Abstract
Background
The high degree of sequence conservation between coding regions in fish and mammals can be exploited to identify genes in mammalian genomes by comparison with the sequence of similar genes in fish. Conversely, experimentally characterized mammalian genes may be used to annotate fish genomes. However, gene families that escape this principle include the rapidly diverging cytokines that regulate the immune system, and their receptors. A classic example is the class II helical cytokines (HCII) including type I, type II and lambda interferons, IL10 related cytokines (IL10, IL19, IL20, IL22, IL24 and IL26) and their receptors (HCRII). Despite the report of a near complete pufferfish (Takifugu rubripes) genome sequence, these genes remain undescribed in fish.
Results
We have used an original strategy based both on conserved amino acid sequence and gene structure to identify HCII and HCRII in the genome of another pufferfish, Tetraodon nigroviridis that is amenable to laboratory experiments. The 15 genes that were identified are highly divergent and include a single interferon molecule, three IL10 related cytokines and their potential receptors together with two Tissue Factor (TF). Some of these genes form tandem clusters on the Tetraodon genome. Their expression pattern was determined in different tissues. Most importantly, Tetraodon interferon was identified and we show that the recombinant protein can induce antiviral MX gene expression in Tetraodon primary kidney cells. Similar results were obtained in Zebrafish which has 7 MX genes.
Conclusion
We propose a scheme for the evolution of HCII and their receptors during the radiation of bony vertebrates and suggest that the diversification that played an important role in the fine-tuning of the ancestral mechanism for host defense against infections probably followed different pathways in amniotes and fish.
Background
The increasing number of sequenced genomes provides molecular explanations for both the unity and diversity of living organisms. The more divergent the organisms, the less they share genes. This explains why annotation of genomes using genes with known functions in other organisms leaves a high number of predicted genes with no predicted function. For some prokaryotes, the percentage of genes with no predicted function rises to 65% but falls to 20% for the closely related vertebrate genomes [1-3].
The majority of genes with no assigned functions are those involved in the recent evolutionary success of the considered taxonomic group. This is both true for prokaryotes that develop original metabolisms allowing growth in special environments and for the vertebrate species that have developed original solutions in response to environmental pressures. Comparison of mammalian proteins show that host defense ligands and receptors make up the group of proteins that diverge the most rapidly [4]. According to the «red queen model» the pressure of pathogens is, at small time scales, the most drastic pressure for the evolution of vertebrate species.
At the genomic level, together with the mutation/modification of regulatory elements, three driving forces are instrumental for the diversification. The first is the emergence of new domain architecture through domain accretion and shuffling, the second is deletion of genes, and the third is the expansion of a gene family either by gene duplications or by retropositions. Lineage specific expansion (LSE) is the proliferation of a given gene family in a given lineage. Its description implies the comparison of sister lineages [5]. Using predicted proteomes, Lespinet et al. have recently performed a systematic comparative analysis of LSEs in the following eukaryotic genomes: Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster and Arabidopsis thaliana. They reached the conclusion that «LSE seems to be one of the most important sources of structural and regulatory diversity in crown-group eukaryotes, which was critical for the tremendous exploration of the morphospace seen in these organisms» [6]. A good example for an LSE is the expansion of immunoglobulin genes in gnathostomes compared to other chordates. But LSEs also exist when comparing the different orders of mammals as exemplified by the expansion of the alpha interferons [7,8].
Vertebrate immunoglobulins (Ig) are built up from modules of one hundred amino acids. These modules are defined both by a common 3-D structure, by conserved disulfide bridges and by conserved amino acid positions. They share the same 3-D structure with the Fibronectin type III repeats (FNIII), but conserved amino acid positions are different in both groups of domains [9,10]. Genes coding for such modules were already present in the genomes of invertebrates [11]. The originality of the gnathostomes is the invention of rearranging antigen receptors by insertion of a transposable element in a gene coding for one of these Ig modules [3,12]. During the further diversification of the vertebrates, the different lineages (for example, condrychtians and osteichtians) have developed the system in different ways but the main difference consists in the maturation of the immune response that mainly takes place in the lymphoid organs. Cytokines that regulate the maturation of the immune response from antigen detection to clonal expansion of the one cell with better affinity mostly belong to the helical cytokine (HC) family [13]. They include interferons, most interleukins, LIF, CNTF, GCSF, GM-CSF, thrombopoïetine. These helical cytokines have no similarities at the level of primary amino acid sequences, but they are all structured around a similar four alpha helix bundle. They share this common 3-D structure with some hormones that for this reason are structurally described as helical cytokines: Growth Hormone (GH), Prolactin (PRL), Erythropoïetin (EPO) and Leptin [14]. These helical cytokines all bind to the extracellular binding domains of their cognate receptors (helical cytokines receptors: HCR) which all contain a 200 amino-acids (D200) domain that is the identification mark of the HCR gene family. These D200 domains are composed of two subdomains of 100 amino-acids (SD100A & SD100B) that are both structured like the basic Ig domains with two β sheets of respectively 3 and 4 strands (C type). Conserved amino-acid positions clearly distinguish these D200 domains from the Ig superfamily and from the FNIII family [9,10].
Whereas Ig and FNIII families have been expanded in invertebrates, a single gene with a D200 has been described in invertebrates: the dome gene in drosophila [15-17]. The HCR family is therefore an interesting example of a vertebrate specific LSE. Like other families of receptors involved in host defense, it mostly consists of highly diverging receptors (28% amino acids identities between the human and chicken IFNAR2 proteins) [4,18]. Together with the difficulty of predicting genes from genomic sequences, this explains why the comparison of the predicted human and Fugu proteomes did not allow the identification of the complete repertoire of HCR in Fugu [1]. Depending on the conserved amino acids residues, HCRs can be divided in two classes: Class I and Class II. Class II consists of the Tissue Factor, the receptors for interferons and the receptors for IL10 and its related cytokines(IL10, IL19, IL20, IL22, IL24 and IL26). Class I consists of all other HCRs [9,10]. Their cognate ligands have been called class I and class II helical cytokines. Genes for HCRI have been described in the major vertebrate groups including fish, birds and mammals, but HCRII have only been described in birds and mammals [10,18-21]. The question is therefore open as to whether the HCRII expansion is amniote specific or not. The recent efforts to sequence genomes from fish offer an interesting opportunity to answer this question.
