Abstract
Cyclin-dependent kinase 5 (cdk5) is found in an active form only in neuronal cells. Activation by virtue of association with the cyclin-like neuronal proteins p35 (or its truncated form p25) and p39 is the only mechanism currently shown to regulate cdk5 catalytic activity. In addition to cyclin binding, other members of the cdk family require for maximal activation phosphorylation of a Ser/Thr residue (Thr160 in the case of cdk-2) that is conserved in all cdks except cdk8. This site is phosphorylated by cdk-activating kinases, which, however, do not phosphorylate cdk5. To examine the possible existence of a phosphorylation-dependent regulatory mechanism in the case of cdk5, we have metabolically labeled PC12 cells with 32Pi and shown that the endogenous cdk5 is phosphorylated. Bacterially expressed cdk5 also can be phosphorylated by PC12 cell lysates. Phosphorylation of cdk5 by a PC12 cell lysate results in a significant increase in cdk5/p25 catalytic activity. Ser159 in cdk5 is homologous to the regulatory Thr160 in cdk2. A Ser159-to-Ala (S159A) cdk5 mutant did not show similar activation, which suggests that cdk5 is also regulated by phosphorylation at this site. Like other members of the cdk family, cdk5 catalytic activity is influenced by both p25 binding and phosphorylation. We show that the cdk5-activating kinase (cdk5AK) is distinct from the cdk-activating kinase (cyclin H/cdk7) that was reported previously to neither phosphorylate cdk5 nor affect its activity. We also show that casein kinase I, but not casein kinase II, can phosphorylate and activate cdk5 in vitro.
Cyclin-dependent kinase 5 (cdk5) is a serine/threonine kinase present at relatively high levels in neurons (1, 2). On the basis of sequence similarity, it is a member of the cyclin-dependent kinase (cdk) family. Full activation of cyclin-dependent kinases characteristically requires combination with one of the cyclins and also phosphorylation of a threonine or serine residue located on the cdk T loop by a cdk-activating kinase. These two events are considered to act cooperatively to open the catalytic site and relieve its obstruction by the T loop (3–6). However, cdk5 is not activated by cyclins, and activation by phosphorylation of its T loop serine has not been demonstrated previously (3). Activation of cdk5 results instead from combination with cdk5-specific activators that include p35 or its truncated form, p25 (7, 8), and p39 (9). Although cdk5 is expressed in many mammalian tissues, the cdk5 activators are largely restricted to neurons. The mechanism of cdk5 interaction with its activators is thought to be analogous to that of cyclin-dependent kinases and forms highly stable and active complexes after prolonged incubation (7–10). Substrates of cdk5 include the neuronal cytoskeletal proteins neurofilament-H (1, 11), tau (12), and MAP (13). However, the embryonic lethality of cdk-5 “knockout” mice suggests additional important functions for this enzyme (14).
Because cdk-activating kinase (cyclin H/cdk7) has been found not to phosphorylate cdk5 or regulate its activity, it was concluded that phosphorylation does not play a role in cdk5 activation (15). However, compared with other cdks, bacterially expressed cdk5 requires prolonged incubation with p25 to achieve maximal activation (N.D.A., R.W.A., and H.C.P., unpublished data). cdk5 substrate phosphorylation rates are relatively low compared with the rates of other cdks, and the T loop phosphorylation site is conserved in cdk5. These observations motivated us to reexamine the possibility that some kinase(s) present in neurons may further activate cdk5.
EXPERIMENTAL PROCEDURES
All fine chemicals were purchased from Sigma. Casein kinase Iδ (CKIδ), CKII, and their peptide substrates were purchased from New England Biolabs. [γ-32 P]ATP and [32P]phosphoric acid were purchased from Dupont/NEN. The glutathione-Sepharose beads are a product of Pharmacia. The cdk5 polyclonal antibody (C-8) was from Santa Cruz Biotechnology, and the cdk5 mAb, Ab-2, was from Oncogene.
Cell Culture and Metabolic Labeling.
PC12 cells were cultured on collagen-coated dishes with DMEM supplemented with 10% heat-inactivated horse serum and 5% FBS as described earlier (16). For metabolic labeling with 32Pi, cells were maintained in a phosphate-free DMEM for 2 hr before adding 2 mCi/ml [32P]phosphoric acid. After a 3-hr incubation the cells were washed three times with cold PBS before lysis.
