Abstract
The variability and adaptability of the amoebae from the class Dictyosteliomycetes greatly complicate their systematics. The nucleotide sequences of the ribosomal internal transcribed spacers and the 5.8S ribosomal DNA gene have been determined for 28 isolates, and their utility to discriminate between different species and genera has been shown.
Dictyostelids are amoebae, frequently found in soils (9), discovered by Oskar Brefeld in 1869 (4) and grouped with myxomycetes and protostelids in the Eumycetozoa (3, 8). A characteristic common to all dictyostelids is the capacity to form multicellular structures by aggregation (6). The morphogenetic process is very plastic, and variations in several characteristics of the structures have been used to classify these organisms. More than 100 species have been described and grouped in three genera: Acytostelium, Dictyostelium, and Polysphondylium.
Systematics in dictyostelids is difficult and, in some cases, controversial due to the relatively few morphological characteristics available for classification and the large variability and adaptability of their fruiting bodies (7, 9). As a consequence, it is often difficult to ascertain whether two samples pertain to the same species. These problems have even led to the description of species complexes, such as Dictyostelium mucoroides or Polysphondylium pallidum, which group specimens with similar (7, 9), but perhaps not identical, characteristics.
Molecular phylogenetic analyses might add significant information for the systematic classification of dictyostelids. A genomic region frequently used in phylogenetic studies, including ribosomal DNAs (rDNA) and their internal transcribed spacers (ITS) (recently reviewed in references 1 and 13), was analyzed in this study.
Dictyostelids were collected from several locations in the south of Europe (Spain and Portugal) or obtained from other collections, as shown in Table 1. Cells were processed and classified according to Raper (9) using the following morphological criteria.
TABLE 1.
Geographic origins of dictyostelid isolates and GenBank accession numbers of the nucleotide sequences
| Species | Origin | Isolate | GenBank accession no. |
|---|---|---|---|
| Acytostelium leptosomum | Algarve, Portugal | 212 | AM282588 |
| Acytostelium sp. | Tennessee | GCE9 | AM282589 |
| North Carolina | DCB6B | AM282590 | |
| Dictyostelium giganteum | North Carolina | 1880 | AM282591 |
| North Carolina | 1889 | AM282592 | |
| Dictyostelium minutum | Sevilla, Spain | 11C | AM282593 |
| Texas | L4B | AM282594 | |
| Dictyostelium mucoroides | Ciudad Real, Spain | 1C | AM282595 |
| Huelva, Spain | 89C | AM282596 | |
| