. 5.
Western blotting analysis of CPB2 expression by selected pig GI disease isolates. (A) Culture supernatant proteins (sup) or total cell (cell) proteins, prepared from each of the C. perfringens isolates specified, were separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. The migrations of the supernatant proteins are shown by arrows. (B) Western blot of the gel shown in panel A. The blot was probed with CPB2 antibodies and developed by chemiluminescence detection to identify immunoreactive species. Results are shown for control strains CWC245 (a cpb2-positive type C strain) and 106527 (a cpb2-negative type C strain) and representative pig GI disease isolates JGS1807 and 106640. Molecular mass markers (in kilodaltons) are shown between the gels.
When similar CPB2-specific Western blotting analyses were performed with culture supernatant proteins prepared from our 29 unknown cpb2-positive pig GI disease isolates, an ∼28-kDa immunoreactive band that comigrated with the 28-kDa band of CWC245 was observed (Table 1; see Fig. 5 for representative results). These results indicate that all of our pig GI disease isolates surveyed express CPB2.
To investigate whether CPB2 is truly a secreted protein and not just a lysed cell product, we compared the results of CPB2-specific Western blotting with culture supernatant proteins and CPB2-specific Western blotting with total cell proteins (Fig. 5) for the detection of CPB. As expected, an ∼28-kDa immunoreactive band was observed with the culture supernatant proteins but not with the total cell proteins of CWC245 (Fig. 5). When similar Western blotting analyses were performed with pig GI disease isolate JGS1807 or 106640, an immunoreactive band that comigrated with the ∼28-kDa immunoreactive band of CWC245 was observed in the assays with the culture supernatant proteins but not in the assays with the total cell proteins (Fig. 5). These results support the fact that the CPB2 detected in the culture supernatant proteins is most likely an excreted product and not a product of cell lysis.
Comparison of cpb2 ORFs present in different cpb2-positive isolates.
To examine whether the cpb2 ORF of pig GI disease isolates encodes a CPB2 protein with an amino acid sequence similar to that of the classical CPB2 protein of pig type C isolate CWC245 (10), the nucleotide sequences of both strands of a 1,373-bp PCR-amplified DNA insert (as shown in Fig. 1) carrying the cpb2 ORFs from five different cpb2-positive pig GI disease isolates (three type A isolates and two type C isolates) were determined. These analyses revealed (data not shown) that the cpb2 ORF sequences present in isolates JGS1817, 120-98, and 170-98 (three U.S. type A isolates) and isolate 97-4029 (a U.S. type C isolate) are identical to those present in isolate 106640 (a French type C isolate). Furthermore, the cpb2 ORF sequences present in five of these isolates exactly match the cpb2 ORF sequence from CWC245 (a type C strain isolated from a diseased pig in France) determined previously (10). Comparison of the cpb2 upstream and downstream sequences present in all five isolates revealed that both the upstream and the downstream sequences are highly conserved in all five isolates and match the cpb2 upstream and downstream sequences of CWC245 determined previously (10).
cpe-specific PCR screening for presence of cpe gene in cpb2-positive isolates.
The cpb2-positive C. perfringens pig GI disease isolates used in this study had been identified earlier as cpe negative by multiplex PCR analysis (9). In this study, we reevaluated whether the cpb2-positive C. perfringens isolates that we surveyed carry the cpe gene by a single cpe-specific PCR analysis with primers 2F and 5R (7, 25) (note that primers 2F and 5R are different from the primers used in the multiplex PCR). These PCR analyses confirmed (data not shown) that none of the isolates examined in this study, regardless of their source, carry the cpe gene; i.e., all type A isolates examined in this survey were classified as cpe-negative type A C. perfringens isolates. The reliability of the cpe-specific PCR assay for the detection of the genes encoding CPE, if they are present, was demonstrated by blinded control studies with reference C. perfringens type A isolates that were cpe negative (isolate ACTC 3624) (18) and cpe positive (isolate NCTC 8239) (18) (data not shown).
Alpha toxin expression by cpb2-positive pig GI disease isolates.
The cpb2-positive C. perfringens pig GI disease isolates used in this study were characterized for their alpha toxin-producing capabilities. By using the reverse CAMP test (12), all cpb2-positive type A or type C isolates surveyed in this study produced (data not shown) the arrow-shaped zone of synergistic hemolysis indicative of alpha toxin expression.
DISCUSSION
The present study offers several significant contributions to our understanding of the pathogenesis of recently discovered CPB2-positive C. perfringens isolates. First, the results of PCR and RFLP analysis presented in this study for 6 isolates from healthy pigs and 29 isolates from pigs with GI diseases previously genotyped by multiplex PCR, which reconfirmed the presence of cpb2 only in the diseased pig isolates and not in the healthy pig isolates, significantly strengthen the hypothesis (9, 10, 17) that cpb2-positive isolates are highly associated with GI diseases in pigs.
