Abstract
Syntheses of metal-containing enzymes often require the participation of accessory proteins. The roles played by many of these accessory proteins are poorly characterized. Klebsiella aerogenes urease, a nickel-containing enzyme, provides an ideal system to study metallocenter assembly. Here, we describe a method for isolating a complex containing urease apoprotein and the UreD, UreF, and UreG accessory proteins. We demonstrate that urease apoprotein in this complex is activated to near wild-type enzyme levels when incubated with nickel ions and high (≈100 mM) concentrations of bicarbonate. Significantly, we also observed nickel-dependent activation at physiologically relevant (≈100 μM) bicarbonate levels, but only in the presence of GTP. Based on studies involving a nonhydrolyzable analog of GTP, we conclude that nucleotide hydrolysis, not just binding, is required for this process. The critical nucleotide-binding site was localized to UreG on the basis of experiments using a variant complex. These studies highlight the relevance of the UreD-UreF-UreG-urease apoprotein complex to nickel metallocenter assembly and explain the previously identified in vivo energy requirement for urease activation.
Urease is a nickel-containing enzyme that acts as a virulence factor in a variety of human pathogens (1). The crystal structure of the heterotrimeric enzyme from Klebsiella aerogenes reveals the presence of a dinuclear metallocenter [i.e., (2 Ni/UreA-UreB-UreC)3], where the nickel ions are 3.6 Å apart and bridged by a carbamylated lysine residue and a hydroxide (2, 3). Assembly of the active site is a surprisingly complicated process (reviewed in ref. 4). In vitro, a portion (typically ≈15%) of urease apoprotein (apourease) is activated by using carbon dioxide and nickel ions (5, 6), with a CO2 molecule being incorporated into the protein to form the lysine carbamate metal ligand. In vivo, however, synthesis of K. aerogenes urease requires three accessory proteins (UreD, UreF, and UreG) and is facilitated by a fourth (UreE). These accessory proteins are encoded in the same gene cluster (ureDABCEFG) as the structural genes (7). UreE is a nickel-binding protein thought to function as a metallochaperone that delivers nickel to urease (8, 9). The three required accessory proteins form complexes with urease apoprotein including UreD-apourease (10), UreD-UreF-apourease (11), and UreD-UreF-UreG-apourease (12). The latter species, present in minute amounts in the cell, was suggested to be the key cellular urease activation machinery. An energy requirement for urease activation has been demonstrated by using intact cells (13). Related to this in vivo finding, the UreG sequence (14) contains a potential nucleotide-binding site, known as a “P-loop” motif that appears to be essential for cellular activation (15). Here, we describe methods to generate UreD-UreF-UreG-apourease, detail its nickel- and bicarbonate-dependent activation properties, and report the presence of a second in vitro activation mechanism that requires GTP hydrolysis. We propose that the GTP-dependent activation process is physiologically relevant and explains the known in vivo energy dependence for urease activation.
MATERIALS AND METHODS
Bacterial Strains and Plasmid Construction.
All molecular biology methods followed the general methods outlined in Sambrook et al. (16). Plasmids pKAUG-1 (to express ureG) and pKAUD2F+ΔureG (to express ureD, ureF, and the urease structural genes) have been described (11, 15). Plasmid pKAUGT21A, containing the ureG gene altered in the region encoding a potential P-loop motif (changing Thr-21 to Ala), was prepared by isolating the 2.9-kb EcoRI–AvrII fragment from pKAU17T21A (15), treating with Klenow enzyme to form blunt ends, and religating the ends together. All plasmids were transformed into Escherichia coli DH5α.
Culture Conditions and Cell Disruption.
All cultures were grown in LB broth supplemented with 100 μg ampicillin per ml. E. coli DH5α cells containing pKAUG-1, pKAUGT21A, or pKAUD2F+ΔureG were grown at 30°C until 3–5 h after reaching stationary phase. The cells were harvested by centrifugation and suspended in HEDG buffer (25 mM Hepes, pH 7.4/1 mM EDTA/1 mM DTT/10% glycerol). Resuspended cells were disrupted by 2–3 passages through a French pressure cell at 18,000 lb/in2, supplemented with 1 mM PMSF, and separated into extracts and pellet fractions by centrifugation at 100,000 × g for 45 min at 4°C.
Preparation of the UreD-UreF-UreG-Apourease Complex.