Interestingly, the intron/exon structure of the vertebrate HCR genes is strictly conserved in all the family: like the exons coding for the Igs and the FNIII, the exons coding each SD100 are bordered by phase 1 introns, but what is specific for D200s is that SD100As are encoded by two exons with an internal phase 2 intron falling at the level of the third β strand and that SD100Bs are encoded by two exons with an internal phase 0 intron falling at the level of the fourth β strand [22-24]. Intron/exon structures can thus be used as a criterion for the identification of homologs in distant species.
We decided to use the genomic data from Tetraodon nigroviridis to look for the genes coding the class II HCR (HCRII) and their ligands. The main interest of T. nigroviridis is both its completely sequenced compact genome and the ease with which it can be maintained in the laboratory and used for experiments. We report here the complete description of the T. nigroviridis class II HCR repertoire and show that its diversification from common ancestral elements has occurred independently in fish and mammalian lineages. We have also characterized two ligands for these receptors.
Results
Identification of HCRII genes in Tetraodon nigroviridis
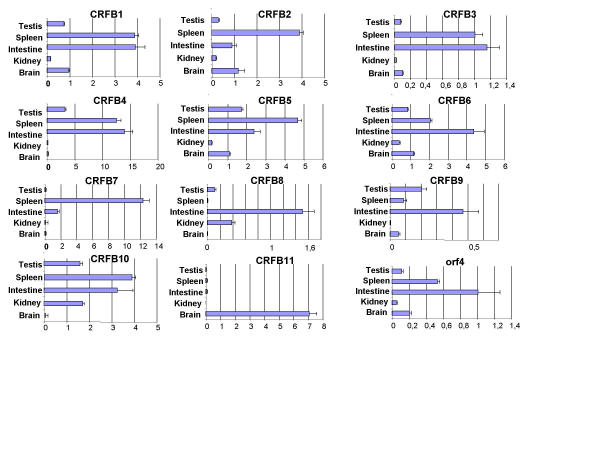
The starting point for the search was the alignment of the classII HCR D200 as reported in Uzé et al. (1995), extended to include the more recently described members IFNAR2 and CRF2-8 to CRF2-12 that allows the definition of a pattern of conserved positions. A conserved tryptophan in exon A1 (the first exon coding for SD100A), a conserved tryptophan and pair of cysteines in exon A2, a conserved serine and pair of cysteines in exon B2. All HCRII were tblastn against the 3 million genomic reads of T. nigroviridis (see methods). Reads with e<0.1 were kept and assembled. Each contig was tested for the presence of correct potential exons: introns of the correct phase and predicted proteins compatible with HCRII. False positives were exons coding for FNIII repeats that do not have the D200 intron/exon structure. All matching contigs were further extended in order to reach sizes of contigs compatible with whole gene size in the compact genome of T. nigroviridis: 5 contigs from 4 to 30 kb were obtained. Gene models were predicted in each contig and most probable exons were used to design oligonucleotides for 3' and 5' RACE. The full-length cDNAs were then aligned against the contigs to deduce gene structures and compared to Genscan predictions http://genes.mit.edu/GENSCAN.html. None of the 11 TnHCRII was correctly predicted by Genscan. The largest contig harbors 6 genes while the 5 others harbour a single HCRII gene. All genes are around 3 kb long. In agreement with the human nomenclature, they were named TnCRFB-1 to TnCRFB-11. All reading frames start with a leader peptide followed by a single D200. Except for TnCRFB-9 they all have a clear transmembrane (TM) domain after the D200. Expression patterns were determined for each of the 11 genes by Q-PCR using cDNAs reverse transcribed from RNAs of brain, spleen, cephalic kidney, gonads and intestine (figure 2). Long open reading frames and high expression in at least one tissue were considered sufficient criteria to state that these genes code for receptors and are not pseudogenes.
Figure 2.
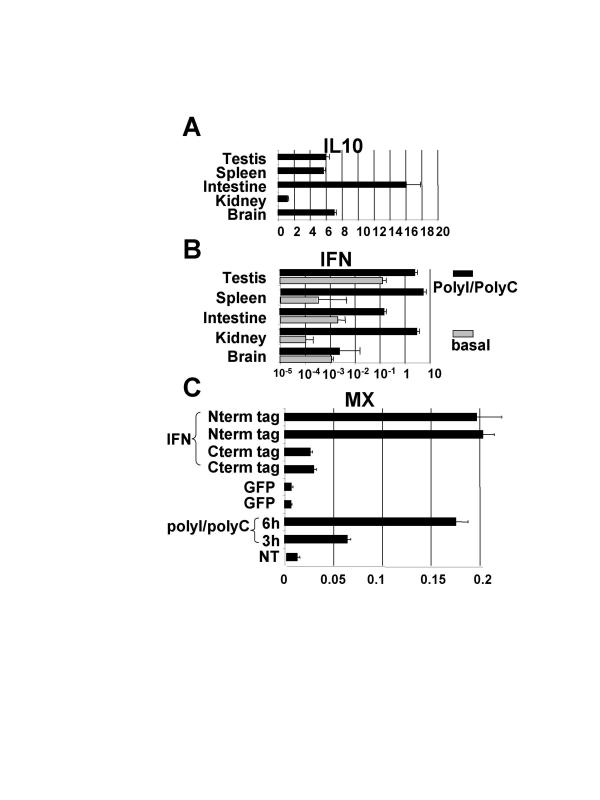
Expression pattern for the classII helical cytokine receptor genes. RNA samples were prepared from tissues, reverse transcribed and abundance of each cDNA was measured by QPCR using oligonucleotides listed in supplementary material. All data were normalized to the level of hnRNPA2 cDNA. 5% confidence in a student T test is shown. Orf4 stands for the T nigroviridis homologue of the human C21orf4 gene.
