Abstract
To evaluate the expression of different forms of a tumor-specific antibody in plants, we adapted a recently described Agrobacterium-mediated transient expression system. A recombinant single-chain Fv antibody (scFvT84.66) and a full-size mouse/human chimeric antibody (cT84.66) derived from the parental murine mAb T84.66 specific for the human carcinoembryonic antigen were engineered into a plant expression vector. Chimeric T84.66 heavy and light chain genes were constructed by exchanging the mouse light and heavy chain constant domain sequences with their human counterparts and cloned into two independent plant expression vectors. In vivo assembly of full-size cT84.66 was achieved by simultaneous expression of the light and heavy chains after vacuum infiltration of tobacco leaves with two populations of recombinant Agrobacterium. Upscaling the transient system permitted purification of functional recombinant antibodies from tobacco leaf extracts within a week. His6-tagged scFvT84.66 was purified by immobilized metal affinity chromatography and cT84.66 by protein A affinity chromatography. Sufficient amounts of recombinant antibodies were recovered for detailed characterization by SDS/PAGE, Western blotting, and ELISA.
Keywords: in vivo antibody assembly, carcinoembryonic antigen
Monoclonal antibodies are essential tools in biology, biochemistry, and medicine. Their high affinity and specificity make them invaluable for diagnostic and therapeutic applications. However, the therapeutic use of murine mAbs is limited because they elicit a human anti-mouse antibody response and large amounts of antibody are required for therapy. These limitations may be overcome by engineering humanized antibodies and by producing these proteins in plants.
The human anti-mouse antibody response can be reduced by using recombinant antibody (rAb) technology to replace the murine light and heavy chain constant domains with the corresponding human domains and the remaining murine variable domains to maintain the antigen specificity and affinity of the original mAb. A second approach is to use single-chain Fv antibody fragments (scFvs) where the constant domains have been removed and the variable domains are joined by a flexible linker (1). Compared with the full-size antibodies, scFvs display better tumor penetration and faster serum clearance but exhibit no effector functions. A critical step in testing the potential therapeutic use of these molecules is the development of a reproducible and efficient method for large-scale antibody production.
Plants are potentially the most economical system for large-scale production of rAbs (2, 3). rAbs are efficiently folded and assembled within the endoplasmic reticulum (ER) of plant cells (4–6) and retain the antigen binding properties of the antibodies produced by plasma or hybridoma cells (2, 5, 7–9). Since the first report of antibody expression in transgenic plants (7), different engineered antibodies have been produced successfully, including full-size antibodies (8–11), Fab fragments (12), scFvs (13–21), and single-domain antibodies (22).
Regenerating transgenic plants from transformed cells is both labor intensive and time consuming. In contrast, transient expression systems allow the rapid evaluation and improvement of plant-expressed antibodies. They present a feasible method for testing antibody expression in vivo before progressing to develop stably transformed plants. In this work, we studied the transient expression of two rAbs specific for the human carcinoembryonic antigen (CEA).
CEA is a cell surface glycoprotein (23) that is widely used as a tumor marker (24). It belongs to the Ig superfamily and consists of seven Ig-like domains (25). Because CEA can be detected in almost all human colon cancers, 50% of all breast cancers, and in other tumors of epithelial origin, anti-CEA antibodies have been used for antibody-mediated cancer therapies and in vivo tumor imaging. Among those, the mAb T84.66, which binds to the A3 domain of CEA with high specificity and affinity (KD = 8 pM) (26), has been used successfully for in vivo imaging and diagnosis of human colorectal carcinoma (27). A recombinant mouse/human chimeric antibody (cT84.66), a minibody (scFv-CH3), and a scFv fragment (scFvT84.66) recently have been engineered, and the expressed proteins were characterized and evaluated for diagnostic and therapeutic applications (28–32). Despite these recent developments, it is evident that treatment of tumor patients will require bulk quantities of the most effective molecules such as scFvs and chimeric rAbs.
Therefore, we evaluated the transient expression and in vivo assembly of a full-size CEA-specific mouse/human chimeric antibody, cT84.66, and a single-chain antibody, scFvT84.66, derived from the parental murine monoclonal mT84.66 in tobacco leaves. Both rAbs were transiently expressed in tobacco leaves by using an Agrobacterium-mediated transient expression system (33). In vivo assembly of cT84.66 was achieved by simultaneous expression of the chimeric heavy and light chain genes. Each gene was encoded by a separate expression plasmid, and each plasmid was carried by a separate population of Agrobacterium. Expressed rAbs were affinity-purified from tobacco leaf extracts by immobilized metal ion affinity chromatography (IMAC) of His6-tagged scFvT84.66 and protein A-based chromatography of cT84.66 and were used for further characterization.
MATERIALS AND METHODS
Construction of cT84.66 Heavy and Light Chain cDNAs in pUC18.