Western Blot Analysis and Immunoprecipitation.
Twenty micrograms of cell lysate (prepared as described below) was electrophoresed on 10–20% denaturing gels (Novex). After transferring the proteins to poly(vinylidene difluoride) membranes, immunoblot analysis was performed by using a mAb for cdk-5 (Ab-2) and alkaline phosphatase-labeled secondary antibodies by using standard procedures. For immunoprecipitation, 100 μg of the cell lysate was incubated with a complex of (protein A+G)-agarose beads and a cdk5 antibody (C-8) for 2 hr at room temperature. The beads were washed with Tris-buffered saline three times for 15 min each before suspending in 100 μl of kinase assay buffer (see below).
Expression and Purification of Proteins.
Glutathione S-transferase (GST)-cdk5 in pGEX 2TK was constructed by putting the BamHI fragment of His-tag-cdk5 (8) into BamHI-cut pGEX 2TK. GST-p25 in pGEX4T-2 was constructed by a PCR method by using the oligonucleotides 5′-GTCCGGATCCGCCCAGCCCCCGCCG-3 as a forward primer, 5′-GTGATGAATTCTGGATCACCGATC-3′ as a reverse primer, and DNA from the His-tag-p35 construct as a template (8). The PCR products were digested with BamHI and EcoRI, and the resulting fragments were cloned into BamHI-EcoRI-cut pGEX 4T-2. The proteins were expressed as described earlier (7, 8). Purification of GST-fusion proteins was carried out by using glutathione-Sepharose affinity chromatography by standard procedures (Pharmacia) as described earlier (7, 8). To remove the GST tags, the fusion proteins were treated with thrombin for 12 h at 27°C. Cleaved proteins were recovered from the supernatants of the GST-Sepharose precipitates. The purity of the proteins was assessed by SDS/PAGE and Coomassie blue staining of the gels. The protein concentration was estimated by densitometric analysis of these gels, by using GST as standard.
Site-Directed Mutagenesis.
The mutations in cdk5 were made by using a commercial kit (Quick Change; Stratagene). In brief, a pair of complementary primers of 30–40 bases were designed with the desired mutations placed in the middle of the sequence. Parental cDNA inserted in pGEX2TK was amplified by using Pfu DNA polymerase with these primers for 15 cycles in a DNA thermal cycler (Perkin–Elmer). After digestion of the parental DNA with DpnI, the mutants were transformed into Escherichia coli (DH5α strain; GIBCO/BRL). The mutations were confirmed by DNA sequencing. The GST-fusion mutant proteins were expressed and purified as described above.
Phosphorylation and Activation of cdk5.
For cdk5-activating kinase assays, PC12 cell lysates were prepared by homogenization of the PC12 cells in a buffer containing 50 mM Tris⋅HCl, pH 7.4/1 mM EDTA/0.1% NP-40 detergent/protease inhibitor mixture (Boehringer)/50 mM β-glycerophosphate/50 mM sodium fluoride/0.1 mM okadaic acid. The lysates were clarified by centrifugation at 20,000 × g for 1 h and used as a source of cdk5-activating kinase. Cell lysate (10 μl), casein kinase I, or casein kinase II each were used to phosphorylate bacterially expressed GST-cdk-5 or GST-S159A in the presence or absence of GST-p25 (1:1.5 molar ratio). In some cases GST-p25 was incubated with GST-cdk5 or the mutant GST-S159A-cdk5 for 3 h before starting the phosphorylation reaction by the addition of ATP. The reaction mixture, which contained 50 mM Tris, pH 7.4/5 mM MgCl2/1 mM EGTA/2 mM ATP (nonradioactive) in a final volume of 50 μl, was incubated for 1 h at 30°C. Modified GST-cdk5 or GST-p25/cdk5 complex was recovered by adsorption on glutathione-Sepharose beads followed by three washes with lysis buffer and two washes with PBS; these glutathione-Sepharose beads were suspended in kinase assay buffer and used for assaying cdk5 activity (see below).
Essentially a similar procedure was followed in experiments in which phosphate incorporation in cdk5 or S159A-cdk5 was monitored except that 100 μM [γ32-P]ATP was used instead of a nonradioactive ATP.