UreD-UreF-apourease complex and UreG (as well as its site-specifically altered form) were purified according to published procedures (11, 15), except that HEDG buffers were used in all purification steps. UreG (4-fold excess, native or altered form) was incubated with UreD-UreF-apourease for 12–24 h at 4°C in HEDG buffer. The incubated samples were concentrated and subjected to Superose 6 column chromatography (1.6 × 49 cm; Amersham Pharmacia Biotech) using HEDG buffer to separate excess UreG monomer from the UreD-UreF-UreG-apourease complex. Aliquots were stored at −70°C to preserve the stability of the complex.
PAGE.
SDS/PAGE was carried out by using the buffers described by Laemmli (17) and included 4.5% and 12.5% polyacrylamide stacking and running gels. Nondenaturing gels used the same buffers without detergent and consisted of 3% and 6% polyacrylamide stacking and running gels. The gels were stained with Coomassie brilliant blue dye. The band intensities of Coomassie blue-stained gels were measured with an ambis gel scanner (San Diego). For calculation of the ratios of UreD, UreF, UreG, and UreC, Mr values of 29,800, 25,200, 21,900, and 60,300 were used.
Urease Activity Assays.
Urease activities were measured by quantitating the rate of ammonia release from urea by formation of indophenol, which was monitored at 625 nm (18). One unit of urease activity was defined as the amount of enzyme required to hydrolyze 1 μmole of urea per min at 37°C. The standard assay buffer consisted of 25 mM Hepes (pH 7.75), 0.5 mM EDTA, and 50 mM urea. Protein concentrations were determined by using a commercial assay (Bio-Rad) with BSA as the standard.
Activation of Urease Apoprotein.
Routine activation buffer consisted of 100 mM Hepes (pH 8.3), 150 mM NaCl, 100 mM NaHCO3, and 100 μM NiCl2 (5, 6). For specific experiments, the conditions were modified by (a) varying the bicarbonate or nickel ion concentrations, (b) adding GTP, GDP, ATP (all from Sigma), or GMP-PNP (Fluka) equilibrated with 2-fold concentrations of Mg+2, or (c) adding other metal ions to the activation buffer.
RESULTS AND DISCUSSION
Isolation of the UreD-UreF-UreG-Apourease Complex.
Using immunological methods, trace levels of UreD-UreF-UreG-apourease complex were previously observed in extracts of E. coli cells containing the wild-type K. aerogenes urease gene cluster (12). Because UreD, UreF, and UreG accessory proteins are essential for cellular synthesis of active urease (7), we proposed that the multicomponent complex is necessary for in vivo activation. We therefore sought a method to enhance the levels of this key activation complex to characterize its properties. Direct overexpression of the ureD, ureF, ureG, and urease structural genes (from plasmid pKAUD2F+, ref. 11) led to the production of a complex containing the desired peptides (M.B.C. Moncrief and R.P.H., unpublished observations); however, the resulting species was clearly distinct from the previously observed UreD-UreF-UreG-apourease complex. For example, the directly overproduced complex appeared to be highly aggregated and failed to migrate into native polyacrylamide gels, in contrast to the defined banding pattern observed for the previously reported species (12). Also consistent with a lack of physiological relevance, the apoprotein in the directly produced complex was less competent for activation than the apoprotein in UreD-UreF-apourease complex (data not shown).
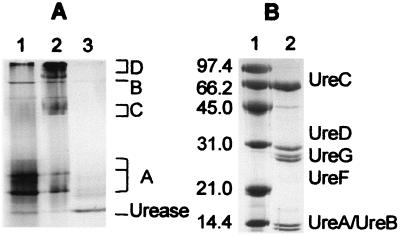
As an alternative approach to obtain the desired UreD-UreF-UreG-apourease complex, we tested whether or not it could be produced by simple incubation of UreD-UreF-apourease complex with excess UreG. The UreD-UreF-apourease complex yielded three major bands during native gel electrophoresis (Fig. 1A, lane 1, labeled A) arising from the interaction of one, two, or three molecules of UreD and UreF per molecule of trimeric urease apoprotein. This sample also had a minor amount of contaminating GroEL (Fig. 1A, band B), as described (11). The addition of UreG afforded new bands (Fig. 1A, lane 2, bands C and D), with those labeled C being identical in position to those reported earlier for UreD-UreF-UreG-apourease (12). Analysis of Coomassie blue-stained bands from denaturing gels (Fig. 1B) revealed a peptide ratio in the UreD-UreF-UreG-apourease complex of 0.74–0.99 UreD, 0.81–1.16 UreG, and 0.72–1.07 UreF per UreC.