Splice overlap extension PCR was used to obtain full-size mouse/human chimeric T84.66 light and heavy chain cDNAs, by in-frame fusion of the variable VL and VH domains of the mouse mAb T84.66 to the human kappa and IgG1 constant domains of the B72.3 mouse/human chimeric antibody cDNAs (34). The human constant domains were amplified from plasmids chiB72.3L and chiB72.3H by using the following primers: 5′-CTG GAA ATA AAA ACT GTG GCT GCA CCA TCT-3′ (chiB72.3L-I), 5′-GCC AAG CTT TTT GCA AAG ATT CAC-3′ (chiB72.3L-II), 5′-ACC GTC TCC TCA GCC TCC ACC AAG GGC CCA-3′ (chiB72.3H-I), and 5′-GCC AAG CTT GGA TCC TTG CAG GGG CCC AGG-3′ (chiB72.3H). The mouse variable domains were amplified from plasmids T84.66L2 (light chain) and T84.66H2 (heavy chain) by using the primers: 5′-GGC GAA TTC ATG GAG ACA GAC ACA CTC-3′ (T84.66L-I), 5′-AGC CAC AGT TTT TAT TTC CAG CTT GGT CCC-3′ (T84.66L), 5′-GGC GAA TTC ATG AAA TGC AGC TGG GTT-3′ (T84.66H), and 5′-GGT GGA GGC TGA GGA GAC GGT GAC TGA GGT-3′ (T84.66H). Chimeric T84.66 light and heavy chain cDNAs obtained by splice overlap extension PCR were cloned as EcoRI/HindIII fragments into pUC18, to give the constructs pUC18-Light and pUC18-Heavy, respectively. All cDNA sequences were confirmed by nucleotide sequencing (ALF, Amersham Pharmacia).
Construction of scFvT84.66 and Full-Size cT84.66 Plant Expression Plasmids.
cDNA fragments encoding the scFv fragment and the chimeric heavy and light chains derived from the anti-CEA antibody T84.66 were amplified by PCR from constructs pUC18-T84.66/212 (32), pUC18-Light, and pUC18-Heavy with 5′-specific primers introducing a NcoI site, and 3′-specific primers introducing a SalI site, for subcloning. The sequences of the primers were: 5′-GCG TCC ATG GAC ATT GAG CTG ACC CAA TC-3′ (scFvT84.66-I), 5′-AGA CGT CGA CTG AGG AGA CGG TGA ACT GA-3′ (scFvT84.66-II), 5′-CAT GCC ATG GAG ACA GAC ACA CTC CTG CTA-3′ (Light-I), 5′-CCG CTC GAG TTT AAC ACT CTC CCC TGT TGA A-3′ (Light-II), 5′-CAT GCC ATG GGA AAA TGC AGC TGG GTT ATC TTC-3′ (Heavy-I), and 5′-ACG CGT CGA CTT TAC CCG GAG ACA GGG AGA G-3′ (Heavy-II).
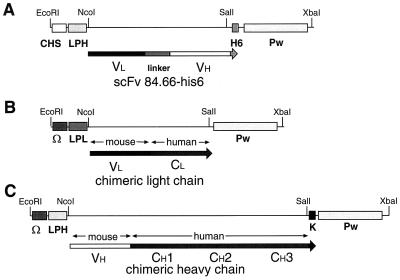
pGEM-3zf was used for cloning the 5′ untranslated region (UTR), either from chalcone synthase (CHS) (9) or the omega leader (Ω) region of tobacco mosaic virus (TMV) (35), followed by one of the two plant codon-optimized leader peptides derived either from the heavy chain (LPH) or from the light chain (LPL) of the murine mAb24 (9), and for cloning the His6 tag or KDEL ER retrieval signal sequence (4) and the 3′ UTR from TMV. PCR-amplified cDNA fragments were digested with NcoI/SalI and subcloned into pGEM3zf vector as shown in Fig. 1. scFvT84.66 was placed downstream from the 5′ UTR of CHS and the LPH, and upstream from a sequence encoding a His6 tag (C-LPH-scFvT84.66-H6-Pw, Fig. 1A); chimeric light chain was inserted downstream from the 5′ Ω region of TMV and the LPL (Ω-LPL-cLightT84.66-Pw, Fig. 1B); chimeric heavy chain was inserted downstream from the 5′ Ω region of TMV and the LPH, and upstream from the KDEL sequence (Ω-LPH-cHeavyT84.66-K-Pw, Fig. 1C). The expression cassettes were cloned between the enhanced 35S promoter and the cauliflower mosaic virus termination region using the EcoRI and XbaI restriction sites of the pSS plant expression vector (9).
Figure 1.
cDNA constructs for transient antibody expression in tobacco leaves. (A) CHS-LPH-scFvT84.66-H6-Pw, (B) Ω-LPL-cLightT84.66-Pw, (C) Ω-LPH-cHeavyT84.66-K-Pw expression cassettes. CHS, 5′ UTR of CHS; Ω, Omega leader region of TMV RNA; LPL, plant codon optimized leader peptide derived from murine light chain of TMV-specific mAb24; LPH, plant codon optimized leader peptide derived from murine heavy chain of mAb24; K, KDEL motif; H6, His6 tag; Pw, 3′ UTR of TMV RNA.
Cultivation of Plants.
Nicotiana tabacum cultivar Petit Havana SR1 was cultivated in the greenhouse in DE73 standard soil. Developing leaves of a uniform size (approximately 12 cm long) were harvested and used for vacuum infiltration.
Agrobacterium-Mediated Transient Expression System.
Agrobacterium tumefaciens strain GV3101 (pMP90RK, GmR KmR RifR) was transformed with each of the plant expression vectors by N2 transformation (36). Growth of recombinant Agrobacterium and vacuum infiltration of tobacco leaves was performed as described (33). After infiltration, leaves were incubated adaxial side down, on wet Whatman paper no. 1 in sealed trays (23°C/16-h photoperiod). After 60 h, leaves were frozen in liquid nitrogen and stored at −80°C until analyzed. For simultaneous expression of chimeric T84.66 light and heavy chains, leaves were infiltrated with equal amounts of two recombinant Agrobacterium cultures independently carrying either the pSS/Ω-LPL-cLightT84.66 (Fig. 1B) or the pSS/Ω-LPH-cHeavyT84.66-K (Fig. 1C) expression vector.
Preparation of Protein Extracts from Infiltrated Leaves.