Phosphorylation Assay for Measuring cdk5 Activity.
Standard assay mixtures contained 50 mM Tris, pH 7.4/2.5 mM MgCl2/1 mM EGTA/10 μl of GST-cdk5 or GST-cdk5/GST-p25 adsorbed to glutathione-Sepharose beads, 100 μM [γ-32P]ATP, and 5 μg of histone H1 in a total volume of 50 μl. Reactions were initiated by adding [γ-32P]ATP and were carried out at 30°C for 40–80 min. The reactions were terminated by boiling samples in Laemmli’s buffer. The phosphate incorporation in histone was visualized by autoradiography of the SDS/PAGE gels. For quantitation of the phosphorylation, densitometric analysis of the autoradiographic films was performed by using nih image software.
RESULTS
Examination of the cdk5 Phosphorylation State in PC12 Cells.
PC12 cells were metabolically labeled with 32Pi for 1 h, and the cdk5 was immunoprecipitated from the labeled cells by using a specific cdk5 antibody.
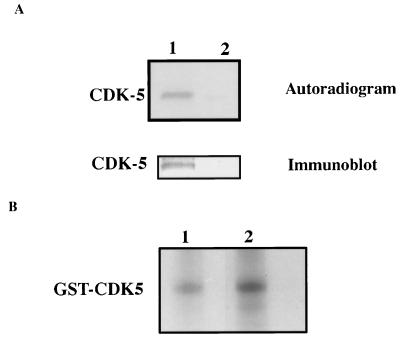
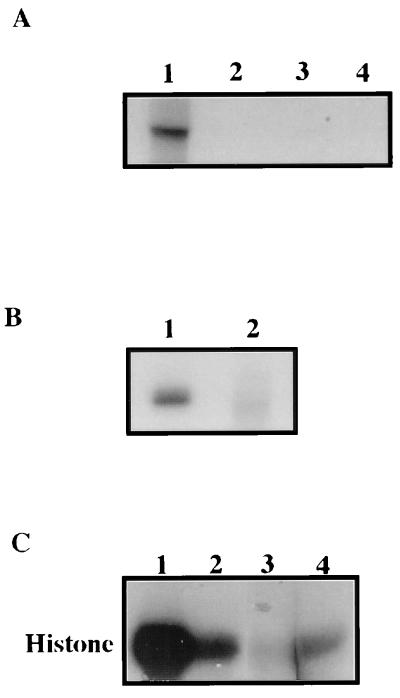
The cdk5 immunoprecipitate was electrophoresed and blotted onto a poly(vinylidene difluoride) membrane. The autoradiogram of the blot revealed a 33-kDa labeled band corresponding to cdk5 (Fig. 1A, lane 1). The band was confirmed to contain cdk5 when the same blot was probed with a specific antibody (Fig. 1A). Thus, cdk5 appears to be phosphorylated in PC12 cells.
Figure 1.
(A) Endogenous cdk5 is phosphorylated in PC12 cells. PC12 cells were metabolically labeled with 32Pi. Immunoprecipitated cdk5 from the 32P-labeled cell lysates of the labeled cells was electrophoresed and blotted onto poly(vinylidene difluoride) membrane. The autoradiogram of the membrane shows a band corresponding to the molecular weight of cdk5 (lane 1). The unbound supernatant of the immunoprecipitate is loaded in lane 2. The same membrane when immunoblotted with the cdk5 antibody recognized the 32P-labeled band. (B) Bacterially expressed GST-cdk5 is phosphorylated by PC12 lysate. Equal amounts of GST-cdk5 were incubated without (lane 1) or with (lane 2) GST-p25 before phosphorylation with the PC12 cell lysate. Glutathione-Sepharose beads were used to separate cdk5 and cdk5/p25 from the lysate after phosphorylation and electrophoresed. The autoradiogram indicates a higher level of cdk5 phosphorylation in the presence of p25 (lane 2).
Phosphorylation of Bacterially Expressed cdk5 by PC12 Cell Lysate.
If, as indicated by the preceding data, cdk5 in PC12 cells can exist in a phosphorylated form in vivo, a cdk5-phosphorylating kinase also may exist in PC12 cells. To examine this possibility, a PC12 cell lysate was assayed for its ability to phosphorylate bacterially expressed cdk5. Fig. 1B confirms that expressed cdk5 can be phosphorylated by a PC12 cell lysate with a stoichiometry of 0.4 pmol phosphate/pmol GST-cdk5. When complexed with p25, cdk5 showed more phosphate incorporation (1.5-fold) under otherwise similar conditions. Similar observations have been made in studies of cdk/cyclin systems (18, 19).
Effect of Phosphorylation on cdk5 Catalytic Activity.
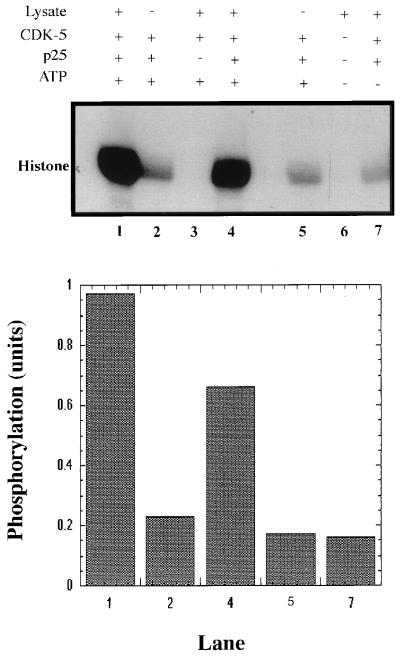
It has been observed in our laboratory (N.D.A., R.W.A., and H.C.P., unpublished data) that cdk5 requires several hours of preincubation with p25 to attain maximal activation. Other cdk/cyclin complexes appear to be more rapidly activated (18, 19). However, these kinases require phosphorylation for stabilization of their cyclin complex as well as for activation of catalytic activity. To probe for a similar effect in the case of p25/cdk5, we compared the activity of p25/cdk5 complexes formed during a 3-h incubation before addition of PC12 lysate and nonradioactive ATP with that of similar p25/cdk5 samples that were not incubated before addition of the cell lysate and ATP by using a histone kinase assay. The phosphorylated cdk5/p25 complex (Fig. 2, lane 1) was 5-fold more active than the unphosphorylated complex (lane 2).This suggests that cdk5 phosphorylation is required for maximal activation of the p25/cdk5 complex.
Figure 2.
Effect of phosphorylation of cdk5 on its catalytic activity. Equal amounts of GST-cdk5 and a molar excess of GST-p25 were incubated with (+) or without (−) PC12 lysate in the presence (+) or absence (−) of 2 mM ATP (nonradioactive) at 30°C for 1 h. GST-cdk5 samples in lanes 1, 2, and 7 were preincubated with GST-p25 for 3 h and were not preincubated in lanes 4 and 5. Control experiments in the absence of GST-p25 (lane 3) and in the absence of GST-cdk5/GST-p25 (lane 6) also were performed. The fusion proteins were recovered on glutathione-Sepharose beads, washed, and assayed for histone kinase activity. The radiolabeled products were electrophoresed and detected by autoradiography. (Upper) Representative of three experiments. (Lower) The quantitation of phosphorylation by densitometric analyses of the corresponding lanes in Upper.
Cdk5 in the absence of p25 does not show catalytic activity (Fig. 2, lane 3) with or without incubation with PC12 lysate and ATP, thus confirming that activator binding is essential for cdk5 activity with or without phosphorylation. This also rules out the possibility that nonspecific effects might occur because of the interaction of PC12 lysate proteins with the GST-cdk5-Sepharose precipitate. Surprisingly, in the presence of p25 but without preincubation, the phosphorylated cdk5 was almost as active (lane 4) as the precomplexed, phosphorylated cdk5/p25 (lane 1). In this case, cdk5 and p25 were incubated together only during the 1-h phosphorylation of cdk-5, whereas the preincubated complex was formed during 3 h of incubation before the phosphorylation reaction. It appears from these data that phosphorylated cdk5 does not require prolonged preincubation with p25 to attain high catalytic activity.
Because the kinase source used to phosphorylate cdk5 was a crude PC12 cell lysate, it was important to rule out the possibility that association of cdk5 with nonkinase cofactors could activate it. Therefore, in a control experiment the cdk5-activating kinase assay was mimicked in the absence of ATP (Fig. 2, lane 7). The cdk5/p25 complex from this experiment showed low catalytic activity similar to unphosphorylated cdk5 (lane 2), which demonstrates that the PC12-lysate activation of cdk5/p25 is, in fact, ATP-dependent.
Effect of cdk5 Phosphorylation on Its Interaction with p25.
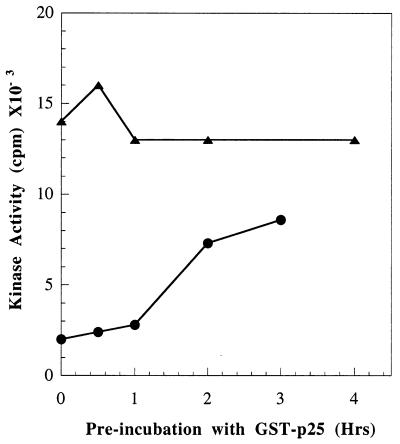
It has been observed that the rate of activation of bacterially expressed cdk5 by p25 is slow (N.D.A., unpublished data). To examine the mechanism that produces the dramatic increase in p25/cdk5 activity described above, we examined the effect of cdk5 phosphorylation on the time course of its interaction with p25. Consistent with the previous observations of Amin et al., p25 must be incubated with cdk5 for more than 3 h to achieve maximal activation of cdk5 (Fig. 3). In contrast, phosphorylated cdk5 is maximally activated without preincubation with p25 before the 40-min kinase assay. When assayed without preincubation, phosphorylated cdk5 is about 8-fold more active than unphosphorylated cdk5. In the experiment of Fig. 2, cdk5 was phosphorylated by the PC12 lysate for 1 h. We later determined that maximal phosphorylation of cdk5 by this lysate required 4 h (data not shown). Therefore, in the experiment of Fig. 3, when we used maximally phosphorylated cdk5, we found even more activation (8-fold). These data suggest that a principal effect of cdk5 phosphorylation is to markedly increase its rate of activation by p25, whereas its Vmax is not markedly changed.
Figure 3.
Effect of cdk5 phosphorylation on the rate of its activation by p25. GST-cdk5 was phosphorylated by a PC12 lysate (4-h incubation time), followed by adsorption on glutathione-Sepharose and washing as described in Experimental Procedures. Parallel control samples were carried through the procedure in the absence of lysate. Aliquots of the GST-coupled phosphorylated (▴) or unphosphorylated (●) cdk5 samples were preincubated with GST-p25 for different times followed by measurement of histone kinase activity as described in Experimental Procedures.
Relevance of Ser159 to cdk5 Activation by Phosphorylation.
Activating phosphorylation of most cdks occurs on a conserved serine/threonine residue in the T loop, which causes, in the case of cdk2, further movement of the T loop. This phosphoserine forms ionic interactions with several basic residues in cdk2, thereby further stabilizing the T loop displacement (6). Because cdk5 has very high sequence homology with other cdks (≈70%), it is reasonable to assume that the structural basis of its activation may be similar.
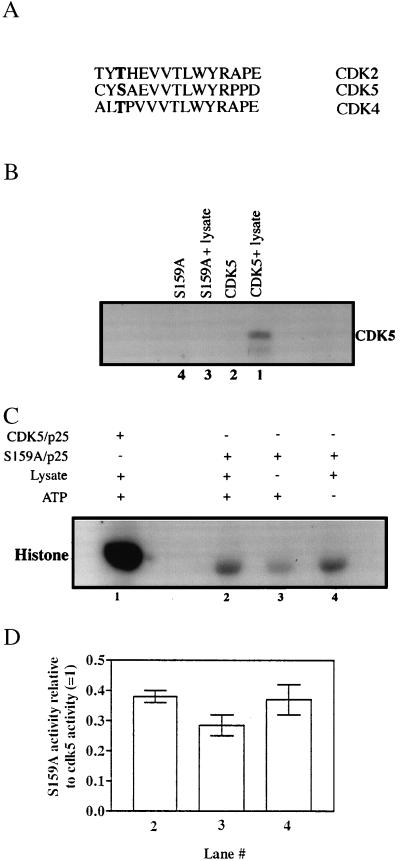
We mutated Ser159 to Ala (Fig. 4A) to produce a cdk5 that cannot be phosphorylated on its T loop (S159A-cdk5). The low level of phosphorylation of S159A-cdk5 compared with wild-type cdk5 suggests that Ser159 is the major site of cdk5 phosphorylation by a PC12 cell lysate (Fig. 4B).
Figure 4.
(A) Alignment of the activation-loop residues of various cdk kinases showing conservation of the phosphorylatable regulatory residue in boldface. (B) Effect on cdk5 phosphorylation of mutation of Ser159 to Ala. Equal amounts of GST-cdk5 (lane 1) or GST-S159A (lane 3) mutant were subjected to phosphorylation by the PC12 cell lysate and the modified proteins were recovered as described in Fig. 1. The GST-fusion tags were cleaved with thrombin and clarified by using glutathione-Sepharose beads before SDS/PAGE analysis. The radiolabeled products were detected by autoradiography. (C) Histone kinase activity of S159A. Equal amounts of GST-cdk5 or GST-S159A were preincubated with GST-p25 (molar excess) for 3 h before incubation with (lanes 1 and 2) or without (lane 3) PC12 cell lysate in presence (+) or absence (−) of nonradioactive ATP. Glutathione-Sepharose beads were used to separate the fusion proteins and used to phosphorylate histone. The radiolabeled histone was detected by autoradiography. Lane 1 is the same experiment done with wild-type GST-cdk5/p25 under similar conditions. (D) Quantitation of the data for experiments described in Fig. 4C by densitometric analyses of the 32P-labeled histone bands. The histogram represents the average of results obtained from two independent experiments (shown as bars) expressed relative to the activity of wild-type cdk5/p25. Lane numbers correspond to those in Fig. 4C. Statistical analysis by using the two-tailed t test did not reveal any significant differences between the data for lanes 2 and lane 4 vs. lane 3 (P > 0.1 and P > 0.2, respectively). The activity of wild-type cdk5 was taken as 1 (lane 1).
As described earlier for wild-type enzyme, S159A-cdk5 also was tested for the effect of incubation with PC12 lysate on histone kinase activity (Fig. 4 C and D). The apparent histone kinase activity of the S159A/p25 incubated with the lysate and ATP (lane 2) was similar to that of S159A/p25 incubated with the lysate in the absence of ATP (lane 4). Correlating with the lack of a phosphorylation site, S159A-cdk5 activity is not regulated by a cdk5-activating kinase in the lysate. A small decrease in the activity of the S159A/p25 not incubated with the lysate (lane 3) was observed. This statistically insignificant decrease (Fig. 4D) may be due to the presence of some endogenous cdk5/p25 complex in the PC12 lysate that bound to the GST-cdk5/p25 and contributed to the slightly higher levels of histone phosphorylation seen in lanes 2 and 4 (Fig. 4C). Phosphorylated cdk5/p25 exhibits much higher activity than S159A/p25 (lanes 1 vs. lanes 2, 3, or 4), confirming that phosphorylation of Ser159 is essential for maximal catalytic activity of cdk-5 in the 60-min histone kinase assay.
Actions of Casein Kinases on cdk5 in Vitro.
The T loop Ser159 of cdk5 is followed by alanine and glutamate residues (SAE, Fig. 5A). This motif is a potential target for both casein kinase I and casein kinase II phosphorylation (20–22). Therefore, we tested the ability of active recombinant CKI and CKII to phosphorylate bacterially expressed cdk5 in vitro. Although CKII did not phosphorylate cdk5 significantly, cdk5 is a good substrate for CKI (Fig. 5A, lane 1) with a stoichiometry of 0.6 pmol PO4/pmol of GST-cdk5. In contrast to the GST-cdk5, no significant phosphorylation of GST-S159A mutant was observed (Fig. 5B). This suggested that Ser159 is also the major phosphorylation site for CKI on CDK5. To see whether CKI-mediated phosphorylation regulates its activity, GST-cdk5 was phosphorylated by CKI as described in Experimental Procedures and separated from CKI by adsorption to glutathione-Sepharose beads. Upon mixing with GST-p25, phosphorylated cdk5 was significantly more active than the unphosphorylated cdk5 (Fig. 5C, lane 1 compared with lane 2). Moreover, CKI phosphorylation did not activate the GST-S159A mutant, suggesting that CKI mediates phosphorylation at Ser159 of cdk5 and is stimulatory (Fig. 5C, lane 4). Thus, CKI can act as a cdk5-activating kinase in vitro.