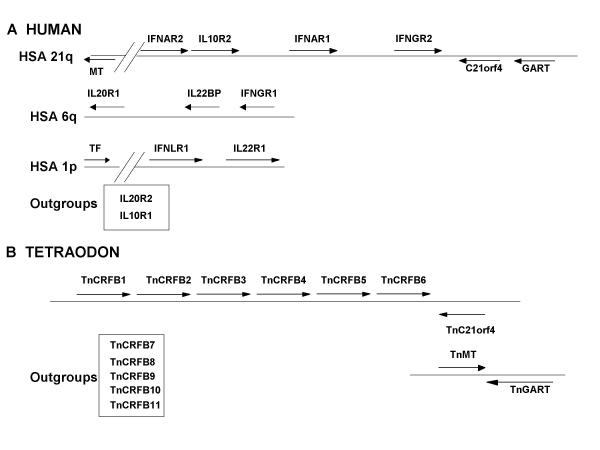
The T. nigroviridis repertoire of HCRII
Figure 3 shows the comparison of the HCRII gene repertoire in human and T. nigroviridis. As already described the HCRIIs are grouped in clusters on the human genome [18,19]. The largest cluster lies on human chromosome 21 (HSA21); it contains four genes (IFNAR2, IL10R2, IFNAR1 and IFNGR2) and is linked to the C21orf4 and GART genes. Two other clusters exist, one on HSA6q containing three genes (IFNGR1, IL22BP and IL20R1) and one on HSA1p containing two genes (IFNLR1 and IL22R2). The TF gene is also located on HSA1p but so distant that it cannot be considered as a member of the same cluster. The IL20R2 and IL10R1 genes are isolated and therefore called outgroups. The T. nigroviridis genome harbors a single HCRII gene cluster. As proved by the presence and similar orientation of the TnC21orf4 gene, this cluster is homologous to the HSA21 gene cluster. It contains six genes instead of the four genes present on the human homologous cluster. Interestingly, the TnGART gene, is not linked to this cluster, but is adjacent to the TnMT gene (T nigroviridis homolog of the yeast YDR140w gene). The same organization of the GART and MT genes, has already been described in the Fugu genome [18]. The human homolog for this MT gene (Acc nb of the cDNA: AF139682) is present on HSA21, 4.5 Mb centromeric to the cluster and transcribed toward the centromere [25]. The respective position of these genes indicates that an inversion has occurred since the divergence of the fish and mammalian ancestors. This inversion has involved a large chromosomal fragment covering genes from C21orf4 to the MT homolog of the yeast YDR140w gene. In one state, the C21orf4 is adjacent to the GART gene (amniotes), but in the other, the MT is next to GART.
Figure 3.
Comparative genomic mapping of the HCR genes in human and T. nigroviridis All genes are represented by an arrow that indicates the orientation of transcription. A) Clusters of HCR in the human genome. Orientation of transcription is relative to the centromere indicated on the left of the figure. MT stands for the human homolog of the S cerevisiae YDR140w gene. All genes are around 30 kb long. The MT gene is approximately 4 Mb centromeric to the IFNAR2 gene. B) The unique T. nigroviridis HCR cluster. TnC21orf4 is for the T. nigroviridis homolog of the human C21orf4 gene. TnMT is for the T. nigroviridis homolog of the S cerevisiae YDR140w gene. All genes are around 3 kb long.
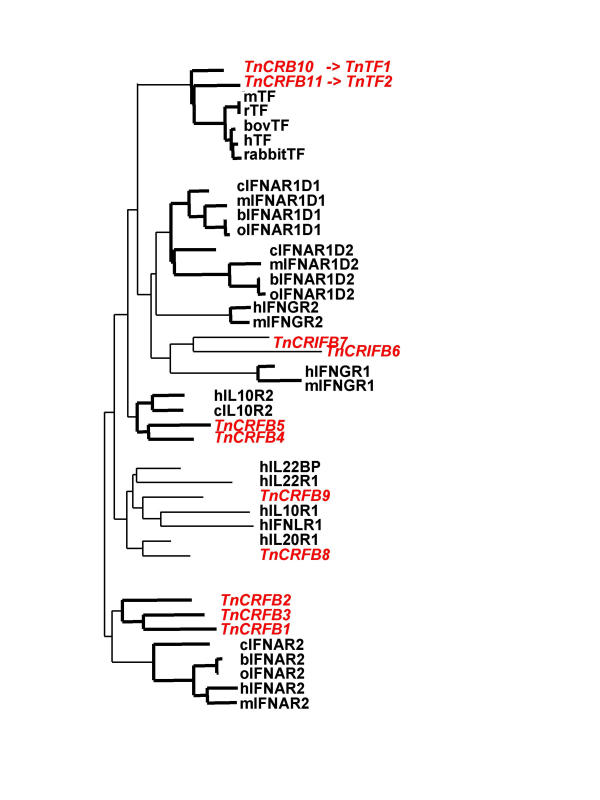
In order to determine if any of the TnHCRII would be the homolog of the functionally characterized human genes, the 13 human D200s (12 genes but IFNAR1 with two D200) together with some of their mammalian or avian orthologs were aligned with the 11 D200s of T. nigroviridis. The alignment was used to draw the phylogenetic tree that is depicted in figure 4. Tree branches with bootstrap values over 80% are indicated in bold. The clearest result is the grouping of TnCRFB10 & 11 with the TFs. TnCRFB10 and TnCRFB11 therefore appear as homologous to the mammalian TFs. This is confirmed by the intron/exon structure of their genes. TnCRFB10 &11, as the mammalian TF genes, are unique among the HCRII genes coding for transmembrane proteins in that the same exon encodes the TM domain and the very short intracellular domain [26]. Except for the genes coding for soluble proteins, all the other HCRII genes have an exon that encode the TM domain plus the first amino acids of the intracellular domain separated from the last exon coding the intracellular domain by a phase 0 intron [27]. Interestingly, TnCRFB10 &11 are not expressed in the same tissues: TnCRFB11 is specifically expressed in the brain (figure 2).
Figure 4.
Phylogenetic tree (NJ) derived from the alignment of the Tn HCR D200 domains together with the human D200s Domains from other species have been included to allow better grouping. T. nigroviridis D200s are written in red italic in order to highlight them. Branching points with bootstrap values over 80% are shown in bold. h, human; Tn, T. nigroviridis; m, mouse; r, rat; b, bovine; o, ovine and c, chicken. Alignment in Additional file: 2.