For the purification of transiently expressed recombinant proteins, approximately 100 g of tobacco leaves was used. Infiltrated leaves were ground in liquid nitrogen to a fine powder with a mortar and pestle. Soluble protein was extracted by using 2 ml of extraction buffer (EB: 200 mM Tris⋅HCl/5 mM EDTA/0.1 mM DTT/0.1% Tween 20, pH 7.5) per gram of leaf material. Cell debris was removed by two rounds of centrifugation (20,000 × g, 30 min, 4°C), and the supernatant was used for expression analyses and protein purification by affinity chromatography. For isolation of scFvT84.66, EDTA and DTT were omitted from EB.
Analyses of scFvT84.66 Expression by ELISA.
Functional scFvT84.66 was detected by competition ELISA (28), using the murine mAb T84.66 (mT84.66) as competitor and recombinant CEA-derived NA3 protein (CEA/NA3) (37) as antigen. Ninety six-well microtiter plates (M129B, Greiner, Nurtingen, Germany) were coated with 50 ng/well of recombinant CEA/NA3 purified from Pichia pastoris (37) and blocked with 1% BSA in saline buffer (0.85% NaCl, pH 7.2). Using siliconized microtiter plates, leaf protein extracts were serially diluted in extracts from noninfiltrated leaves. All samples were supplemented with mT84.66 to a final concentration of 25 ng/ml and transferred to CEA/NA3-coated ELISA plates. Bound mT84.66 was detected with alkaline phosphatase (AP)-conjugated Fc-specific goat anti-mouse IgG (GAM-Fc) followed by incubation in substrate buffer (1 mg⋅ml−1 p-nitrophenyl phosphate in 0.1 M diethanolamine, 1 mM MgCl2, pH 9.8). Substrate reaction was carried out for 1 h at 37°C, and absorption at 405 nm was measured in a Spectra Max 340 spectrophotometer (Molecular Devices). The data were fitted to the equation R = ODbg + (ODmax − ODbg)/(1 + ODsample/I50) by using Microcal Origin 5.0 (where ODbg is the background, ODmax the reactivity of mT84.66 without competitor, and ODsample the reactivity of mT84.66 in the samples; R is the reactivity of the sample and I50 represents the dilution at which the reactivity of mAb T84.66 is reduced to 50%). The I50 was used to determine the scFvT84.66 concentration, neglecting the bivalency of mT84.66 and assuming that at this dilution both scFvT84.66 and mT84.66 are present in equimolar concentrations.
Analyses of Chimeric T84.66 Expression by ELISA.
Mouse/human chimeric T84.66 antibody was detected by ELISA with an AP-conjugated Fc-specific goat anti-human IgG (GAH-Fc) by using recombinant CEA/NA3 for coating. Serial dilutions of leaf extracts were made in siliconized microtiter plates by using DB buffer (2% polyvinylpyrrolidone K25/0.2% BSA/0.05% Tween 20 in PBS, pH 7.4) and transferred to CEA/NA3-coated and BSA-blocked ELISA plates as described above. AP-conjugated GAH-Fc was used to detect bound cT84.66 as described above.
Purification of scFvT84.66.
Total soluble protein was extracted from infiltrated tobacco leaves expressing the His6-tagged scFvT84.66. After homogenization and centrifugation, leaf extracts were filtered through Whatman 3M paper. A 0.5 × 20-cm column (Bio Cart, Kronlab, Sinsheim, Germany) was packed with 2 ml of ProSep Chelating (BioProcessing, Consett, U.K.), charged with 5 column volumes (CV) of 50 mM NiSO4 and equilibrated with 10 CV of binding buffer (PBS, pH 7.4/1 M NaCl). Filtered leaf extract was applied to the column at a flow rate of 2 ml/min. After sample application, the column was washed with 5 CV of binding buffer. Nonspecifically bound proteins were removed with binding buffer containing 25 mM imidazole. His6-tagged scFvT84.66 was eluted by using 2 CV of binding buffer containing 250 mM imidazole.
Purification of Chimeric T84.66.
Tobacco leaves were infiltrated with a mix of two Agrobacterium cultures independently carrying the chimeric T84.66 heavy and light chain constructs. Leaves were frozen in liquid nitrogen, ground, and homogenized in extraction buffer to extract the total soluble proteins. The extract was filtered through Whatman 3M paper and the pH was adjusted to 8.3, before application on an equilibrated Prosep A-column (BioProcessing). Then, the matrix was extensively washed with 10 bed volumes of washing buffer (PBS, pH 7.4/100 mM NaCl). Bound antibodies were eluted with citrate buffer, pH 3.0. Eluted fractions were neutralized immediately by addition of 1/6 vol of 1 M Tris. When necessary, the pH was further adjusted by using 0.1 M NaOH. Elution fractions were combined, dialyzed against PBS, pH 7.4, and stored at 4°C.
Preparation of Anti-scFvT84.66 IgY Antibodies.
The scFvT84.66 cDNA fragment was subcloned in the pET22b expression vector, which provides a C-terminal His6 tag. Escherichia coli strain BL21 (DE3) was transformed with the expression construct, and scFvT84.66 production was induced by the addition of 1 mM isopropyl β-d-thiogalactoside at 25°C. Inclusion bodies were recovered as described (38). For affinity purification, inclusion bodies were resuspended in 8 M urea, 10 mM Tris⋅HCl (pH 8.0), 100 mM NaH2PO4, and used for IMAC.
For immunization of Brown Leghorn chickens (Achternbosch, Hückelhoven, Germany) 100 μg of affinity purified bacterial scFvT84.66 was used per injection. Eggs from immunized animals were collected 2 weeks after the second immunization. IgY was purified as described (39) and characterized by using bacterially expressed scFvT84.66 and mT84.66 (data not shown).