Second, the present study reports the first PFGE genotypic characterization of cpb2-positive isolates. Our PFGE and Southern blotting results for 14 of the cpb2-positive type A pig GI disease isolates indicates that 14 of these type A isolates carry their cpb2 gene on a large plasmid. When these results are coupled with similar results for type C isolates obtained in this study and a previous study (10), it is now clear that the cpb2 gene has a plasmid location in C. perfringens pig GI disease isolates, regardless of their toxinotypes or geographical origins. Furthermore, the PFGE and RFLP analyses performed in this study demonstrated no clonal relationship among the pig GI disease isolates surveyed. Collectively, these PFGE results suggest that the acquisition of the putative cpb2-containing plasmid may be a critical step for C. perfringens isolates to become enteropathogenic for animals. Further studies are needed to determine (i) whether this cpb2-containing plasmid, like the cpe-containing plasmid (2), can be mobilized between C. perfringens isolates and (ii) whether virulence genes other than cpb2 might be encoded on the cpb2-containing plasmid.
Third, the present study's most significant finding is the presentation of the first evidence that CPB2 is expressed by many or all cpb2-positive type A isolates originating from pigs with CPB2-associated GI diseases. Our CPB2-specific Western blotting results provide direct evidence that the CPB2 produced by cpb2-positive type A isolates from pigs with GI diseases is a secreted protein, as it is for type C isolate CWC245 (10). Furthermore, the results of the present study also show that type A isolates secrete a CPB2 protein of the same size as that from type C isolates and that the sequences of the CPB2 proteins from the two types are identical (10; this study). Collectively, these findings offer important evidence supporting the virulence potential of type A isolates and suggest that the cpb2 present in many, if not all, type A isolates is regulated similarly to that present in type C isolates.
The similarity between the CPB2 protein produced by the isolates surveyed and the CPB2 protein produced by original cpb2-positive strain CWC245 (10) receives further support from the nucleotide sequencing data obtained in this study, which reveal that the cpb2 ORF sequences present in five different isolates (originating from different geographic sources) are identical to each other and to a previously determined (10) cpb2 ORF sequence from CWC245. These sequencing results also provide the strongest evidence to date that the cpb2 ORF is highly conserved, if not identical, in most or all cpb2-positive pig GI disease isolates.
The involvement of CPB2-positive C. perfringens type C isolates in GI diseases in piglets raises an important, but still unanswered, question about the pathogenesis of CPB2-associated GI diseases in animals; i.e., is CPB2 responsible for most or all GI symptoms of CPB2-associated GI diseases, or do these symptoms also result from the expression of another C. perfringens toxin(s)? All of the type A pig GI disease isolates surveyed in this study were classified as cpe-negative type A isolates, arguing that the expression of CPE or the beta, iota, or epsilon toxin is not required for a CPB2-producing type A isolate to cause GI diseases in piglets. Since the present study confirms that cpb2-positive type A porcine disease isolates can produce both CPB2 and alpha toxin, it is possible that the enteropathogenicities of these CPB2-producing type A isolates are augmented by either alpha toxin (particularly since alpha toxin has been implicated in some veterinary enteric illnesses [11]) or their ability to produce one or more of the minor C. perfringens toxins (21). Further gene knockout studies should address the relative contributions of CPB2 versus those of other toxins in CPB2-associated GI diseases in animals.
Acknowledgments
This research was supported by grant from the N. L. Tartar Foundation of Oregon State University, a grant from the Medical Research Foundation of Oregon Health Science University, and USDA grant 2002-02281 from the Ensuring Food Safety Research Program.