The phylogenetic tree derived from the alignment also reveals an interesting grouping of the TnCRFB4 and TnCRFB5 with the amniotes IL10R2. We can therefore postulate that the adjacent corresponding genes are derived from a recent tandem duplication and are homologs of the amniotes IL10R2. Furthermore the alignment derived grouping of TnCRFB1, 2 & 3 most probably reflects recent tandem duplications of their cognate genes but with no obvious amniote homologs. The other TnHCRII genes do not appear robustly linked to mammalian genes.
Ligands for TnHCRII
The presence of so many HCRII raises the question of their ligands and more specifically, that of the existence of an interferon system in fish [1]. Clearly, as shown in figure 4 (see also Additional file: 2, tetraodons, contrarily to amniotes have no IFNAR1 receptor with its typical double D200 that has certainly been instrumental to the diversification of the type I IFNs [27]. But the question remains open whether or not fish have interferon related molecules with similar functions.
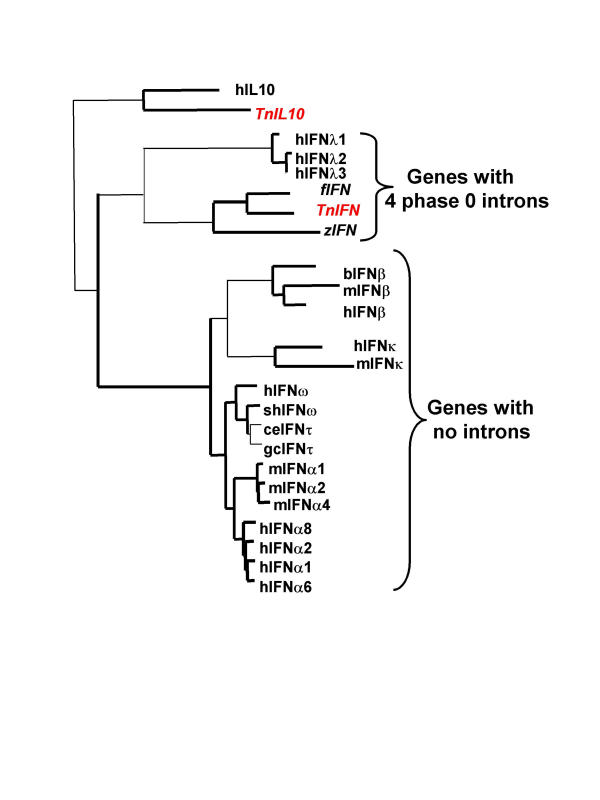
The T. nigroviridis reads were searched for exons capable of coding for molecules structurally related to the IFNs and IL10 related cytokines (tblastn). Contrary to their receptors, these cytokines are not encoded by genes with similar intron/exon structures. Genes for type I IFNs (IFNI) have no introns, those for IFNII have three introns.[28], those for IFN lambda have four introns [29,30]and those for IL10 related cytokines have four common introns [19]. Therefore the intron/exon structure could not be used as a criterion for the search of homologs in distant species. Potential exons were used for 3' and 5' RACE and the genes for four helical cytokines could be cloned: three genes coding IL10 related cytokines with the four conserved phase 0 introns and an interesting TnIFN gene also interrupted by four phase 0 introns. This gene codes for an interferon structurally related to IFNI and IFN lambda. For this reason, we call it TnIFN. The same full-length cDNA was cloned both from a wild animal and from an animal from breeders.
In order to establish that this fish IFN gene is not specific for tetraodons, we also cloned the orthologous gene from Danio rerio (zebrafish) using the trace repository reads to design oligonucleotides on potential exons for 3' and 5' RACE. The first four exons could easily be identified in silico using the T. nigroviridis sequence, but the last exon could be identified only by 3' RACE. The corresponding gene was called zIFN. Full-length cDNAs were cloned from two individuals from different breeders: alleles A & B. Both zIFN sequences differed at four silent positions, two non silent positions and differed at their COOH terminus; allele B codes for two extra amino acids. Despite the report from Aparicio et al. (2002) that they could not identify a Fugu IFN gene, reexamination of the Fugu genomic data allowed the identification of a Fugu IFN gene.
Of the three IL10 related cytokines, one is clearly the homolog of mammalian IL10, it is therefore called TnIL10. The two others are so divergent that it is difficult to identify them as clear orthologs of mammalian genes. However, according to the identity of the most similar genes, one is called TnIL20, the other is called TnIL24 (not shown). Interestingly TnIL10 and TnIL20 are in tandem (Acc Number AY294557)
An alignment of amino acid sequences of some IFNI and IFN lambda with the T. nigroviridis and D rerio IFNs was used to draw a phylogenetic tree of these IFNs (Figure 5, see also Additional file: 3). Branchings with bootstrap values over 80% are shown in bold. This tree shows a clear grouping of IFNI with fish IFNs and IFN lambda. This is illustrated using the outgroup genes hIL10 and TnIL10. Similar results are obtained whichever IL10 related cytokine or IFNII is used as an outgroup. The trees with more of these IL10 related cytokines are not shown because the bootstrap values are too low to state phylogenetic relationships between them. This tree illustrates very well the independent diversification of alpha IFNs in the different mammalian orders [7,8]. Expression patterns for TnIL10 mRNA (figure 6A) and for TnIFN mRNA in five tissues from animals treated or not by PolyI/PolyC intraperitoneal injection (figure 6B) have been determined. PolyI/polyC injection induces a very high induction of TnIFN from more than 10 times in testis to more than 104 times in kidney.
Figure 5.
Phylogenetic tree (NJ) derived from the alignment of the fish interferons with human IFN lambda and some typeI IFNs Number and phase of introns in the corresponding genes are indicated. Symbols same as in figure 4 plus: sh, sheep; ce, Cervus elaphus (red deer); f, Fugu; gc, Giraffa camelopardalis (giraffe) and z: Danio rerio (zebrafish). Alignment in Additional file: 3.