Analyses of Purified scFvT84.66 and cT84.66 by SDS/PAGE, Western Blot, and ELISA.
Purified chimeric T84.66 (cT84.66) and scFvT84.66 antibodies and samples from different antibody purification steps were analyzed by SDS/PAGE and immuno-blotting. scFvT84.66 and cT84.66 were detected by using 1:1,000 diluted anti-scFvT84.66 IgY. ScFvT84.66 also was detected by using 1:2,000 diluted anti-His6 mAb (Qiagen, Hilden, Germany). Chimeric cT84.66 heavy chain was detected by using 1:1,000 diluted anti-KDEL mAb (StressGen Biotechnologies, York, U.K.).
For Western blotting and ELISA analyses, cT84.66 and mT84.66 were detected with AP-conjugated GAM-Fc, GAH-Fc, and F(ab′)2-specific goat anti-human IgG [GAH-F(ab′)2] and biotinylated κ-specific rabbit anti-mouse IgG (RAM-κ) (Dianova, Hamburg, Germany). All secondary antibodies were used at 1:5,000 dilution, and bound biotinylated RAM-κ was revealed by additional AP-conjugated streptavidin.
RESULTS
Our aim was to engineer and express a CEA-specific recombinant scFv and a mouse/human chimeric antibody in tobacco leaves, using Agrobacterium-mediated transient expression (33). The antibody expression constructs derived from mT84.66 were engineered and cloned into a plant expression vector and transiently expressed in tobacco leaves upon vacuum infiltration with recombinant Agrobacterium. Chimeric T84.66 (cT84.66) was constructed by exchanging the mouse light and heavy chain constant domains with their human counterparts. In vivo assembly of cT84.66 antibody was achieved by simultaneous expression of the light and heavy chains after vacuum infiltration of leaves with two populations of recombinant Agrobacterium.
scFvT84.66 Transient Expression in Tobacco Leaves and Purification.
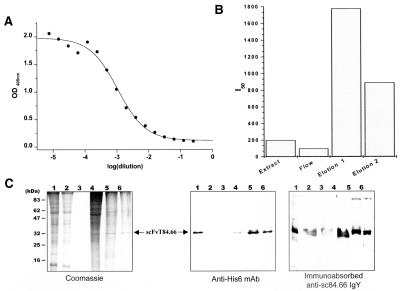
An scFvT84.66 expression construct was cloned into the pSS plant expression vector. In this vector the scFv gene was flanked by the 5′ UTR of CHS and the sequence encoding the LPH leader peptide, and by the 3′ His6-tag coding sequence (Fig. 1A). ScFvT84.66 was expressed in tobacco leaves after infiltration with recombinant Agrobacterium. Initially, a small number of leaves were infiltrated and processed to determine whether functional recombinant scFvT84.66 was produced. Plant expressed scFvT84.66 was functional because protein extracts from infiltrated leaves inhibited the binding of the murine mAb mT84.66 to the CEA/NA3 in competition ELISA (Fig. 2A). Time-course analyses showed that maximum accumulation of functional protein was reached at 60–72 h postinfiltration (data not shown). Therefore, soluble protein extracts for analyses were prepared from tobacco leaves at 60 h postinfiltration. The scFvT84.66 accumulated to ≈ 5 mg/kg fresh weight leaf material as deduced from the I50 values determined by competition ELISA.
Figure 2.
Transient expression and purification of scFvT84.66. (A) Protein extracts from infiltrated tobacco leaves were assayed for functional scFvT84.66 expression by competition ELISA 60 h after vacuum infiltration. The inhibition curve was obtained by plotting OD405 versus the dilution factor. IMAC purification of recombinant scFvT84.66 was analyzed by (B) competition ELISA and (C) Coomassie-stained SDS/PAGE and immunoblotting using the detection antibodies indicated. (B) Bars represent I50 values of the inhibition curves of the IMAC purification fractions. (C) Samples correspond to leaf extract (lane 1), column flow-through (lane 2), washing step (lane 3), 25 mM imidazole wash (lane 4), and 250 mM imidazole elution (lanes 5 and 6).
Upscaling to 100 g of leaf material and larger volumes of recombinant Agrobacterium culture permitted purification of recombinant His6-tagged scFvT84.66 by IMAC. The purification of scFvT84.66 was monitored by competition ELISA, SDS/PAGE, and Western blotting (Fig. 2 B and C). Competition ELISA data showed that there was a significant enrichment in scFvT84.66 activity in the elution fractions, indicating the efficient binding of the recombinant protein to the ligand (Fig. 2B). Coomassie-stained SDS/PAGE showed that nonspecifically bound plant proteins were preferentially removed by washing the column with 25 mM imidazole (Fig. 2C, lane 4). His6-tagged scFvT84.66 then was eluted with 250 mM imidazole. The elution fraction consisted of a major band with an electrophoretic mobility of ≈32 kDa (Fig. 2C, lane 5 and 6), which was recognized by an scFvT84.66-specific antisera (IgY) and anti-His6 tag mAb (Fig. 2C).
In Vivo Assembly of Chimeric Full-Size cT84.66 in Tobacco Leaves.
Chimeric light and heavy chain cDNAs were constructed by fusing the variable regions of mT84.66 in-frame with the human constant regions from cB72.3 (34). For plant expression, the light chain DNA fragment was placed downstream of the Ω 5′ UTR and the sequence encoding the codon optimized leader peptide LPL (Fig. 1B). The heavy chain fragment was inserted downstream of the Ω 5′ UTR and the sequence encoding the plant codon optimized leader peptide LPH, and upstream of the C-terminal KDEL ER retrieval signal (4) (Fig. 1C). The expression cassettes were placed under the control of the enhanced cauliflower mosaic virus 35S promoter in the pSS plant expression vector (9) and used for Agrobacterium transformation.
