Abstract
The active sites of aminopeptidase A (PepA) from Escherichia coli and leucine aminopeptidase from bovine lens are isostructural, as shown by x-ray structures at 2.5 Å and 1.6 Å resolution, respectively. In both structures, a bicarbonate anion is bound to an arginine side chain (Arg-356 in PepA and Arg-336 in leucine aminopeptidase) very near two catalytic zinc ions. It is shown that PepA is activated about 10-fold by bicarbonate when l-leucine p-nitroanilide is used as a substrate. No activation by bicarbonate ions is found for mutants R356A, R356K, R356M, and R356E of PepA. In the suggested mechanism, the bicarbonate anion is proposed to facilitate proton transfer from a zinc-bridging water nucleophile to the peptide leaving group. Thus, the function of the bicarbonate ion as a general base is similar to the catalytic role of carboxylate side chains in the presumed mechanisms of other dizinc or monozinc peptidases. A mutational analysis shows that Arg-356 influences activity by binding the bicarbonate ion but is not essential for activity. Mutation of the catalytic Lys-282 reduces kcat/Km about 10,000-fold.
Leucine aminopeptidase (LAP) is a prototypic dizinc peptidase that has been studied intensely by kinetic and crystallographic investigations on the enzyme isolated from bovine lens (1–5). The enzyme is present in animals, plants, and bacteria and has different tissue-specific physiological roles in the processing or degradation of peptides. Human LAP has been shown to catalyze postproteasomal trimming of the N terminus of antigenic peptides for presentation on major histocompatibility complex class I molecules (6). Here, interferon-γ not only promotes proteasomal cleavage but also induces LAP for N-terminal processing of the peptides. LAP has also been implicated in HIV pathophysiology (7). Bestatin, an inhibitor of LAP, significantly decreases HIV infection of human lymphocytic cells.
X-ray structures have been determined for the native enzyme from bovine lens (8, 9) and for complexes with the transition-state analogue inhibitors bestatin (8), amastatin (10), l-leucinal (9), and l-leucinephosphonic acid (11) bound to the active site. On the basis of these structures and of kinetic studies, a detailed reaction mechanism has been proposed, with a metal-bridging water molecule as the nucleophile (9). A surprising result of the 1.6-Å structure of unliganded LAP was the presence of a carbonate or bicarbonate ion in the active-site pocket next to Arg-336. In cocrystal structures with the inhibitors leucinal (9) and leucinephosphonic acid (11), this bicarbonate ion is replaced by two water molecules, which are at a very close distance of 2.3 Å. It has been speculated that the two water molecules are deprotonated to form a bihydroxide ion (H3O2−) (9). Because neither activation nor inhibition of bovine lens LAP by bicarbonate was observed, binding of the bicarbonate ion to the active site of native LAP was initially considered an artifact of the crystallization conditions. Besides the dimetal cluster and the arginine side chain, Lys-262 is proposed to have an important function in binding the substrate and in transition-state stabilization (8–11).
Aminopeptidase A (PepA) from Escherichia coli has similar biochemical properties to LAP (12). The two aminopeptidases share 31% sequence identity (52% in the catalytic domain) (13). All catalytic active-site residues are conserved between the two enzymes. In contrast to LAP, PepA also has a function as a DNA-binding protein in Xer site-specific DNA recombination. Here PepA acts in a complex with three other proteins, XerC, XerD, and arginine repressor (14). This function is independent of the aminopeptidase activity of PepA (15).
Here, we report on a mutational analysis of the active-site lysine and arginine residues in PepA and on kinetic studies on the wild-type enzyme. By these studies, the contribution of the two positively charged residues to catalysis is analyzed. Interestingly, wild-type PepA is significantly activated by bicarbonate ions at physiological concentrations. Together with the crystallographic data, these results indicate a functional role of the active-site carbonate or bicarbonate ion as a general base. In this article, we will use the residue numbering of PepA. The corresponding numbers of the LAP active-site residues can be obtained by subtracting 20 from these numbers.