Figure 6.
Expression pattern of the TnIFN and TnIL10 genes and accumulation of the TnMX mRNA after IFN treatment. Results are amounts of mRNA relative to the hnRNPA2 mRNA. 5% confidence in a student T test is shown. A) TnIL10 in different tissues. B) TnIFN in different tissues in animals injected by PolyI/PolyC or PBS(basal). C) TnMX in primary kidney cells treated either with PolyI/PolyC, recombinant GFP (Green Fluorescent Protein) or recombinant TnIFN with either Nterm or Cterm 6His tag.
Activity of the newly discovered fish interferon
To test the biological activity of this interferon we decided to produce recombinant IFN in order to treat cells and to use quantitative RT-PCR to test for the induction of an interferon inducible gene. For this purpose, we looked for the MX genes both in T. nigroviridis and in zebrafish. MX genes code for mechanoenzymes of the Dynamin family [31] and are typical interferon induced genes. We have identified seven MX genes in zebrafish (zMXA to zMXG) and have found evidence of expression for all of them except zMXF. The zMXA gene corresponds to the already reported zebrafish MX gene [32]. In contrast T. nigroviridis has a single MX gene. The amount of TnMX mRNA was therefore used as a test for the biological activity of TnIFN. The TnIFN orf was cloned in either pIVEX2.3-MCS or pIVEX 2.4bNde (6HIS Cterm and Nterm fusions respectively) and the resulting plasmids were used to produce recombinant TnIFN. The recombinant protein was used to challenge primary cultures of cephalic kidney T. nigroviridis cells. After a 6 hours treatment, cells were harvested for total RNA preparations and quantitative RT PCR was used for measuring the amount of TnMX mRNAs. The T. nigroviridis mRNA for hnRNPA2, a house keeping splicing regulator, was used as a reference. PolyI/PolyC treatment was used as a control of interferon induction. Results shown in figure 6C show that the recombinant TnIFN molecule with a Nterm HIS tag can induce the expression of the TnMX mRNA to a level similar to PolyI/PolyC treatment.
We verified that, in contrast to the PolyI/PolyC treatment, recombinant TnIFN and GFP do not induce the TnIFN mRNA (not shown). Similar result were obtained with zebrafish cell lines ZF4/7 using zMXE as a reporter mRNA as it is the zMX gene the more induced by IFN (not shown). PKR is an other very well characterized IFN induced gene [33]. T. nigroviridis has two PKR genes (PKR1 and PKR2) which were used as reporters to confirm the results with TnMX: both are induced like the single TnMX gene (not shown).
While this manuscript was in preparation, Altmann et al (2003) [32] have reported the molecular and functional analysis of zIFN and shown its antiviral activity and Yap et al. (2003) [34] have shown that the promoter of the single Fugu MX gene can be induced by human interferon when transfected in human cells.
Discussion
Rapidly evolving lineage specific gene families are intrinsically difficult to analyze by large scale comparative genomic analysis [35]. A good example of this difficulty is the recent report of the near complete sequence of the Fugu genome [1]. This report clearly stated that IFNs and their related IL10 family cytokines and most of their receptors could not be identified in the Fugu genomes. We show here that reexamination of the data leads to opposite conclusions. Careful analysis of the HCR family in amniotes reveals features that can be used as criteria for a specific search of their homologs in distant species. Both conserved positions in the amino acid sequence of the protein and conserved phase and positions of introns are instrumental for this search.
Receptor diversification
Using the strategy described in figure 1, we have been able to describe 11 TnHCRII genes in T. nigroviridis. Primary amino acid sequences are so divergent that the phylogenetic tree is poorly reliable. Only a limited number of branches have good bootstrap values. For this reason, we cannot make estimates of divergence time for the different family members. It also explains why the tree differs from that of Kotenko et al [19] and why it is difficult to distinguish between paralogy and orthology for those receptors. The clearest homology is for TnCRFB10 &11 that are paralogous and represent the homologs of the mammalian TFs. TF is an interesting member of the HCRII family in that it does not bind a helical cytokine, but a coagulation factor (VIIa) whose 3D structure is similar to that of a helical cytokine. It clearly is not involved in host defense against pathogens and as such is not diverging as rapidly as the other HCRII family members from one species to the other. Interestingly, the two T. nigroviridis genes are not expressed in the same tissues, TnCRFB11 being specific for the brain. We see two reasons to postulate that these two genes have not been duplicated recently. The first is that the encoded proteins show only 35% amino acid identity and the second is that they do not lie in cluster on the genome of T. nigroviridis. This could lead us to postulate that other teleost fish species also have two TF genes. Curiously the Fugu proteome deduced from the near complete sequence of the Fugu genome contains only the ortholog of TnCRFB10 (Scaffold8956, protein 61906). A Oncorhynchus mykis TF gene has been cloned (Acc nb CAC82787) that is also the ortholog of TnCRFB10. For this reason we propose that TnCRFB10 would be called TnTF1 and TnCRFB11 be called TnTF2. The absence of TF2 in other species could simply reflect lack of detection of the paralogous TF2 gene. We therefore re-examined the Fugu genome for potential exons coding for a FuguTF2. Such exons could be found on Fugu-Scaffold 5445, but the Scaffold seems badly assembled as the exons are scrambled, therefore the correct gene model has escaped automatic detection. This proves that the Fugu genome has the TF2 gene. The O. mikis TF2 gene has probably escaped cloning by classical means just by chance as investigators were probably looking for just one gene.
Figure 1.
Strategy for the characterization of the T. nigroviridis HCR genes
The other possible homology is between TnCRFB4 & 5 and IL10R2. The IL10R2 receptor is in fact a "common chain" as it is a necessary component of the receptors for IL10, IL22 and IFN lambda and it is (apart from TF) the HCRII with the lowest sequence divergence in amniotes. Its gene lies in the center of the HSA21 HCRII gene cluster. Interestingly, TnCRFB4 &5 also lie in a similar central position on the homologous TnC21orf4 linked gene cluster. Finally, TnCRFB1, 2 & 3 are closely related to each other. Careful inspection of the data indicates that the three genes are also present in the Fugu genome, but because of assembly problems they appear on different contigs (Fugu Scaffolds 3897 & 6320). They seem to code for receptors distantly related to amniote's IFNAR2. This is in accordance with their mapping at the extremity of the C21orf4 linked cluster and could suggest a common ancestry. The successive tandem duplications that lead to the three fish genes could be fish specific. The vicinity of the unassigned outgroups TnHCRII did not reveal genes whose homologs would be linked to human HCRII genes, it is therefore not possible to state any clear homology to mammalian genes. Careful inspection of the Fugu genome reveals that the 11 TnHCRIIs have homologs in Fugu.