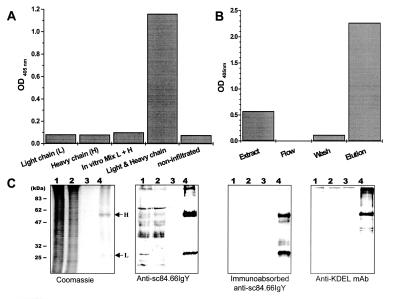
Light, heavy, or both the light and the heavy chains of cT84.66 were transiently expressed after vacuum infiltration of tobacco leaves with the corresponding recombinant Agrobacterium culture or a mix of both cultures, as described in Materials and Methods. Functional full-size antibody was detected only by ELISA in extracts from leaves simultaneously infiltrated with a mix of two Agrobacterium cultures that carried either the light or the heavy chain expression plasmid (Fig. 3A). No CEA/NA3 binding activity was detected in leaves infiltrated with light or heavy chain alone (Fig. 3A). Moreover, CEA/NA3 binding activity was not detected by in vitro mixing of protein extracts from two sets of infiltrated leaves expressing either the cT84.66 light chain or the cT84.66 heavy chain 2 h before analyses.
Figure 3.
Transient expression and purification of cT84.66. (A) Protein extracts from tobacco leaves infiltrated with the light chain (L), the heavy chain (H), and leaves simultaneously infiltrated with both light and heavy chain constructs were analyzed by ELISA using CEA/NA3 as antigen. An in vitro mixture of extracts from leaves infiltrated with only light or heavy chain constructs and noninfiltrated leaf extract were used as controls. Bound antibodies were detected with AP-conjugated GAH-Fc. Bars represent OD405 values of 1:64 diluted samples. Protein A affinity purification of recombinant cT84.66 from tobacco leaves simultaneously expressing both light and heavy chains was analyzed by (B) ELISA and (C) by Coomassie-stained SDS/PAGE and Western blotting using the detection antibodies indicated. Protein samples correspond to leaf extract (lane 1), column flow-through (lane 2), washing step (lane 3), and elution fraction (lane 4).
Purification and Characterization of in Vivo Assembled Chimeric T84.66.
Soluble protein extracts were prepared from infiltrated tobacco leaves simultaneously expressing the chimeric heavy and light chains of cT84.66 at 60 h postinfiltration. This extract was used for affinity purification of chimeric antibody on a protein A column. cT84.66 purification was monitored by ELISA, SDS/PAGE, and Western blotting (Fig. 3 B and C).
ELISA analyses showed that no functional antibody was detected in the flow-through of the protein A column, whereas CEA/NA3 binding activity was detected in the starting leaf homogenate and was significantly enriched in the elution fraction (Fig. 3B). Furthermore, eluted cT84.66 competed with murine T84.66 for binding to CEA/NA3 in competition ELISA. These results indicate that assembled full-size cT84.66 was produced within tobacco leaf cells.
SDS/PAGE and immunoblotting analyses showed the integrity of the antibody upon protein A affinity purification (Fig. 3C). The affinity-purified antibody preparation consisted of two distinct major bands (Figs. 3C and 4B) of approximately 53 and 26 kDa corresponding to the cT84.66 heavy and light chains. The identity of these protein bands was confirmed by Western blot using a chicken-derived IgY specific for scFvT84.66. The IgY antiserum nonspecifically bound to plant proteins in the total extract and protein A column flow-through. Preadsorption of the IgY sera with leaf protein extract from N. tabacum plants removed this background binding. Both untreated and preabsorbed anti-scFvT84.66 serum detected the cT84.66 heavy and the light chains in the protein A affinity-purified cT84.66 fraction. However, the cT84.66 chimeric antibody concentration in the starting material was too low for detection with anti-scFvT84.66 IgY (Fig. 3C, lane 1). The purified cT84.66 heavy chain also was detected by a KDEL-specific mAb (Fig. 3C). The yield and enrichment of cT84.66 during purification was estimated from Coomassie-stained SDS/PAGE gels and by ELISA. The concentration of cT84.66 in the affinity-purified antibody preparation was determined to be ≈ 15 μg/ml, resulting in a yield of approximately 1 mg cT84.66 per kg of fresh weight leaf material.
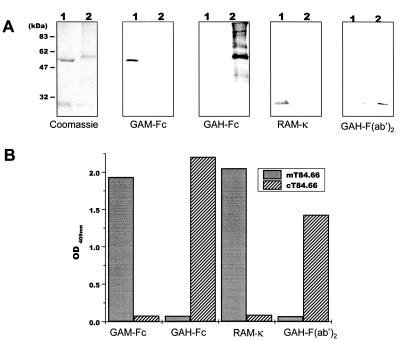
Figure 4.
Characterization of affinity-purified plant-expressed cT84.66. (A) One microgram of murine mAb T84.66 (lane 1) or 10 μl of Protein A purified cT84.66 (lane 2) was analyzed by SDS/PAGE and Coomassie staining. For Western blotting 250 ng of murine mAb T84.66 (lane 1) and 5 μl of cT84.66 (lane 2) was used. Blots were probed with indicated Abs. (B) For ELISA analyses 1.5 ng of mT84.66 and a 1:64 dilution of affinity-purified cT84.66 were applied to microtiter plates coated with 50 ng of CEA/NA3 and detected by the secondary antibodies indicated. Bars represent OD405 readings.