Figure 5.
CKI phosphorylates and activates cdk5 in vitro. (A) Equal amounts of GST-cdk5 were subjected to phosphorylation by CKI (lane 1) and CKII (lane 3). GST-cdk5 was separated from the assay mix by using glutathione-Sepharose beads. SDS extracts of these beads were subjected to electrophoresis and autoradiography. Lanes 2 and 4 show that cdk5 is not phosphorylated in absence of CKI or CKII. (B) Equal amounts of CKI were used to phosphorylate equal amounts of either GST-cdk5 (lane 1) or GST-S159A (lane 2) and detected as above. (C) Samples of GST-cdk5 (lanes 1 and 2) and GST-S159A (lane 4) after phosphorylation by CKI (lane 1) (using nonradioactive ATP) each were adsorbed onto glutathione-Sepharose beads and incubated with GST-p25 for 10 min before performing histone phosphorylation assays. Lane 3 represents a control without any GST-cdk5. The radiolabeled histone was visualized by autoradiography of the protein gels.
DISCUSSION
Cyclin binding to cdk kinases results in some kinase activity, but full activation is achieved only when cdks also are phosphorylated by a cdk-activating kinase (CAK) at a single threonine residue on the T loop (reviewed in ref. 3). The activities of complexes formed by cdc2 or cdk2 with cyclin A or B are regulated both positively and negatively by phosphorylation of the bound catalytic subunits (3). Some cyclin pairs, such as human cdc2-cyclinB, cdk2-cyclin A, and cdk2-cyclin E, interact with high affinity in the absence of other modifications (19). In contrast, cdc2-cyclin A and cdk7-cyclin H do not bind tightly unless the cdk subunit is phosphorylated at the T loop threonine residue (18). Cyclin D binding to cdk4 is not influenced by phosphorylation, but phosphorylation by a CAK is required for catalytic activity (23).
The cdk5 protein is widely expressed but cdk5 activity occurs principally in brain and few other tissues. This tissue specificity has been attributed to the restricted expression of its cyclin-like activators (7, 8). Because the cdk7/cyclin H complex (CAK), which phosphorylates most other cdk kinases, does not phosphorylate cdk5, it was suggested that phosphorylation does not play a role in cdk5 activation (15). The present findings demonstrate that phosphorylation dramatically increases cdk5 activation.
Cdk2 phosphorylated at the conserved phosphorylation site (Thr160) is structurally similar, in this region, to phosphorylated PKA (6). This region shows a further movement of the T loop toward the kinase C lobe as a result of interaction between the phosphate and some acidic residues, and the catalytic cleft opens even more, thereby increasing the kinase activity. This regulatory phosphorylation site is well conserved in cdks and other protein kinases. The lack of significant phosphorylation of the cdk5 S159A mutation suggests that this serine residue is also the major phosphorylation target for cdk5-activating kinases. It appears that cdk5 follows a regulatory mechanism similar to other cdk kinases, although no studies of the rates of cyclin binding to cdk kinases have been reported.
We show here that CKI, but not CKII, can phosphorylate and activate cdk5 in vitro. It is possible that the shape of the T loop (containing Ser159) is more conducive for interaction with CKI than with CKII. However, CKI phosphorylation and regulation of cdk5 in vivo remains to be established. CKI and cdk5 have been found to be constituents of a multimeric complex in squid axon (24). CKI also is associated with neurofilaments (25) that are also substrates for cdk5 (1, 11, 17).
Clearly, these observations challenge the previously held belief that phosphorylation does not play a role in cdk5 activation. It seems cdk5 requires both p35 binding and phosphorylation at Ser159 for maximal rates of activation. The abnormal lethality of cdk5 “knockout” mice suggests important functions of this kinase system (14). We currently are investigating the identity of cdk5-phosphorylating kinase(s) in neurons, which will be helpful in understanding the regulatory mechanism of this kinase system at the cellular level.
Acknowledgments
We thank Dr. S. Fenella DasGupta for help in cell labeling experiments and Drs. Ram Sihag, Phil Grant, and Hal Gainer for their suggestions regarding the manuscript. DNA sequencing done by Jim Neagle, DNA-sequencing facility, National Institute of Neurological Disorders and Stroke, National Institutes of Health, is acknowledged.