MATERIALS AND METHODS
Mutagenesis and Protein Purification.
Plasmid derivatives were obtained by PCR mutagenesis by using the following primers: K282A, 5′-GGTATCTCGATCGCGCCTTCAGAAGGCAT and 5′-GCCTTCTGAAGGCGCGATCGAGATACC; R356X, GACGCTGAAGGCxxxCTGGTACTGTGC and 5′-GCACAGTACCAGyyyGCCTTCAGCGTCGG (with GCC, ATG, AAG, and GAG for xxx and GGC, CAT, CTT, and CTC for yyy of mutants R356A, R356M, R356K, and R356E, respectively). PCR reactions were performed by using Taq DNA polymerase (Perkin–Elmer Cetus). Vector pCA9 (obtained from David Sherratt, Oxford University, UK) was used as template. This plasmid contains pepA cloned into pTrc99A (Amersham Pharmacia) between the NcoI and BamHI sites. The PCR products were subcloned into pCA9 between the BglII and EcoNI sites (K282A mutant) or between the EcoNI and BamHI sites (R356 mutants). All mutations were verified by nucleotide sequence analysis of the subcloned fragments by using the dideoxy method (Thermo Sequenase kit, Amersham Pharmacia).
Wild-type and mutant PepA were purified from E. coli strain DS957 (DS941 pepA∷Tn5) containing plasmid pCA9 or the derived mutant plasmids, which express PepA from the trc promoter, as described previously (15). The precipitate obtained by dialysis against 20 mM KCl/50 mM Tris⋅HCl, pH 8.0/1 mM Mg(OAc)2 was dissolved in a buffer containing 1 M KCl, 50 mM Tris, pH 8.0, and 1 mM Mg(OAc)2.
Activity Assay.
All assays were carried out spectrophotometrically with l-leucine p-nitroanilide as substrate by continuously monitoring the appearance of p-nitroaniline at 405 nm with a Shimadzu UV-160A spectrophotometer. l-leucine p-nitroanilide was obtained from Sigma. The incubation was done at 22°C and the absorbance converted to a concentration scale by a molar absorption coefficient of 9,800 M−1 cm−1 for p-nitroaniline. The kinetic parameters kcat and Km were determined from double-reciprocal plots (16). The value of kcat refers to a monomer of the hexameric PepA (kcat = Vmax/6⋅[E]). For the assays of the activation by bicarbonate, all buffers except the protein stock solution were repeatedly degassed and flushed with nitrogen gas. Degassing of the protein stock solution proved impracticable because degassing of the concentrated protein solution resulted in excessive formation of foam and loss of activity, whereas sufficiently diluted solutions also showed a slow irreversible loss of enzyme activity under the conditions used for the degassing procedure. Nevertheless, attempts to degas the protein solutions in addition to the other buffers indicated that the resulting activity is not significantly lower than the one determined without degassing of the protein stock solution. During the about 2-min time span of mixing of the buffers in the cuvette and recording of the released p-nitroaniline, the cuvettes were in contact with air. No deviation from linearity of product formation over time was noticed during this time interval. For the kinetic analysis of activation by bicarbonate, a nonlinear least-squares regression algorithm was used to determine Ka, Vmax, and V0 in the equation V = V0 + (Vmax − V0)⋅[A]/(Ka + [A]), where V is the velocity that corresponds to activator concentration [A] (16).
Protein Structures, Structure Analysis, and Figure Generation.
Protein structures for PepA (to be submitted), bovine lens LAP (1LAM), the LAP–leucinal complex (1LAN), and Aeromonas aminopeptidase (1AMP) are deposited in the Protein Data Bank. Electron density maps were calculated with programs x-plor and cns (17) by using crystallographic data described elsewhere (9, 18). Least-squares structure superpositions and structure analysis were done with program o (19) at a Silicon Graphics Workstation (Mountain View, CA). Fig. 1C was generated with molscript (20), Fig. 1 A and B with bobscript (21) and Fig. 2A with grasp (22).