REFERENCES
- 1.Bacciarni, L. N., O. Pagan, J. Frey, and A. Grone. 2001. Clostridium perfringens beta2-toxin in an African elephant with ulcerative enteritis. Vet. Rec. 149:618-620. [DOI] [PubMed] [Google Scholar]
- 2.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collie, R. E., J. F. Kokai-kun, and B. A. McClane. 1998. Phenotypic characterization of enteropathgenic Clostridium perfringens isolates from non-food-borne human gastrointestinal diseases. Anaerobes 4:69-79. [DOI] [PubMed] [Google Scholar]
- 5.Collins, J. E., M. E. Bergeland, D. Bouley, A. L. Ducommun, D. H. Francis, and P. Yeske. 1989. Diarrhea associated with Clostridium perfringens type A enterotoxin in neonatal pigs. J. Vet. Diagn. Investig. 1:351-353. [DOI] [PubMed] [Google Scholar]
- 6.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 7.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrada-Correa, A. E., and D. J. Taylor. 1989. Porcine Clostridium perfringens type A spores, enterotoxin and antibody to enterotoxin. Vet. Rec. 124:606-610. [DOI] [PubMed] [Google Scholar]
- 9.Garmory, H. S., N. Chanter, N. P. French, D. Bueschel, J. G. Songer, and R. W. Titball. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibert, M., C. Jolivet-Reynaud, and M. R. Popoff. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203:56-73. [DOI] [PubMed] [Google Scholar]
- 11.Ginter, A., E. D. Williamson, F. Dessy, P. Coppe, H. Bullifent, A. Howells, and R. W. Titball. 1996. Molecular variation between the alpha-toxins from the type strain (NCTC 8237) and clinical isolates of Clostridium perfringens associated with disease in man and animals. Microbiology 142:191-198. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, M. V., and L. P. Elliott. 1980. New presumptive identification test for Clostridium perfringens: reverse CAMP test. J. Clin. Microbiol. 12:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herholz, C., R. Miserez, J. Nicolet, J. Frey, M. R. Popoff, M. Gibert, H. Gerber, and R. Straub. 1999. Prevalence of β2-toxigenic Clostridium perfringens in horses with intestinal disorders. J. Clin. Microbiol. 37:358-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jestin, A., M. R. Popoff, and S. Mahe. 1985. Epizootiologic investigations of a diarrheic syndrome in fattening pigs. Am. J. Vet. Res. 46:2149-2151. [PubMed] [Google Scholar]
- 15.Johannen, U., P. Arnold, B. Kohler, and H. J. Selbitz. 1993. Studies into experimental Clostridium perfringens type A enterotoxaemia of suckled piglets: experimental provocation of the disease by Clostridium perfringens type A intoxication and infection. Monash Vet. Med. 48:29-136. [Google Scholar]
- 16.Jolivet-Reynaud, C., M. R. Popoff, M. A. Vinit, P. Ravisse, H. Moreau, and J. E. Alouf. 1986. Enteropthogenicity of Clostridium perfringens beta toxin and other clostridial toxins. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. Reihe S 15:145-151. [Google Scholar]
- 17.Klaasen, H. L. B. M., M. J. C. H. Molkenboer, J. Bakker, R. Miserez, H. Hani, J. Frey, M. R. Popoff, and J. F. Van den Bosch. 1999. Detection of the β2 toxin gene of Clostridium perfringens in diarrhoeic piglets in The Netherlands and Switzerland. FEMS Immunol. Med. Microbiol. 24:325-332. [DOI] [PubMed] [Google Scholar]
- 18.Kokai-kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manteca, C., G. Daube, T. Jauniaux, A. Linden, V. Pirson, J. Detilleux, A. Ginter, P. Coppe, A. Kaeckenbeeck, and J. G. Mainil. 2002. The role of Clostridium perfringens beta2-toxin in bovine enterotoxaemia? Vet. Microbiol. 86:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClane, B. A. 2001. Clostridium perfringens, p. 351-372. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 21.McDonel, J. L. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p. 477-517. In F. Dorner and H. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom.
- 22.Nabuurs, M. J. A., J. Haagsma, E. J. van der Molen, and P. J. van der Heijden. 1983. Diarrhea in one to three week-old piglets associated with Clostridium prfringens type A. Ann. Rech. Vet. 14:408-411. [PubMed] [Google Scholar]
- 23.Niilo L. 1988. Clostridium perfringens type C enterotoxaemia. Can. Vet. J. 29:658-664. [PMC free article] [PubMed] [Google Scholar]
- 24.Okewole, P. A., A. E. Itodo, I. L. Oyetunde, J. C. Chima, E. A. Irokanulo, and R. A. Ocholi. 1991. Clostridium perfringens type A enterotoxaemia in pigs: a report of five cases. Br. Vet. J. 147:484-485. [DOI] [PubMed] [Google Scholar]
- 25.Popoff, M. R., and A. Jestin. 1985. Enteropathogenicity of purified Clostridium perfringens enterotoxin in the pig. Am. J. Vet. Res. 46:2147-2148. [PubMed] [Google Scholar]
- 26.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 27.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songer, J. G. 1996. Clostridial enteric disease of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiede, S., R. Goethe, and G. Amtsberg. 2001. Prevalence of beta2-toxin gene in Clostridium perfringens type A from diarrheic dogs. Vet. Rec. 149:273-274. [DOI] [PubMed] [Google Scholar]
- 32.Van Damme-Jongsten, M., J. Haagsma, and S. Notermans. 1990. Testing strains of Clostridium perfringens type A isolated from diarrheic piglets for the presence of the enterotoxin gene. Vet. Res. 126:191-192. [PubMed] [Google Scholar]