ABBREVIATIONS
- cdk
cyclin-dependent kinase
- GST
glutathione S-transferase
- CK
casein kinase
References
- 1.Shetty K T, Link W T, Pant H C. Proc Natl Acad Sci USA. 1993;90:6844–6848. doi: 10.1073/pnas.90.14.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellmich M R, Pant H C, Wada E, Battey J F. Proc Natl Acad Sci USA. 1992;89:10867–10871. doi: 10.1073/pnas.89.22.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 4.De Bondt H L, Rosenblatt J, Jancarik H D, Morgan D O, Kim S H. Nature (London) 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 6.Russo A A, Jeffrey P D, Pavletich N P. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 7.Lew J, Huang Q Q, Qi Z, Winkfein R J, Aebersold R, Hunt T, Wang J H. Nature (London) 1994;371:423–426. doi: 10.1038/371423a0. [DOI] [PubMed] [Google Scholar]
- 8.Tsai L H, Delalle I, Caviness V S, Chae T, Harlow E. Nature (London) 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- 9.Tang D, Yeung J, Lee K Y, Matasushita M, Matsui H, Tomizawa H, Hatase O, Wang J H. J Biol Chem. 1995;270:26897–26903. doi: 10.1074/jbc.270.45.26897. [DOI] [PubMed] [Google Scholar]
- 10.Nikolic M, Dudek H, Kwon Y T, Ramos Y F, Tsai L H. Genes Dev. 1996;10:816–825. doi: 10.1101/gad.10.7.816. [DOI] [PubMed] [Google Scholar]
- 11.Pant A C, Veeranna, Pant H C, Amin N. Brain Res. 1997;765:259–266. doi: 10.1016/s0006-8993(97)00561-1. [DOI] [PubMed] [Google Scholar]
- 12.Paudel H K, Lew J, Ali Z, Wang J H. J Biol Chem. 1993;268:23512–23518. [PubMed] [Google Scholar]
- 13.Pigino G, Ulloa L, Avila J, Caceres A. J Cell Sci. 1997;110:257–270. doi: 10.1242/jcs.110.2.257. [DOI] [PubMed] [Google Scholar]
- 14.Ohshima T, Ward T M, Huh C G, Longenecker G, Veeranna, Pant H C, Brady R O, Martin L J, Kulkarni A B. Proc Natl Acad Sci USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon R Y C, Lew J, Hunter T. J Biol Chem. 1997;272:5703–5708. doi: 10.1074/jbc.272.9.5703. [DOI] [PubMed] [Google Scholar]
- 16.Yan G Z, Ziff E B. J Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma M, Sharma P, Pant H C. J Neurochem. 1999;73:79–86. doi: 10.1046/j.1471-4159.1999.0730079.x. [DOI] [PubMed] [Google Scholar]
- 18.Desai D, Gu Y, Morgan D. Mol Cell Biol. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Z Q, Amin A, Hurwitz J. J Biol Chem. 1993;271:1538–1543. [Google Scholar]
- 20.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Fihol O, Cochet C, Brickey D A, Soderling T R, Bartheson C, Graves D J, DeMaggio A J, et al. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreegipuu A, Blom N, Brunak S, Jarv J. FEBS Lett. 1998;430:45–50. doi: 10.1016/s0014-5793(98)00503-1. [DOI] [PubMed] [Google Scholar]
- 22.Marin O, Meggio F, Sarno S, Andretta M, Pinna L A. Eur J Biochem. 1994;223:647–653. doi: 10.1111/j.1432-1033.1994.tb19037.x. [DOI] [PubMed] [Google Scholar]
- 23.Kato J-Y, Masaaki M, Storm D K, Sherr C J. Mol Cell Biol. 1994;14:2713–2721. doi: 10.1128/mcb.14.4.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takshashi M, Amin N, Grant P, Pant H C. J Neurosci. 1995;15:6222–6229. doi: 10.1523/JNEUROSCI.15-09-06222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Link W T, Dosemeci A, Floyd C C, Pant H C. Neurosci Lett. 1993;151:89–93. doi: 10.1016/0304-3940(93)90053-n. [DOI] [PubMed] [Google Scholar]