Compared with the murine T84.66, affinity-purified plant-derived cT84.66 heavy chain migrated with a higher molecular weight in SDS/PAGE, whereas the light chains had comparable sizes (Fig. 4A). This finding was in good agreement with the calculated theoretical molecular masses of 53 kDa for the chimeric heavy chain, including the KDEL motif, and 49 kDa for the murine T84.66 heavy chain.
Immunoblot and ELISA analyses were performed to confirm the chimeric nature of purified rAb cT84.66 (Fig. 4 A and B). Antibodies specific for the murine Ig constant domains (GAM-Fc and κ) recognized only the control murine T84.66 heavy and light chain but not the chimeric T84.66 chains, because of the replacement of the mouse constant domain sequences with human sequences. Human IgG constant domain-specific antibodies (GAH-Fc and κ) recognized only the purified plant-expressed cT84.66 heavy and light chains and not murine T84.66 (Fig. 4A). Furthermore, the chimeric nature of the assembled heavy and light chains of purified rAb cT84.66 was confirmed by CEA-specific ELISA where functional antibody was detected only by GAH antibodies (Fig. 4B).
DISCUSSION
To characterize diagnostic and therapeutic antibodies expressed in plants, we adapted a recently described Agrobacterium-mediated transient expression system (33). The expression of two rAbs, a scFv (scFvT84.66) and a full-size mouse-human chimeric antibody (cT84.66), specific for the CEA were studied. Both antibodies were functionally expressed in tobacco leaves after transformation with recombinant Agrobacterium. We concluded that cT84.66 assembly occurred after simultaneous expression of the cT84.66 heavy and light chains within the same cell, because of multiple transformation events. Furthermore, we demonstrated that the system is suitable for the upscaling of production and subsequent purification of transiently expressed rAbs. His6-tagged scFvT84.66 was purified by IMAC and cT84.66 by protein A affinity chromatography.
Efficient expression was achieved by directing the CEA/NA3-specific antibodies to the secretory pathway. Within the plant ER, proper folding and assembly of the rAbs is mediated by foldases and chaperones that are homologous to those found in mammalian cells (BiP/GRP78 and GRP94; refs. 40–42). Our results show that tobacco leaves transiently expressed both functional scFv and full-size chimeric antibodies. ELISA data revealed that plant-expressed scFvT84.66 and full-size cT84.66 antibody displayed the same specificity as the hybridoma-produced mT84.66 and bound to the A3 domain of the CEA.
The production of full-size mouse/human chimeric antibody was achieved by simultaneous expression of the heavy and light chains. The genes were transferred into tobacco leaf cells by two independent Agrobacterium populations. Multiple transformation events and a high number of transformed cells led to the accumulation of considerable amounts of assembled cT84.66. This result demonstrates that multimeric proteins can be analyzed by using this system. Because the genes for the individual chains can be encoded on separate plasmids no further cloning steps, e.g., making tandem expression constructs, are required, facilitating genetic engineering of multimeric proteins. The number of genes that can be simultaneously transiently expressed within the same cell may well reach up to five or six, allowing for complex multimeric proteins to be analyzed.
Transient gene expression systems offer several advantages over analyses of stable expression, for example initiation of gene expression, and synthesis of the protein can be analyzed within a very short time and is not affected by position effects (33). Our experiments underline the use of the Agrobacterium-mediated transient gene expression system for rapid testing of both the gene construct and the expressed protein. The results are obtained within a few days and can be evaluated before initiating stable transformation of plants. Therefore, this system is suitable for the development and initial characterization of new or improved engineered rAbs. Furthermore, because the system can be upscaled and transiently expressed proteins can be purified, a more detailed analysis can be carried out. Other advantages of this system are that the experimental procedure does not require sophisticated equipment and is inexpensive, compared with other transient systems such as microinjection, particle bombardment, or electroporation. Our data demonstrate that Agrobacterium-mediated transient expression in tobacco leaves is a suitable method for efficient expression of functional CEA-specific rAbs.
CEA is the best-characterized tumor antigen and is widely used in the diagnosis of colon cancer. The CEA-specific mAb T84.66 has a very high affinity constant (KD = 8 pM) and does not crossreact with other members of the CEA family (26, 43). These properties make this antibody an ideal candidate for tumor detection and immunotherapy. However, the use of the murine mAb T84.66 is compromised by the human anti-mouse antibody response (44), which is associated with serious clinical complications. Antibodies where the murine constant domains have been removed or humanized have been shown to be of lower immunogenicity. Molecules with these properties have been generated by protein engineering and were tested for in vivo tumor targeting and therapy (28–32). However, evaluating the performance of these reagents in tumor imaging and therapy requires significant amounts of protein.
Several reports have demonstrated that plants can produce full-size antibodies and antibody fragments and potentially are the most economical production system for functional rAbs. To date, bacteria (45) and mammalian cell cultures (46) are the most established systems for antibody production, whereas yeast and baculovirus-infected insect cell systems play only a minor role (47). Bacteria do not produce glycosylated full-size antibodies, contaminating endotoxins are difficult to remove, and recombinant proteins often form inclusion bodies, making labor- and cost-intensive in vitro refolding necessary. Because of the delicate nature of mammalian cells, cultivation can be difficult and requires expensive equipment and media. During downstream processing, care must be taken to remove oncogenic sequences or viral contaminants, in particular for in vivo therapeutic applications of antibodies.
Transgenic plants offer a practicable approach for mass production of recombinant proteins and antibodies. They can be easily grown, stored, and distributed. Protein secretion, folding, and posttranslational modification are similar in plant and animal cells (4, 40–42). Antibodies accumulate to high levels in plant cells and are essentially indistinguishable from those produced by hybridomas (7, 8). Copurification of blood-borne pathogens and oncogenic sequences is entirely avoided during downstream processing of plant-expressed recombinant proteins.
The successful application of the Agrobacterium-mediated transient expression system in intact tobacco leaves for characterizing the tumor-specific scFvT84.66 and cT84.66 prompted us to proceed with stable transformation. Although the transient system allows for upscaling and purification, large-scale production of rAbs can be achieved only through use of transgenic plants. Approximately 650,000 new cases of colon cancer are diagnosed in the U.S. every year, and it is estimated that 10–200 mg of rAbs per patient are required for tumor therapy. That would result in a demand for 6.5–130 kg of purified rAbs each year. We feel that transgenic plants are the only feasible approach to meet this demand for safe, therapeutic antibodies.
The results presented here demonstrate the suitability of plants for production of antibodies as an alternative to hybridoma or microbial production systems. Our long-term goal is the use of engineered antibodies produced by transgenic plants for diagnostic and therapeutic applications. These studies will help pinpoint the remaining hurdles that have to be overcome for the establishment of plant-derived antibodies as therapeutic agents.
Acknowledgments
We thank Dr. John E. Shively and Dr. Anna Wu for kindly providing mAb T84.66, T84.66/212 scFv cDNA as well as cDNAs encoding T84.66L2 light chain and T84.66H2 heavy chain (Beckman Research Institute of the City of Hope, Duarte, CA). We also thank Stephan Hellwig for the production of CEA/NA3 and Anna Kowol and Ulrike Sommer for their contribution to this paper. This work was supported by a European Community Training and Mobility of Researchers fellowship (FAIRCT96–5068) awarded to C.V. and a European Community grant (FAIRCT96–3110) awarded to R.F.
ABBREVIATIONS
- AP
alkaline phosphatase
- GAH
goat anti-human
- GAM
goat anti-mouse
- RAM
rat anti-mouse
- rAb
recombinant antibody
- scFv
single-chain Fv antibody
- UTR
untranslated region
- CEA
carcinoembryonic antigen
- IMAC
immobilized metal ion affinity chromatography
- CHS
chalcone synthase
- TMV
tobacco mosaic virus
- LPH
leader peptide derived from the heavy chain
- LPL
leader peptide derived from the light chain
- ER
endoplasmic reticulum
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bird R E, Hardman K D, Jacobson J W, Johnson S, Kaufman B M, Lee S M, Lee T, Pope S H, Riordan G S, Whitlow M. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 2.Whitelam G C, Cockburn W, Owen M R. Biochem Soc Trans. 1994;22:940–944. doi: 10.1042/bst0220940. [DOI] [PubMed] [Google Scholar]
- 3.Hiatt A, Ma J K. Int Rev Immunol. 1993;10:139–152. doi: 10.3109/08830189309061691. [DOI] [PubMed] [Google Scholar]
- 4.Denecke J, De Rycke R, Botterman J. EMBO J. 1992;11:2345–2355. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiatt A. Nature (London) 1990;344:469–470. doi: 10.1038/344469a0. [DOI] [PubMed] [Google Scholar]
- 6.Ma J K, Hiatt A, Hein M, Vine N D, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 7.Hiatt A, Cafferkey R, Bowdish K. Nature (London) 1989;342:76–78. doi: 10.1038/342076a0. [DOI] [PubMed] [Google Scholar]
- 8.Düring K, Hippe S, Kreuzaler F, Schell J. Plant Mol Biol. 1990;15:281–293. doi: 10.1007/BF00036914. [DOI] [PubMed] [Google Scholar]
- 9.Voss A, Niersbach M, Hain H J, Hirsch R, Liao Y C, Kreuzaler R, Fischer R. Mol Breeding. 1995;1:39–50. [Google Scholar]
- 10.Baum T J, Hiatt A, Parrot W A, Pratt L H, Hussey R S. Mol Plant-Microbe Interact. 1996;9:382–387. [Google Scholar]
- 11.De Wilde C, De Neve M, De Rycke R, Bruyns A-M, De Jaeger G, Van Montagu M, Depicker A, Engler G. Plant Sci. 1996;114:233–241. [Google Scholar]
- 12.De Neve M, De Loose M, Jacobs A, Van Houdt H, Kaluza B, Weidle U, Van Montagu M, Depicker A. Transgenic Res. 1993;2:227–237. doi: 10.1007/BF01977353. [DOI] [PubMed] [Google Scholar]
- 13.Artsaenko O, Peisker M, zur Nieden U, Fiedler U, Weiler E W, Muntz K, Conrad U. Plant J. 1995;8:745–750. doi: 10.1046/j.1365-313x.1995.08050745.x. [DOI] [PubMed] [Google Scholar]
- 14.Bruyns A M, De Jaeger G, De Neve M, De Wilde C, Van Montagu M, Depicker A. FEBS Lett. 1996;386:5–10. doi: 10.1016/0014-5793(96)00372-9. [DOI] [PubMed] [Google Scholar]
- 15.Fecker L F, Kaufmann A, Commandeur U, Commandeur J, Koenig R, Burgermeister W. Plant Mol Biol. 1996;32:979–986. doi: 10.1007/BF00020494. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler U, Phillips J, Artsaenko O, Conrad U. Immunotechnology. 1997;3:205–216. doi: 10.1016/s1380-2933(97)00014-6. [DOI] [PubMed] [Google Scholar]