Figure 1.
PAGE analysis of UreD-UreF-UreG-apourease complex. (A) Native gel electrophoresis. Samples include UreD-UreF-apourease complex (lane 1), UreD-UreF-UreG-apourease complex (lane 2), and the same sample as shown in lane 2 after activation (lane 3) for 1.5 h at 37°C in the presence of 100 μM NiCl2 and 100 mM bicarbonate in standard activation buffer. (B) SDS/PAGE analysis of UreD-UreF-UreG-apourease complex. Samples include molecular weight markers (phosphorylase b, Mr 97,400; BSA, Mr 66,200; ovalbumin, Mr 45,000; carbonic anhydrase, Mr 31,000; soybean trypsin inhibitor, Mr 21,000; lysozyme, Mr 14,400) (lane 1) and UreD-UreF-UreG-urease apoprotein complex (lane 2).
Nickel- and Bicarbonate-Dependent Activation of UreD-UreF-UreG-Apourease Complex.
When incubated in typical activation conditions containing 100 μM NiCl2 and 100 mM bicarbonate, the accessory proteins dissociated from the UreD-UreF-UreG-apourease complex to yield the faster migrating free urease (Fig. 1A, lane 3). This transformation was accompanied by the generation of active enzyme, with a specific activity ranging from 800 to 1,500 units/mg. For comparison, activation of urease apoprotein in the UreD-UreF-apourease complex using these conditions results in a specific activity of 800 ± 100 units/mg (11). The wide variability in activation competence for the different UreD-UreF-UreG-apourease samples likely reflected instability of this complex, shown to decompose to form UreD-UreF-apourease and free UreG.
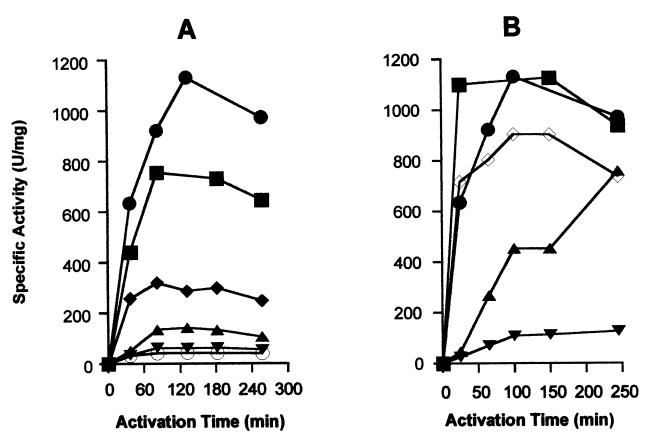
The bicarbonate and nickel ion concentration dependencies for urease activation using the UreD-UreF-UreG-apourease complex were determined, as shown in Fig. 2. The shapes of these plots closely resembled those reported for UreD-UreF-apourease, UreD-apourease, and urease apoproteins (5, 11), but the levels and rates of activation were generally enhanced. For example the activation rate using UreD-UreF-UreG-apourease complex incubated with 200 μM NiCl2 (44 units/mg per min when calculated by using the zero and 25-min time points of Fig. 2B) was found to be the fastest in vitro urease activation rate yet reported. Activation of urease apoprotein within the UreD-UreF-UreG-apourease complex was not significantly inhibited by incubation with nickel in the absence of bicarbonate, similar to results obtained for UreD-UreF-apourease and in contrast to those obtained by using UreD-apourease and urease apoprotein (6, 11). Inclusion of zinc, copper, or cobalt ions in the activation mixture led to an inhibition of the activation process.
Figure 2.
Bicarbonate- and nickel-dependent activation of urease apoprotein in the UreD-UreF-UreG-apourease complex. (A) Samples of UreD-UreF-UreG-apourease complex (0.2 μM) were incubated at 37°C in standard activation buffer containing 100 μM NiCl2 and 0.1 (○), 0.5 (▾), 5 (▴), 25 (⧫), 75 (■), or 100 (●) mM NaHCO3. (B) Similar incubations used standard activation buffer containing 100 mM NaHCO3 and 20 (▾), 50 (▴), 100 (●), 200 (■), or 400 (◊) μM NiCl2. For both sets of studies, aliquots were removed at the indicated times and the samples were assayed for urease activity.
GTP-Dependent Activation of UreD-UreF-UreG-Apourease Complex.
Although the UreG sequence includes a potential nucleotide-binding site in the protein, prior studies had failed to detect nucleotide hydrolysis by this protein or association between UreG and ATP or GTP (15). Notably, however, a urease-free complex containing UreD, UreF, and UreG was shown to bind to an ATP-linked agarose resin (15). These results suggested that the nucleotide-binding properties of UreG may be modulated by the presence of other accessory proteins, and led to studies involving the UreD-UreF-UreG-apourease complex.
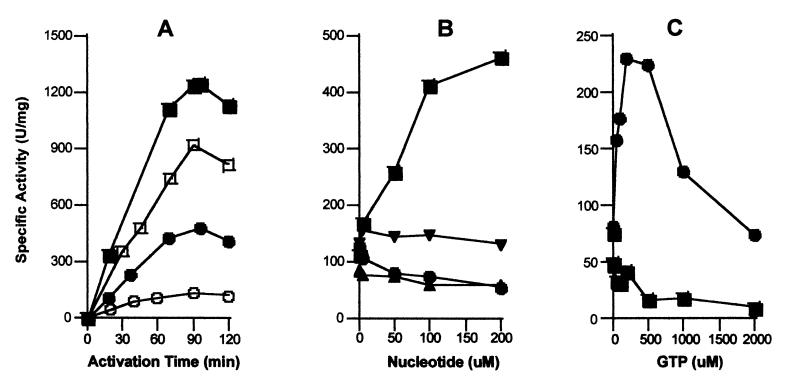
As shown in Fig. 3, GTP significantly affects urease apoprotein activation in the UreD-UreF-UreG-apourease complex. In Fig. 3A, the time course for activating urease apoprotein in the complex was compared in the presence and absence of 200 μM GTP and using either 100 mM or 100 μM bicarbonate. In this experiment, and others to follow, magnesium ions were provided at concentrations 2-fold that of the nucleotides. The presence of GTP had a small, but reproducible, positive effect on urease apoprotein activation when examined at high bicarbonate levels. Of greater significance, inclusion of GTP led to a dramatic enhancement of activity in the low bicarbonate sample. Other nucleotides failed to enhance activation of urease apoprotein in the complex in 100 μM bicarbonate after 1.5 h (Fig. 3B). Whereas 200 μM GTP resulted in 4-fold enhancement of urease apoprotein activation to reach a specific activity of about 460 units/mg, this concentration of ATP or GDP caused a diminishment in activation competence. Significantly, GMP-PNP (a nonhydrolyzable analogue of GTP with the β and γ phosphates linked by NH) failed to enhance the activation of urease apoprotein in the complex. To establish that GMP-PNP binds to the protein complex, its effect on GTP-dependent activation was assessed (data not shown). Although the presence of 100 μM GTP enhanced activation so as to increase the urease activity from ≈100 units/mg to about 400 units/mg, the additional presence of 100 μM GMP-PNP dropped the final activity to approximately 250 units/mg. Thus, GTP hydrolysis, not just binding, is needed to enhance apoprotein activation in the UreD-UreF-UreG-apourease complex.
Figure 3.
GTP-dependence of urease apoprotein activation for the UreD-UreF-UreG-apourease complex. (A) The time dependence of urease apoprotein activation using UreD-UreF-UreG-apourease (0.2 μM) was studied at 37°C in the presence (closed symbols) or absence (open symbols) of 200 μM Mg2GTP in 100 mM Hepes buffer (pH 8.3) containing 150 mM NaCl, 100 μM NiCl2, and 100 mM (squares) or 100 μM (circles) bicarbonate. (B) The nucleotide specificity and concentration dependence was examined by monitoring the extent of activation after 1.5 h in buffer containing 100 μM NiCl2, 100 μM NaHCO3, and the indicated concentrations of Mg2GTP (■), Mg2ATP (▴), Mg2GDP (●), and Mg2GMP-PNP (▾). (C) The effect of using a P-loop defective UreG variant (T21A UreG) was assessed. The urease apoproteins in UreD-UreF-UreG-apourease complexes containing the variant protein (0.17 μM; ■) or native UreG (0.2 μM; ●) were activated for 1.5 h in the presence of 100 μM NiCl2, 100 μM NaHCO3, and GTP at the concentrations indicated. In all cases, urease activities were measured as previously described. All results are representative of three separate protein preparations.