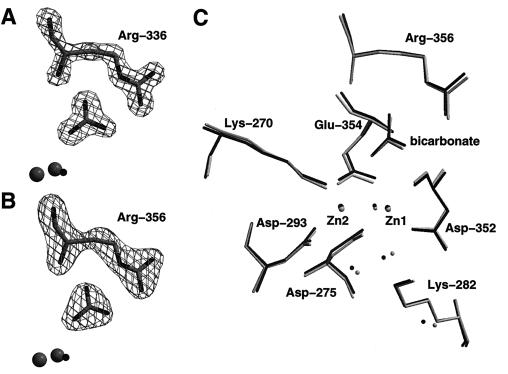
Figure 1.
Active-site structures of PepA and LAP. (A) Fo − Fc omit electron density map at 1.6-Å resolution of Arg-336 and the bicarbonate ion in the active site of LAP. Also shown are the two zinc ions and the metal-bridging water molecule. (B) Fo − Fc (2.5 Å) omit electron density map of the analogous residues of PepA. (C) Superposition of active-site residues of PepA and LAP (see text).
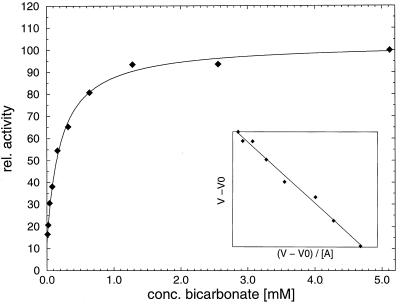
Figure 2.
Activation of PepA by bicarbonate ions. The curve shown is calculated according to equation V = V0 + (Vmax − V0)⋅[A]/(Ka + [A]) with Ka, Vmax and V0 derived from the experimental data. V is given in relative units. Ka was determined as 0.21 mM, and Vmax/ V0 = 8.2.
Continuum Electrostatic Calculations.
Electrostatic potential maps were calculated by solution of the Poisson–Boltzmann equation in a continuum electrostatic model as implemented in the program delphi (23). A monomer of LAP was used for the calculations. Using the complete hexamer structure did not significantly influence the distribution shown in Fig. 3B, including the potentials of the active-site water molecules. A box fill of 60% and a probe radius of 1.8 Å was used, and the dielectric constant was set to 2 for the protein region and to 80 for the solvent. Crystallographically determined water molecules were included into the high dielectric region. The atomic charges were obtained by using the charmm24(b1) parameter set (24).
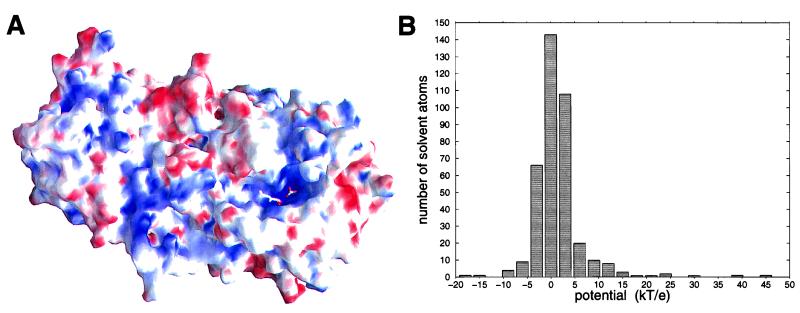
Figure 3.
Electrostatic potential of LAP. (A) Molecular surface of an LAP monomer colored by the surface electrostatic potential at the protein surface. Positive potential is shown in blue and negative potential in red. Dark red and dark blue colors mark potentials lower than −13 kT/e and higher than 13 kT/e, respectively. (B) Distribution of the number of crystallographically determined solvent molecules (waters and the bicarbonate ion) over the potential at the oxygen atoms.
RESULTS AND DISCUSSION
Active-Site Structures of PepA and LAP.