Ligand diversification
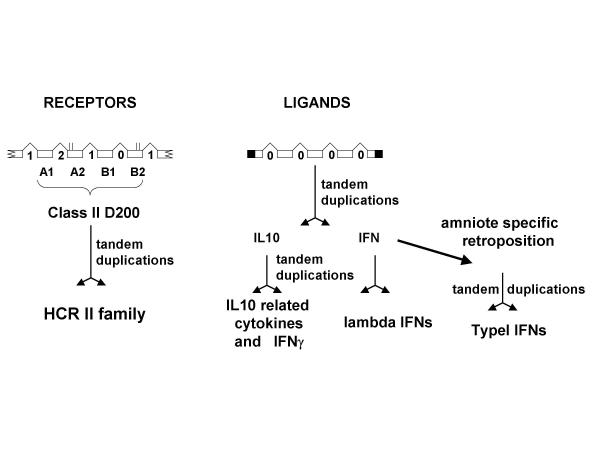
The search for ligands has revealed only four classII helical cytokines (HCII): IFN and three IL10 related cytokines (IL10, IL20 and IL24). The present work allows the definition of two categories of classII cytokines. One category is made up in mammals of lambda IFN and typeI IFN, the other would be made up of IL10 related cytokines and gamma IFN (see below). In fish, the first category would be made up of only an ancestral interferon gene with four phase 0 introns that is homologous to the human IFN lambda genes. The three human IFN lambda genes are located on human chromosome 19 and their four phase 0 are perfectly conserved both amongst them [30] and with TnIFN and zIFN. The key element during the evolution of the IFN genes has therefore been a retroposition event that occurred during evolution after the separation of sarcopterygians from actinopterygians and that created an intronless type I IFN gene. In the mammalian lineage, this gene then underwent successive duplications to generate first the alpha and the beta interferons and then during the mammalian radiation, the numerous alpha IFN genes [7,8,36]. This key retroposition event has probably been associated with duplication of receptor genes that generated the IFNAR1 and IFNAR2 genes. The IFNAR1 gene that was present in amniote ancestors already had two D200 allowing the building of receptor complexes with different binding sites that could accommodate the diversification of the ligands [18,27,37,38]. The "IL10 related cytokines" category is represented in Tetraodon by three genes with four phase 0 introns (IL10, 20 &24). If some have extra-introns, all genes coding for IL10 related cytokines have the same four phase 0 introns. The high expression of the TnIL10 gene in the intestine (figure 2) suggests that the encoded fish cytokine could play a role similar to that of mammalian IL10 whose function is to keep under strict control the balance between immune and inflammatory response especially in the bowels [39,40].
Interestingly these two categories of HCII, despite having no similarity at the amino acid level, are both encoded by genes with exactly the same intron/exon structure. The four phase 0 introns fall at similar positions. This suggests that both genes derive from a common ancestor harboring the same four phase 0 introns. As in figure 7, we therefore postulate that the ancestor for classII helical cytokine was encoded by a gene with this intron/exon structure. The first duplications would have taken place before the osteichtian radiation and would have generated the ancestral IFN and an ancestor for the IL10 related genes. The different lineages having then expanded this gene family by different means of retrotransposition and duplication. In this context, the gene for amniote type II IFN (gamma IFN) poses an interesting problem. It has only three phase 0 introns that correspond to the first three introns of both the ancestral IFN and IL10 related genes. Is it derived from an IFN gene or from an IL10 related gene? The location of the human IFN gamma gene on a cluster of classII cytokine genes including two other genes for the IL10 related cytokines (IL22 and IL26) [19] suggests that it is in fact derived from an IL10 related gene. Despite the functional similarities of type I and type II IFNs, receptor binding characteristics of the dimeric forms of IL10 and IFN gamma also favor a closer relationship between these ligands [41,42].
Figure 7.
Schematic drawing for the diversification of the helical cytokines and their receptors during the evolution of the osteichthians. Open boxes are for coding exons, black parts for 3' and 5' non coding regions. Broken lines are for introns; their phase is indicated. For the receptors, broken boxes indicate that all D200 part of larger proteins. Exons are numbered A1 (for the first exon coding the SD100A) to B2 (second exon coding the SD100B). Conserved cysteines are indicated as vertical bars over the exon boxes. The retroposition event leading to typeI IFNs has only been observed in amniotes and is therefore labeled "amniote specific". Data from other sarcopterygians could lead to a revision.
Conclusions
To the origins of a diversified ligand/receptor system
This work provides an interesting perspective on the evolution and diversification of the classII helical cytokines and their receptors during the radiation of the osteichtians. It shows that the fine tuning of the main mechanisms for host defense against infections has been performed independently in the different vertebrate lineages. This is both true for the non specific antiviral defenses mediated by interferons and for the regulation of the immune response invented by ancestors of the gnathostomes in which IL10 related cytokines play a major role.
The question remains open of what happened in other vertebrate lineages, but the most fascinating question is the origin of this ligand/receptor system. Both genetic (intron/exon structures) and structural data (conserved amino acid positions and/or common 3D structures) argue in favor of a common ancestry for all classII ligand/receptor systems. The fascinating quest is now to find organisms that would have retained this single ancestral ligand/receptor pair and the central question will be: what is its function? In this perspective, the dome gene of drosophila is intriguing. Primary amino acid sequence clearly indicates that it harbors a D200 domain but the intron/exon structure is not that of the vertebrate HCR genes. Thedome gene has one D200 domain plus FNIII repeats in its extracellular domain, but none of the canonical introns that border such domains in vertebrate genes is conserved [17]. The dome gene has lost the intron/exon «memory». Interestingly, it is the only invertebrate gene with a D200 described so far; the expansion of the HCR family has occurred only in vertebrates. The dome gene could therefore testify for the presence of an HCR ancestor in invertebrates. The first step in the diversification of this gene family in deuterostomes or chordates has therefore been the duplication to generate class I and class II ancestors. We have started the quest for these classI and classII ancestors by searching homologs of these genes in animal groups branching close to the vertebrate ancestors. In the genome of Ciona Intestinalis, we have found just two HCR genes, one coding for a classI and the other for a classII receptor. We have started experiments in order to determine in which biological functions they are involved.