Two experiments provide evidence that GTP-dependent effects were associated with UreG within the complex. First, GTP failed to provide any enhancement of activation for uncomplexed apoprotein or apoprotein in the UreD-apourease and UreD-UreF-apourease complexes. Indeed, the presence of GTP led to a reduction in activation competence for these species (data not shown) similar to what was seen for the ATP effect on UreD-UreF-UreG-apourease (Fig. 3B). Second, GTP-dependent activation was observed only in the presence of UreG with a functional P-loop motif (Fig. 3C). A UreG variant (T21A) altered at a critical residue in the nucleotide-binding site was used to generate UreD-UreF-UreG (T21A)-apourease. Whereas a functional P-loop was not required for formation of this complex, the GTP-dependent enhancement of activation was abolished.
Very high levels of GTP are inhibitory to urease apoprotein activation using the UreD-UreF-UreG-apourease complex (Fig. 3C). This diminishment in activity was not caused by inhibition by contaminating GDP as shown by the absence of effect when using a GTP recycling system. More likely is the possibility that the nucleotide acts by binding nickel ions and reducing the effective nickel concentrations. Consistent with this notion, GTP and other nucleotides have been shown to impair activation of urease apoprotein when uncomplexed and when present in the UreD-apourease or UreD-UreF-apourease complexes. Elevated levels of magnesium ions partially alleviated the inhibition by high levels of GTP (data not shown), buttressing this nickel chelation hypothesis. Inhibition of apourease activation by various nucleotides results in the need for an effective delivery system for nickel ions within the cell. In the case of urease metallocenter assembly, the UreE accessory protein appears to serve this function (8, 9).
Summary and Perspectives.
UreD-UreF-UreG-apourease complex is formed by the simple incubation of UreG with UreD-UreF-apourease, and two distinct processes can be used for its activation. In the presence of nickel ions and very high bicarbonate levels (a source of CO2), direct activation converts urease apoprotein in this complex to highly active urease with a specific activity of up to 1,500 units/mg. This activity matches that typically observed for enzyme purified from recombinant E. coli cells containing the wild-type K. aerogenes urease gene cluster and grown in the presence of 1 mM nickel ions (1,500–1,900 units/mg; refs. 7 and 19). Furthermore, this value approaches that for wild-type urease isolated from K. aerogenes (2,500 units/mg; ref. 20). This direct activation process is unlikely to be biologically significant because of the necessity for high bicarbonate levels. In contrast, a separate GTP-dependent process overcomes this requirement. By coupling the energy of GTP hydrolysis to apourease activation, the UreD-UreF-UreG-apourease complex forms active enzyme at low levels of bicarbonate. We suggest that this GTP-dependent activation process is physiologically relevant and could explain the previously described in vivo energy dependence of urease activation (13). The role of nucleotide hydrolysis in urease activation remains unclear, but two alternatives can be considered. Nucleotide-dependent protein conformational changes are well known (21), and similar GTP-dependent structural changes of the urease apoprotein complex could be important for increasing nickel ion or carbon dioxide accessibility to the developing active site. Alternatively, the UreD-UreF-UreG-apourease complex might use GTP and bicarbonate (not CO2) to synthesize carboxyphosphate. Although this molecule possesses a half-life of less than 70 ms when free in solution (22), if generated within the complex near the lysine undergoing carbamylation it could function as an excellent CO2 donor. We used established methods (5) to try to distinguish if CO2 or bicarbonate was the true substrate for the GTP-dependent activation of urease apoprotein in the UreD-UreF-UreG-apourease complex. Unfortunately, clear results were not obtained for two reasons. First, the apoprotein in UreD-UreF-UreG-apourease complex is activated at low bicarbonate levels (presumably by CO2) even in the absence of GTP, and this rate is nearly half that observed in the presence of nucleotide (Fig. 3A). Second, the sample of UreD-UreF-UreG-apourease complex always included some UreD-UreF-apourease complex (Fig. 1A). Urease apoprotein from the latter complex is activated at low levels of bicarbonate (11), also presumably by reaction with CO2.