Determination of the structure of PepA recently showed that not only are the active-site residues well conserved and similarly oriented compared with LAP, but also a bicarbonate ion is present next to Arg-356 (Fig. 1) in the same orientation as in the native structure of LAP (18). Binding of the trigonal planar ion to the active sites is clearly demonstrated by the 1.6 Å and 2.5 Å electron density maps of LAP and PepA, respectively (Fig. 1 A and B). In Fig. 1C, we have superimposed the two enzyme active sites on the basis of protein residues shown in this figure. A root-mean-square deviation of 0.16 Å for the Cα-atoms of these residues demonstrates that the active sites are identical within the error limits of the atomic coordinates. In addition to the side chain conformations and the orientation of the bicarbonate ion, many active-site water molecules are “conserved” between the two enzymes. This includes the zinc-bridging water molecule, which is the proposed nucleophile in the enzyme mechanism.
Activation of Wild-Type PepA by Bicarbonate.
When the kinetic properties of wild-type PepA were determined, different values for the rate constant of the enzyme were obtained depending on whether freshly prepared buffer was used for the assay or whether the buffer had been in contact with air for several hours before the assay. Determination of the turnover number (kcat) in degassed buffer and in the presence of various concentrations of sodium bicarbonate showed that this activation of PepA is caused by carbonate or bicarbonate ions derived from dissolved carbon dioxide (Fig. 2). An 8-fold increase of kcat in the presence of sodium bicarbonate is observed relative to the assay in degassed buffer. An apparent binding constant of Ka = 0.21 ± 0.03 mM was determined for the nonessential activator NaHCO3. Determination of the Michaelis–Menten kinetic parameters of PepA in degassed buffer and in the presence of 5 mM NaHCO3, however, showed that Km is increased about 4-fold in the presence of bicarbonate (Table 1), such that the second-order rate constant (kcat/Km) is increased only about 2-fold (from 8.3 mM−1⋅s−1 to 13.6 mM−1⋅s−1) for the nonnatural l-leucine p-nitroanilide substrate. The bicarbonate ion binds near to the substrate, and it is hydrogen bonded to the gem-diol group of the transition-state analogue inhibitor l-leucinal in the LAP-leucinal complex. It might therefore influence substrate binding directly by hydrogen bonding and van der Waals contacts. However, this argument does not explain why the Arg-356 mutants, which presumably contain no bound bicarbonate ion (see below), have Km values that are comparable to or higher than the activated wild-type enzyme (Table 1).
Table 1.
Michaelis–Menten kinetic data of wild-type and mutant PepA
| PepA | Km, mM | kcat, s−1 |
|---|---|---|
| Wild type (degassed buffer) | 0.056 ± 0.003 | 0.464 ± 0.007 |
| Wild type (5 mM NaHCO3) | 0.23 ± 0.01 | 3.13 ± 0.06 |
| R356A | 0.49 ± 0.03 | 0.227 ± 0.007 |
| R356K | 0.29 ± 0.03 | 0.209 ± 0.008 |
| R356M | 0.56 ± 0.04 | 0.024 ± 0.001 |
| R356E | ND | <3·10−4 |
| K282A | 3.9 ± 1.3 | 0.005 ± 0.001 |
Possibly, the influence of the bicarbonate ion on kcat is stronger in the hydrolysis of natural peptide substrates than it is in cleaving l-leucine p-nitroanilide, which contains an activated leaving group. Nevertheless, these data show that the turnover number of PepA is increased at least 8-fold in the presence of bicarbonate. This value might well be larger, because a complete absence of bicarbonate was not achieved in these activity assays.
Mutations of Arg-356.
