If you walk through a laboratory at the Salk Institute, all the people working in it look fairly similar: two legs, a head and so on. However, it is only through a more detailed analysis that one could reveal the differences between the first year graduate student (nervously pipetting) and the well-respected principal investigator (PI) (editing manuscripts). Analogous to this, how can we discriminate diverse neuronal populations (e.g. graduate student versus PI) and even understand their function in the adult brain?
Genome-wide, unbiased gene expression studies using DNA microarrays or serial analyses of gene expression have created a tremendous amount of data defining tissue-specific and even region-specific gene expression in mammals (Lockhart et al, 1996; Datson et al, 2001; Lewandowski and Small, 2005). In other words, the ‘laboratory' (in analogy to organs, e.g., the brain) and maybe even the ‘job description' (e.g., the substructure of the brain) of many genes have been already identified.
In a study recently published in Nature, Lein et al (2007) went one step further and have provided a genome-wide gene expression atlas of the adult mouse brain (the Allen Brain Atlas) using fine-resolution in situ hybridization with which they studied the expression patterns of more than 20 000 genes on a quasi-cellular level. Will this new tool help us to understand the mammalian brain, which shows a tremendous degree of diversity (Muotri and Gage, 2006) and consists, in the case of humans, of approximately 100 billion neurons that represent about 10 000 different neuronal subtypes?
Visualizing unbiased genome-wide gene expression with a cellular resolution will promote the identification of neuronal subtypes, based on specific sets of genes that are either on or off in those neurons. The Allen Brain Atlas will shape our views about traditional, functional neuroanatomy, which have relied almost exclusively on the spatial proximity of cells or on the similarity of neuronal structure. Gene expression analysis on a cellular level has already been instrumental in shifting the common understanding of brain anatomy (Lein et al, 2004). The Allen Brain Atlas will help to identify new, functionally connected cellular entities. The major strength of the Allen Brain Atlas compared to earlier approaches is its unbiased nature and the sheer number of genes it analyzes. Furthermore, web-enabled image analysis tools provided by the Allen Institute will make it possible for every scientific user to search for genes, brain areas, and cell types in a three-dimensional template (www.brain-map.org; Figure 1).
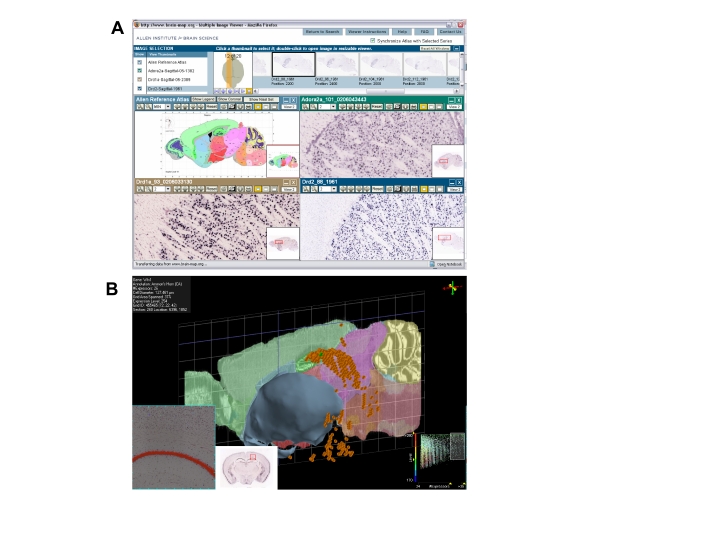
Figure 1.
The Allen Brain Atlas. (A) Screen shot of the web-enabled gene browser (http://www.brain-map.org). In this example, the expression patterns of the adenosine 2A (top right), dopamine D1 (botton left), and dopamine D2 receptors (bottom right) are shown in striatum. (B) Three-dimensional view of the expression of Wfs1 (orange) in hippocampus as provided by the Brain Explorer tool.
The identification of marker genes entails the possibility of using their genomic promoter sequences (e.g., by BAC technology or tissue-specific virus expression) to label or manipulate specific cell types. For instance, one can envisage neuronal subtype-specific promoter-driven marker expression (such as GFP), thus enabling electrophysiological analyses of neuronal subtypes. Cell-type-specific loss- or gain-of-function studies to understand the functional significance of genes of interest (Le and Sauer, 2001) or the specific electrical silencing of neuronal subtypes (Tan et al, 2006) will promote our understanding of brain circuitries and neuronal diversity. The identification of highly specific expressed genes and the use of their promoters are much needed because most of the current approaches deal with a certain degree of nonspecificity that might strongly influence the obtained results.
However, the strategy presented by Lein and colleagues has its drawbacks. The data analyses used by Lein et al require the setting of a certain threshold of gene expression to extract signal from background noise. Even though this threshold appears to be lower than with traditional DNA microarrays, important genes could still be missed owing to their low abundance. The detection of low-abundant but functionally important genes might be better obtained with BAC-based transgenic libraries, such as the NINDS Gensat Bac Transgenic Project (www.gensat.org). The in situ signal needed for the Lein et al strategy also depends on the quality of the probe used to detect the mRNA. This strategy could result in false results (owing to probe nonspecificity or mix ups) and could also—at least in its current form—not systematically discriminate between splice variants. In this regard, it is helpful that primer and gene sequences are available on the website. There is also growing evidence that many genes are functionally regulated on a post-transcriptional level, and not every mRNA is necessarily translated into protein. Therefore, future projects aiming to generate proteomics with a cellular resolution will complement the Allen Brain Atlas. In general, connecting the existing data sets that are designed to generate more functional conclusions, such as the QTL-based Genenetwork (www.genenetwork.org), with the Allen Brain Atlas could potentially lead to a synergistic increase in knowledge. Evidently, the wish list could be endlessly continued, for example to gene and protein expression atlases with a subcellular resolution, expression atlases of splice variants, expression data over the lifespan, or expression atlases of short, uncoding RNAs.
Ultimately, the new genome-wide expression atlas presented by Lein and colleagues will cause a run on genes with exciting expression patterns and will hopefully boost our understanding of connectivity and neuronal diversity. Thus, in the analogy used earlier, we will not only be able to see if the person in the lab is a graduate student or the PI, but also get an idea of the project the person is actually doing right now. However, the visualization of gene and protein expression in the mammalian brain should not stop there. Rather, new strategies will have to be developed that spur on the visualization of genes and gene products, as ‘Science is nothing but perception' (Plato).
Websites: www.brain-map.org—Genome-wide expression atlas of the adult mouse brain www.gensat.org—Quickly growing gene expression database using transgenic mice www.genenetwork.org—QTL-based gene network database
References
- Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E (2001) Expression profile of 30 000 genes in rat hippocampus using SAGE. Hippocampus 11: 430–444 [DOI] [PubMed] [Google Scholar]
- Le Y, Sauer B (2001) Conditional gene knockout using Cre recombinase. Mol Biotechnol 17: 269–275 [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chi Chin M, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Feng Yuan X, Zhang B, Zwingman TA Jones AR (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176 [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH (2004) Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci 24: 3879–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski NM, Small SA (2005) Brain microarray: finding needles in molecular haystacks. J Neurosci 25: 10341–10346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Muotri AR, Gage FH (2006) Generation of neuronal variability and complexity. Nature 441: 1087–1093 [DOI] [PubMed] [Google Scholar]
- Tan EM, Yamaguchi Y, Horwitz GD, Gosgnach S, Lein ES, Goulding M, Albright TD, Callaway EM (2006) Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron 51: 157–170 [DOI] [PubMed] [Google Scholar]