The closest parallel to the GTP-dependent process reported here is found in the activation of hydrogenase, another nickel-containing enzyme. Hydrogenase maturation in E. coli requires a suite of accessory proteins including HypB (23, 24), a protein sharing 25% identity with UreG. HypB uses a P-loop sequence to bind GTP, whose hydrolysis is required for hydrogenase activation. Similarly, ATP binding to dinitrogenase reductase is needed for incorporating the iron-molybdenum cofactor into Azotobacter vinelandii nitrogenase (25–27). Rhodospirillum rubrum CooC (needed for activating carbon monoxide dehydrogenase) (28) and Pseudomonas stutzeri NosF (involved in the synthesis of nitrous oxide reductase) (29) also possess P-loop motifs, suggesting a nucleotide requirement. Further characterization of the GTP-dependent activation of urease may provide useful insight into the maturation processes in these other nucleotide-dependent metallocenter assembly systems.
Acknowledgments
We thank our lab colleagues for editorial comments. Ths work was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases (DK45686).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Mobley H L T, Island M D, Hausinger R P. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jabri E J, Carr M B, Hausinger R P, Karplus P A. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 3.Pearson M A, Michel L O, Hausinger R P, Karplus P A. Biochemistry. 1997;36:8164–8172. doi: 10.1021/bi970514j. [DOI] [PubMed] [Google Scholar]
- 4.Taha T, Brayman T G, Karplus P A, Hausinger R P. In: Transition Metals in Microbial Metabolism. Winkelmann G, Carrano C J, editors. Amsterdam: Harwood; 1997. pp. 391–413. [Google Scholar]
- 5.Park I-S, Hausinger R P. Science. 1995;267:1156–1158. doi: 10.1126/science.7855593. [DOI] [PubMed] [Google Scholar]
- 6.Park I-S, Hausinger R P. Biochemistry. 1996;35:5345–5352. doi: 10.1021/bi952894j. [DOI] [PubMed] [Google Scholar]
- 7.Lee M H, Mulrooney S B, Renner M J, Markowicz Y, Hausinger R P. J Bacteriol. 1992;174:4323–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colpas G J, Brayman T G, McCracken J, Pressler M A, Babcock G T, Ming L J, Colangelo C M, Scott R A, Hausinger R P. J Biol Inorg Chem. 1998;3:150–160. [Google Scholar]
- 9.Colpas G J, Brayman T G, Ming L-J, Hausinger R P. Biochemistry. 1999;38:4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]
- 10.Park I-S, Carr M B, Hausinger R P. Proc Natl Acad Sci USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moncrief M B C, Hausinger R P. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park I-S, Hausinger R P. J Bacteriol. 1995;177:1947–1951. doi: 10.1128/jb.177.8.1947-1951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M H, Mulrooney S B, Hausinger R P. J Bacteriol. 1990;172:4427–4431. doi: 10.1128/jb.172.8.4427-4431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulrooney S B, Hausinger R P. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moncrief M B C, Hausinger R P. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Weatherburn M W. Anal Chem. 1967;39:971–974. [Google Scholar]
- 19.Park I-S, Hausinger R P. Protein Sci. 1993;2:1034–1041. doi: 10.1002/pro.5560020616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd M J, Hausinger R P. J Biol Chem. 1989;264:15835–15842. [PubMed] [Google Scholar]
- 21.Sprang S R. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 22.Sauers C L, Jencks W P, Groh S. J Am Chem Soc. 1975;97:5546–5553. [Google Scholar]
- 23.Maier T, Jacobi A, Sauter M, Böck A. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier T, Lottspeich F, Böck A. Eur J Biochem. 1995;230:133–138. [PubMed] [Google Scholar]
- 25.Robinson A C, Chun T W, Li J-G, Burgess B K. J Biol Chem. 1989;264:10088–10095. [PubMed] [Google Scholar]
- 26.Allen R M, Homer M J, Chatterjee R, Ludden P W, Roberts G P, Shah V K. J Biol Chem. 1993;268:23670–23674. [PubMed] [Google Scholar]
- 27.Rangaraj P, Shah V K, Ludden P W. Proc Natl Acad Sci USA. 1997;94:11250–11255. doi: 10.1073/pnas.94.21.11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerby R L, Ludden P W, Roberts G P. J Bacteriol. 1997;179:2259–2266. doi: 10.1128/jb.179.7.2259-2266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zumft W G, Viebrock-Sambale A, Braun C. Eur J Biochem. 1990;192:591–599. doi: 10.1111/j.1432-1033.1990.tb19265.x. [DOI] [PubMed] [Google Scholar]