Arg-356 is located next to the bicarbonate ion (Fig. 1). Mutations of Arg-356 demonstrate that this residue contributes to catalysis, but it is not essential (Table 1). Interestingly, the mutants R356A and R356K have approximately the same rate constant as the wild-type enzyme in the absence of activation by bicarbonate ions. This result indicates that the role of Arg-356 is indeed to bind an exogenous bicarbonate ion in the positively charged pocket next to the arginine side chain. Replacement of the arginine by a lysine side chain preserves the positive charge of this pocket, but this mutation destroys the optimal hydrogen bonding pattern between the guanidinium group of Arg-356 and the bicarbonate ion. These interactions may be necessary for binding of the anion. Replacement of the arginine side chain by a methionine has a larger effect on kcat, which is reduced about 130-fold compared with the activated wild-type enzyme. Presumably, the relatively bulky and hydrophobic methionine side chain distorts the active-site structure. Replacement of Arg-356 by a glutamate side chain yielded an enzyme variant whose activity is greatly diminished, but in the range of substrate concentration used for the activity tests (0.1–2 mM) almost independent of the substrate concentration. This behavior indicates a very low apparent binding constant, which may be caused by an unnatural nonproductive substrate binding mode. We prepared this mutant also to test for the possibility that the carboxylate group of the glutamate side chain might take over the function of the bicarbonate ion and lead to an active enzyme. Possibly, the glutamate side chain is not properly oriented to replace the bicarbonate ion, or the more negative charge of the active site of the R356E variant inactivates the enzyme.
Mutation of Lys-282.
The catalytic activity of PepA is greatly diminished by mutation of Lys-282 to an alanine (Table 1). In addition, the binding affinity is reduced about 20-fold. Thus, the second-order rate constant (kcat/Km) is reduced about 10,000-fold from 13.6 mM−1⋅s−1 to 1.3⋅10−3 mM−1⋅s−1. In the structure of LAP with the transition-state analogue leucinal, this residue is hydrogen bonded to the Zn1-coordinated oxygen atom of the gem-diol group of the inhibitor. In the presumed catalytic mechanism, Lys-282 binds to the carbonyl oxygen of the scissile peptide bond and also stabilizes the transition state (9, 11). The kinetic data are in agreement with a direct interaction of Lys-282 with the substrate and with an important contribution of this residue to the catalytic steps and to transition-state stabilization.
In contrast to the wild-type enzyme, the mutant variants are not activated by concentrations of 1 mM or 10 mM NaHCO3 relative to the activity in degassed buffer. To test the possibility that the wild-type enzymes or the mutants can be activated by acetate ions, which also contain a carboxylate group, the activity of wild-type PepA and the enzyme variants was determined in the presence of 1 mM and 10 mM sodium acetate. None of the enzymes was activated by sodium acetate under these conditions.
Binding of Inhibitors.
In the presence of the transition-state analogue inhibitors l-leucinal and l-leucine phosphonic acid, the bicarbonate ion was found to be replaced by three water molecules (9, 11). Two of these water molecules were found to be at a close distance of only 2.3 Å in both protein-inhibitor complexes. In the light of the well defined densities of the 1.6 and 1.9 Å resolution x-ray structures, this short distance is considered as significant with respect to the coordinate error. The occurrence of such a short oxygen–oxygen distance, which is inconsistent with the presence of two neutral water molecules, can be rationalized by the assumption that one water molecule is deprotonated by the positive potential of the cavity next to Arg-356, such that a bihydroxide ion (H3O2−) is formed. However, this hypothesis might prove difficult to confirm experimentally.
Electrostatic Potential.
Calculation of the electrostatic potential at the enzyme surface shows that the active site has a distinct positive potential (Fig. 3A). This includes the region near the two zinc ions where the gem-diol group of the transition-state analogue inhibitor leucinal binds as well as the binding site for the bicarbonate ion. Fig. 3B shows the distribution of the electrostatic potential at the positions of all crystallographically determined water molecules and the oxygen atoms of the bicarbonate ion. With potentials of 29.1 kT/e (for OA), 38.1 kT/e (OB), and 21.4 kT/e (OC), all three oxygen atoms of the bicarbonate ion are at a potential that is significantly more positive than that of most water molecules. Thus, the negatively charged bicarbonate ion probably has a large binding affinity to this pocket next to Arg-356.