Methods
Fish samples and sequences
T nigroviridis imported from Thailand (wild animals) or from Indonesia (breeders) were purchased at local dealers. Average animal weight was 3 grams. Phenoxy 2 ethanol was used as an anesthetic prior injections or dissections. PolyI/PolyC treatment was an IP injection of 0.1 ml of a 2.2 mg/ml solution in PBS. RNAs were prepared using the High Pure RNA Tissue Kit from Roche. Primary cultures of Cephalic Kidney cells were prepared by scratching the organ in a 200 micron mesh nylon in DMEM/F12 medium supplemented with 10% fetal calf serum. The primary cells were either used as such or separated in heavy and light populations by centrifugation on a Ficoll cushion. Primary cultures were kept up to 4 days with 5% CO2 at 30°C.
D. rerio from breeders were purchased at local dealers. The ZF4/7 (ATCC: CRL-2050) cell line was maintained in the same conditions as the T. nigroviridis primary cells.
Genomic sequences for the T. nigroviridis genes are assemblies of shotgun reads produced by the Genoscope http://www.genoscope.cns.fr and the Whitehead Institute Center for Genome Research http://www.genome.wi.mit.edu. The shotgun reads are available through the Trace Repository at http://trace.ensembl.org/. They represent a 8.3X genome coverage Assemblies of small sets of reads (up to a few hundred) were done using cap http://www.infobiogen.fr. Sequence of the resulting contigs were finished by designing oligonucleotides and resequencing regions of problems.
Cloning and Quantitative-PCR
Oligonucleotides are listed in Additional file: 1. 3' and 5'RACE were performed using the GeneRacer Kit from Invitrogen. Amplified products were tested for the presence of specific products using internal oligonucleotides, cloned using the Topo TA cloning kit from Invitrogen. Bacterial colonies were screened using the internal oligonucleotide and the plasmids were entirely sequenced. Oligonucleotides TnIFN.52 and TnIFN.32 were used to amplify the complete ORF of TnIFN. The resulting fragment was digested by NdeI and SacI and cloned in the pIVEX2.3-MCS vector (Roche) digested by the same enzymes for the production of recombinant TnIFN with a Cterm 6His tag. For the Nterm 6His tagging, amplification was with TnIFN52 and TnIFN33; cloning was in pIVEX2.4bNde. In vitro production of recombinant IFN was done using the RTS100 E. coli HY kit from Roche using 300 ng of CsCl purified plasmid per 10 μl of reaction (3 h incubation at 30°C). Production was checked using SDS-PAGE.
Conventional PCR were performed using Platinium Taq DNA polymerase in a MJ Research PTC200 thermocycler. Real time Quantitative PCR (Q-PCR) were performed using SYBR GREEN technology in a LightCycler Instrument from Roche. RNA samples were reverse transcribed using Spl2XhoT18 as primer and M-MuLV Reverse Transriptase as an enzyme [18]. First strand cDNAs were purified using Quiaquick purification columns (Quiagen).
Phylogenetic analysis
Alignments were performed using Clustal and phylogenetic trees were calculated using the Phylo_win package (distance, PAM) [43]. Drawing of trees was done using TREEVIEW.
Authors' contributions
GL and DM did the search, assembly, predictions, cloning and sequencing of the cDNAs coding for the receptors and their ligands. They also finished the sequencing of the genes. GL did the biological work with Tetraodons and zebrafish and drafted the manuscript. KM did the analysis of the protein structures. HRC, NST and OJ did the shotgun sequencing of the Tetraodon genome and performed searches and assemblies. All authors read and approved the final manuscript.
Supplementary Material
Oligonucleotides. All sequences are 5' to 3'
Acknowledgments
Acknowledgements
We are indebted to Gilles Uzé for his constant support and exciting discussions. We thank Dr C. Bonnerot and B. Philippi for their help and critical reading of the manuscript. We would like to thank the Genoscope and the Whitehead Institute MIT Center for Genome Research for their joined efforts in the Tetraodon nigroviridis sequencing program and for providing unpublished sequence data. We more specifically want to thank Jean Weissenbach and Eric S. Lander for their constant support. Many thanks to Christine Dambly-Chaudière and Nicolas Cubedo for their help at the fish facilities. We want to thank Mark Ekker for providing the zebrafish cell line. This work was supported by CNRS.
Contributor Information
Georges Lutfalla, Email: lutfalla@infobiogen.fr.
Hugues Roest Crollius, Email: hrc@genoscope.cns.fr.
Nicole Stange-thomann, Email: sthomann@genome.wi.mit.edu.
Olivier Jaillon, Email: ojaillon@genoscope.cns.fr.
Knud Mogensen, Email: knud@athos.igm.cnrs-mop.fr.
Danièle Monneron, Email: monneron@igm.cnrs-mop.fr.