- 17.Firek S, Draper J, Owen M R, Gandecha A, Cockburn B, Whitelam G C. Plant Mol Biol. 1993;23:861–870. doi: 10.1007/BF00021540. [DOI] [PubMed] [Google Scholar]
- 18.Owen M, Gandecha A, Cockburn B, Whitelam G. Bio/Technology. 1992;10:790–794. doi: 10.1038/nbt0792-790. [DOI] [PubMed] [Google Scholar]
- 19.Rosso M N, Schouten A, Roosien J, Borst-Vrenssen T, Hussey R S, Gommers F J, Bakker J, Schots A, Abad P. Biochem Biophys Res Commun. 1996;220:255–263. doi: 10.1006/bbrc.1996.0428. [DOI] [PubMed] [Google Scholar]
- 20.Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. Nature (London) 1993;366:469–472. doi: 10.1038/366469a0. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann S, Schillberg S, Liao Y-C, Fischer R. Mol Breeding. 1998;4:369–379. [Google Scholar]
- 22.Benvenuto E, Ordas R J, Tavazza R, Ancora G, Biocca S, Cattaneo A, Galeffi P. Plant Mol Biol. 1991;17:865–874. doi: 10.1007/BF00037067. [DOI] [PubMed] [Google Scholar]
- 23.Gold P, Freedman S O. J Exp Med. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd C W, Shively J E. In: Glycoproteins and Glycolipids in Disease Processes. Walborg E F, editor. Washington, DC: Am. Chem. Soc.; 1978. pp. 342–356. [Google Scholar]
- 25.Paxton R J, Mooser G, Pande H, Lee T D, Shively J E. Proc Natl Acad Sci USA. 1987;84:920–924. doi: 10.1073/pnas.84.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hefta L J, Neumaier M, Shively J E. Immunotechnology. 1998;4:49–57. doi: 10.1016/s1380-2933(98)00004-9. [DOI] [PubMed] [Google Scholar]
- 27.Williams L E, Beatty B G, Beatty J D, Wong J Y, Paxton R J, Shively J E. Cancer Res. 1990;50:1029s–1030s. [PubMed] [Google Scholar]
- 28.Hu S, Shively L, Raubitschek A, Sherman M, Williams L E, Wong J Y, Shively J E, Wu A M. Cancer Res. 1996;56:3055–3061. [PubMed] [Google Scholar]
- 29.Neumaier M, Shively L, Chen F S, Gaida F J, Ilgen C, Paxton R J, Shively J E, Riggs A D. Cancer Res. 1990;50:2128–2134. [PubMed] [Google Scholar]
- 30.Williams L E, Lewis M R, Bebb G G, Clarke K G, Odom-Maryon T L, Shively J E, Raubitschek A A. Bioconjugate Chem. 1998;9:87–93. doi: 10.1021/bc970137n. [DOI] [PubMed] [Google Scholar]
- 31.Wong J Y, Williams L E, Yamauchi D M, Odom-Maryon T, Esteban J M, Neumaier M, Wu A M, Johnson D K, Primus F J, Shively J E, et al. Cancer Res. 1995;55:5929s–5934s. [PubMed] [Google Scholar]
- 32.Wu A M, Chen W, Raubitschek A, Williams L E, Neumaier M, Fischer R, Hu S Z, Odom-Maryon T, Wong J Y, Shively J E. Immunotechnology. 1996;2:21–36. doi: 10.1016/1380-2933(95)00027-5. [DOI] [PubMed] [Google Scholar]
- 33.Kapila J, De Rycke R, Van Montagu M, Angenon G. Plant Sci. 1996;122:101–108. [Google Scholar]
- 34.Primus F J, Pendurthi T K, Hutzell P, Kashmiri S, Slavin D C, Callahan R, Schlom J. Cancer Immunol Immunother. 1990;31:349–357. doi: 10.1007/BF01741406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz J, Prufer D, Rohde W, Tacke E. Nucleic Acids Res. 1996;24:257–263. doi: 10.1093/nar/24.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höfgen R, Willmitzer L. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You H, Hefta L J, Yazaki P J, Wu A M, Shively J E. Anticancer Res. 1998;18:3193–3201. [PubMed] [Google Scholar]
- 38.Vaquero C, Liao Y C, Nähring J, Fischer R. J Gen Virol. 1997;78:2095–2099. doi: 10.1099/0022-1317-78-8-2095. [DOI] [PubMed] [Google Scholar]
- 39.Polson A, Coetzer T, Kruger J, von Maltzahn E, van der Merwe K J. Immunol Invest. 1985;14:323–327. doi: 10.3109/08820138509022667. [DOI] [PubMed] [Google Scholar]
- 40.Boston R S, Viitanen P V, Vierling E. Plant Mol Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- 41.Denecke J, Carlsson L E, Vidal S, Hoglund A S, Ek B, van Zeijl M J, Sinjorgo K M, Palva E T. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnick J, Aviel S, Argon Y. J Biol Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- 43.Esteban J M, Paxton R, Mehta P, Battifora H, Shively J E. Hum Pathol. 1993;24:322–328. doi: 10.1016/0046-8177(93)90044-h. [DOI] [PubMed] [Google Scholar]
- 44.Morton B A, O’Connor-Tressel M, Beatty B G, Shively J E, Beatty J D. Arch Surg. 1988;123:1242–1246. doi: 10.1001/archsurg.1988.01400340068012. [DOI] [PubMed] [Google Scholar]
- 45.Skerra A. Curr Opin Immunol. 1993;5:256–262. doi: 10.1016/0952-7915(93)90014-j. [DOI] [PubMed] [Google Scholar]
- 46.Bebbington C R, Renner G, Thomson S, King D, Abrams D, Yarranton G T. Bio/Technology. 1992;10:169–175. doi: 10.1038/nbt0292-169. [DOI] [PubMed] [Google Scholar]
- 47.Taticek R A, Lee C W, Shuler M L. Curr Opin Biotechnol. 1994;5:165–174. doi: 10.1016/s0958-1669(05)80031-x. [DOI] [PubMed] [Google Scholar]