In the presence of the inhibitor leucinal bound to LAP, the three water molecules that replace the bicarbonate ion have even more positive potentials of 33.9 kT/e, 45.8 kT/e and 17.3 kT/e (for the waters next to OA, OB, and OC in the bicarbonate-bound uncomplexed enzyme, respectively). These large positive potentials might lower the pKa value of water molecules A and B to about 7 such that they are deprotonated to a bihydroxide ion at neutral pH. Continuum electrostatic calculations indicate that such large pKa-shifts of about 7 pKa units are possible for these water molecules. However, the magnitude of the calculated pKa shifts for these residues close to the interface between low and high dielectric regions shows a very strong dependence on the dielectric constant assigned to the protein region. Therefore, these data are not included here.
Comparison to Other Dizinc Peptidases.
The two related aminopeptidases of Aeromonas proteolytica (25) and Streptomyces griseus (26) also contain a dizinc center in their active sites. The catalytic sites of these aminopeptidases are not homologous to LAP. However, the active sites may be superimposed based on the coordinates of the two metal ions and the metal-bridging water molecules (superposition not shown). Interestingly, this superposition not only maps the hydrophobic binding pockets for the N-terminal side chains of the substrates but also the carboxylate group of Glu-151 in Aeromonas aminopeptidase onto the bicarbonate ion of LAP. Possibly, these two residues have similar functions in the catalytic mechanisms of the two nonhomologous aminopeptidases. Glu-151 in A. proteolytica is proposed to function as a general base in accepting a proton from the zinc-bound nucleophile (27).
A Functional Role for the Bicarbonate Ion.
Although the presence of a bicarbonate ion in the crystal of unliganded LAP was initially believed to be an artifact of the crystallization conditions (9), there is now convincing evidence that this ion indeed has a functional role in the enzyme mechanism. First, PepA is significantly activated by physiological concentrations of bicarbonate, e.g., which are present in the cell from dissolved carbon dioxide. However, the presence of the bicarbonate ion is not essential for the proteolytic activity. Second, this ion has been observed in the active sites of PepA and LAP in almost identical orientations. This finding argues against the notion that the binding of the bicarbonate ion is an artifact caused by the crystallization conditions. Third, continuum electrostatic calculations showed that the binding site of the bicarbonate ion has a characteristic positive potential, favoring binding of an anion. Hydrogen bond donor groups from the guanidinium group of Arg-356 and a main chain NH group form a perfect binding pocket for the trigonal planar ion. Fourth, a comparison to other dinuclear and mononuclear zinc peptidases shows that these have proteinogenous carboxylate side chains that are proposed to serve as general bases in the enzyme mechanism. The bicarbonate ion in the active site of LAP is positioned to have a similar role in PepA and LAP.
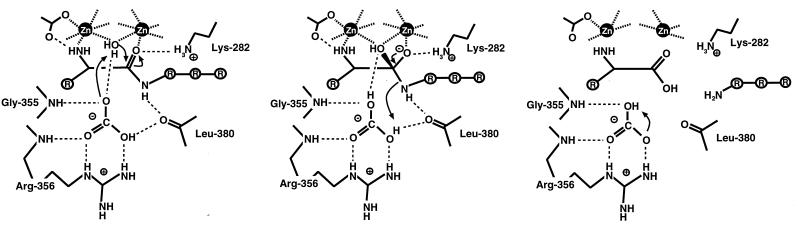
Enzyme Mechanism.
On the basis of results presented in this article, we propose a revised mechanism for peptide hydrolysis by leucine aminopeptidase, in which the active-site bicarbonate ion has the function of a general base that accepts a proton from the zinc-bridging water nucleophile and shuttles the proton to the leaving group (Fig. 4). The two zinc ions are involved in the deprotonation of the nucleophile and together with Lys-282, in the polarization of the substrate carbonyl group and in transition-state stabilization.
Figure 4.
Proposed enzyme mechanism of PepA and LAP. The bicarbonate ion next to Arg-356 (numbering according to PepA) acts as a general base and accepts a proton from the metal-bridging water nucleophile. The proton is shuttled to the leaving peptide amino group to facilitate breakdown of the tetrahedral gem-diol intermediate. However, proton movements may not be unique. For example, water molecules may be involved in the proton transfer paths.
The function of the bicarbonate ion in PepA and LAP is equivalent to the role of glutamate side chains in the active sites of other peptidases containing one or two catalytic zinc ions. In the A. proteolytica aminopeptidase, residue Glu-151 is proposed to accept a proton from the zinc-bound water nucleophile and shuttle it to the leaving amino group of the peptide (27). This mechanism has similarities to our mechanism proposed for LAP and PepA, but differs in the substrate binding mode, where the terminal amino group is not coordinated to one of the zinc ions, but is hydrogen bonded to Glu-151. Furthermore, Chen et al. (27) assume that before nucleophilic attack the bridging water becomes terminally coordinated to one of the zinc ions. Aeromonas aminopeptidase is 80% active in the presence of only one metal ion. The binding mode of transition-state analogue inhibitors to LAP, showing a symmetrical bridging of the two zinc ions by one of the gem-diol oxygens, which is thought to be derived from the attacking nucleophile, argues against the possibility that the water nucleophile becomes terminally coordinated before the nucleophilic attack. Binding of the nucleophile to two metal ions might be expected to reduce its nucleophilicity. However, the type and charge of the coordinating ligands as well as the dielectric constant and electrostatic potential of the protein environment will also influence the pKa and nucleophilicity of the water nucleophile. The net charge of the metal-ligand complex in thermolysin (28) and carboxypeptidase (29) (both Zn2+, 2 His, 1 Glu) is +1 compared with a net charge of +2 in LAP. If one further takes into account that trivalent metal ions serve as efficient catalytic centers in enzymes, such as purple acid phosphatase (30), as well as in nonenzymatic hydrolytic catalysts, it appears likely that other factors from the protein environment and the influence of the metal ligands compensate for this small difference in the net charge. Alternatively, the bridging water ligand might be a hydroxide ion, which is deprotonated to an oxide ion on nucleophilic attack. This has also been discussed for the catalysis of phosphate ester hydrolysis by symmetrical dinuclear metal complexes (31).
Mononuclear zinc peptidases, including carboxypeptidase A (28) and thermolysin (29), also have a glutamate residue near the catalytic zinc ion. Again, the carboxylate group of the side chain is proposed to act as a general base and deprotonate the zinc-bound water nucleophile.
We have previously speculated that the bihydroxide ion, which is proposed to be present in the structures of LAP complexed with leucinal or leucine phosphonic acid, might serve as a general base (2). This now appears to be less likely on the basis of the kinetic results reported in this article. Whereas wild-type PepA is activated by bicarbonate ions, the bicarbonate binding site is destroyed in the mutants R356A and R356M, and these variants are not activated by bicarbonate. However, it also appears unlikely that one of the active-site water molecules is deprotonated in the absence of the positive charge of Arg-356. This suggestion is strongly supported by continuum electrostatic calculations of pKa shifts in the absence of the arginine side chain (data not shown). Because the R356A mutant has an activity that is comparable to that of the wild-type enzyme in the absence of bicarbonate, the possible presence of a bihydroxide ion in the bicarbonate-free active site of wild-type PepA is unlikely to significantly contribute to the activity of this enzyme form.
In conclusion, we have characterized an example for the versatility of enzyme catalysis, here in the catalytic use of an exogenous bicarbonate anion, derived from dissolved carbon dioxide. A similar use of carbon dioxide or bicarbonate ions as catalytic groups in an enzyme mechanism is not known, to our knowledge. Related examples are the carbamylation of lysine side chains as metal ligands and binding of bicarbonate to the iron carrier protein transferrin.
Acknowledgments
We thank David Sherratt for making an overexpression system for PepA and strain DS957 available. Javier Lopez-Jaramillo is acknowledged for many helpful comments. This work was supported by National Institutes of Health grant GM06920 to W.N.L. and by a grant from the Deutsche Forschungsgemeinschaft to N.S.
ABBREVIATIONS
- PepA
aminopeptidase A
- LAP
leucine aminopeptidase
References
- 1.Kim H, Lipscomb W N. Adv Enzymol. 1994;68:153–213. doi: 10.1002/9780470123140.ch4. [DOI] [PubMed] [Google Scholar]
- 2.Sträter N, Lipscomb W N. Chem Rev. 1996;96:2375–2433. doi: 10.1021/cr950042j. [DOI] [PubMed] [Google Scholar]
- 3.Sträter N, Lipscomb W N, Klabunde T, Krebs B. Angew Chem Int Ed Engl. 1996;35:2024–2055. [Google Scholar]
- 4.Sträter N, Lipscomb W N. In: Handbook of Proteolytic Enzymes. Barrett A J, Rawlings N D, Woessner J F, editors. New York: Academic; 1998. pp. 1384–1389. [Google Scholar]
- 5.Hanson H, Frohne M. Methods Enzymol. 1976;45:504–521. doi: 10.1016/s0076-6879(76)45045-0. [DOI] [PubMed] [Google Scholar]
- 6.Beninga J, Rock K L, Goldberg A L. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 7.Pulido-Cejudo G, Conway B, Proulx P, Brown R, Izaguirre C A. Antiviral Res. 1997;36:167–177. doi: 10.1016/S0166-3542(97)00052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burley S K, David P R, Sweet R M, Taylor A, Lipscomb W N. J Mol Biol. 1992;224:113–140. doi: 10.1016/0022-2836(92)90580-d. [DOI] [PubMed] [Google Scholar]
- 9.Sträter N, Lipscomb W N. Biochemistry. 1995;34:14792–14800. doi: 10.1021/bi00045a021. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Lipscomb W N. Biochemistry. 1993;32:8465–8478. doi: 10.1021/bi00084a011. [DOI] [PubMed] [Google Scholar]
- 11.Sträter N, Lipscomb W N. Biochemistry. 1995;34:9200–9210. doi: 10.1021/bi00028a033. [DOI] [PubMed] [Google Scholar]
- 12.Vogt V M. J Biol Chem. 1970;245:4760–4769. [PubMed] [Google Scholar]
- 13.Stirling C J, Colloms S D, Collins J F, Szatmari G, Sherratt D J. EMBO J. 1989;8:1623. doi: 10.1002/j.1460-2075.1989.tb03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stirling C J, Stewart G, Sherratt D J. Mol Gen Genet. 1988a;214:80–84. doi: 10.1007/BF00340183. [DOI] [PubMed] [Google Scholar]
- 15.McCulloch R, Burke M E, Sherratt D J. Mol Microbiol. 1994;12:241–251. doi: 10.1111/j.1365-2958.1994.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 16.Brooks S P J. BioTechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 17.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Sträter N, Sherratt D J, Colloms S D. EMBO J. 1999;18:4513–4522. doi: 10.1093/emboj/18.16.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 21.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls A, Sharp K, Honig B H. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 23.Gilson M K, Sharp K A, Honig B H. J Comp Chem. 1987;9:327–335. [Google Scholar]
- 24.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 25.Chevrier B, Schalk C, D’Orchymont H, Rondeau J M, Moras D, Tarnus C. Structure (London) 1994;15:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 26.Greenblatt H M, Almog O, Maras B, Spungin-Bialik A, Barra D, Blumberg S, Shoham G. J Mol Biol. 1997;265:620–636. doi: 10.1006/jmbi.1996.0729. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Edwards T, D’souza V M, Holz R C. Biochemistry. 1997;36:4278–4286. doi: 10.1021/bi9618676. [DOI] [PubMed] [Google Scholar]
- 28.Christianson D W, Lipscomb W N. Acc Chem Res. 1989;22:62–69. [Google Scholar]
- 29.Matthews B W. Acc Chem Res. 1988;21:333–340. [Google Scholar]
- 30.Sträter N, Klabunde T, Tucker P, Krebs B. Science. 1995;268:1489–1492. doi: 10.1126/science.7770774. [DOI] [PubMed] [Google Scholar]
- 31.Williams N H, Takasaki B, Wall M, Chin J. Acc Chem Res. 1999;32:485–493. [Google Scholar]