References
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Murphy PM. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., A Makarova, K.S., A Spouge, J.L., A Wolf, Y.I., A Koonin, E.V. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Research. 2001;11:555–565. doi: 10.1101/gr.GR-1660R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of the type I interferon gene family in mammals. J Mol Evol. 1995;41:539–548. doi: 10.1007/BF00175811. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Liu L, Guo Q, Leaman D, Bixby J. The evolution of the type I interferons. J Interferon Cytokine Res. 1998;18:805–816. doi: 10.1089/jir.1998.18.805. [DOI] [PubMed] [Google Scholar]
- Thoreau E, Petridou B, Kelly PA, Djiane J, Mornon J-P. Structural symmetry of the extracellular domain of the cytokine/growth hormone/prolactin receptor family and interferon receptors revealed by hydrophobic cluster analysis. FEBS Lett. 1991;282:26–31. doi: 10.1016/0014-5793(91)80437-8. [DOI] [PubMed] [Google Scholar]
- Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier L. The immune system of invertebrates and vertebrates. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:1–15. doi: 10.1016/S1096-4959(01)00306-2. [DOI] [PubMed] [Google Scholar]
- Du Pasquier L. Several MHC-linked Ig superfamily genes have features of ancestral antigen-specific receptor genes. Curr Top Microbiol Immunol. 2002;266:57–71. doi: 10.1007/978-3-662-04700-2_5. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Nicola NA. An introduction to the cytokines. In: NA Nicola, editor. Guidebook to cytokines and their receptors. Oxford, Oxford University Press; 1994. pp. 1–7. [Google Scholar]
- Bazan JF. Haemopoietic receptors and helical cytokines. Immunol Today. 1990;11:350–354. doi: 10.1016/0167-5699(90)90139-Z. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medioni C, Cerezo D, Noselli S. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/S0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Reboul J, Gardiner K, Monneron D, Uze G, Lutfalla G. Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res. 1999;9:242–250. [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV. The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine Growth Factor Rev. 2002;13:223–240. doi: 10.1016/S1359-6101(02)00012-6. [DOI] [PubMed] [Google Scholar]
- Wang T, Secombes CJ. Cloning and expression of a putative common cytokine receptor gamma chain (gammaC) gene in rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2001;11:233–244. doi: 10.1006/fsim.2000.0310. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner J, Duval H, Chesnel F, Boeuf G, Perez-Sanchez J, Boujard D. Fish growth hormone receptor: molecular characterization of two membrane-anchored forms. Endocrinology. 2001;142:3269–3273. doi: 10.1210/en.142.7.3269. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Gardiner K, Proudhon D, Vielh E, Uzé G. The structure of the human interferon alpha/beta receptor gene. J Biol Chem. 1992;267:2802–2809. [PubMed] [Google Scholar]
- Nakagawa Y, Kosugi H, Miyajima A, Arai K, Yokota T. Structure of the gene encoding the alpha subunit of the human granulocyte-macrophage colony stimulating factor receptor. J Biol Chem. 1994;269:10905–10912. [PubMed] [Google Scholar]
- Uzé G, Lutfalla G, Mogensen KE. alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, Groner Y, Soeda E, Ohki M, Takagi T, Sakaki Y, Taudien S, Blechschmidt K, Polley A, Menzel U, Delabar J, Kumpf K, Lehmann R, Patterson D, Reichwald K, Rump A, Schillhabel M, Schudy A, Zimmermann W, Rosenthal A, Kudoh J, Schibuya K, Kawasaki K, Asakawa S, Shintani A, Sasaki T, Nagamine K, Mitsuyama S, Antonarakis SE, Minoshima S, Shimizu N, Nordsiek G, Hornischer K, Brant P, Scharfe M, Schon O, Desario A, Reichelt J, Kauer G, Blocker H, Ramser J, Beck A, Klages S, Hennig S, Riesselmann L, Dagand E, Haaf T, Wehrmeyer S, Borzym K, Gardiner K, Nizetic D, Francis F, Lehrach H, Reinhardt R, Yaspo ML. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Mackman N, Morrissey JH, Fowler B, Edington TS. Complete sequence of the human tissue factor gene, a highly regulated cellular receptor that initiates the coagulation protease cascade. Biochemistry. 1989;28:1755–1762. doi: 10.1021/bi00430a050. [DOI] [PubMed] [Google Scholar]
- Mogensen KE, Lewerenz M, Reboul J, Lutfalla G, Uze G. The type I interferon receptor: structure, function, and evolution of a family business. J Interferon Cytokine Res. 1999;19:1069–1098. doi: 10.1089/107999099313019. [DOI] [PubMed] [Google Scholar]
- Taya Y, Devos R, Tavernier J, Cheroutre H, Engler G, Fiers W. Cloning and structure of the human immune interferon-gamma chromosomal gene. Embo J. 1982;1:953–958. doi: 10.1002/j.1460-2075.1982.tb01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr Opin Cell Biol. 2001;13:454–460. doi: 10.1016/S0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol. 2003;77:1992–2002. doi: 10.1128/JVI.77.3.1992-2002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- Yap WH, Tay A, Brenner S, Venkatesh B. Molecular cloning of the pufferfish (Takifugu rubripes) Mx gene and functional characterization of its promoter. Immunogenetics. 2003;54:705–713. doi: 10.1007/s00251-002-0525-x. [DOI] [PubMed] [Google Scholar]
- Fahrer AM, Bazan JF, Papathanasiou P, Nelms KA, Goodnow CC. A genomic view of immunology. Nature. 2001;409:836–838. doi: 10.1038/35057020. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Roberts RM. Independent origin of IFN-alpha and IFN-beta in birds and mammals. J Interferon Cytokine Res. 2000;20:737–739. doi: 10.1089/10799900050116444. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C, Uzé G, Lutfalla G, Mogensen KE. Hydrophobic cluster analysis reveals duplication in the external structure of human alpha interferon receptor and homology with gamma interferon receptor external domain. FEBS Lett. 1990;269:1–3. doi: 10.1016/0014-5793(90)81103-U. [DOI] [PubMed] [Google Scholar]
- Lewerenz M, Mogensen KE, Uzé G. Shared receptor components but distinct complexes for alpha and beta interferons. J Mol Biol. 1998;282:585–599. doi: 10.1006/jmbi.1998.2026. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, Sordat B, Gibbs VC, Aguet M. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, Zauodny PJ, Narula SK. Crystal structure of a complex between interferon gamma and its soluble high-affinity receptor. Nature. 1995;376:230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- Walter MR, Nagabhushan TL. Crystal structure of interleukin 10 reveals an interferon gamma-like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring phylogenies from DNA sequences of unequal base compositions. Proc Natl Acad Sci U S A. 1995;92:11317–11321. doi: 10.1073/pnas.92.24.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides. All sequences are 5' to 3'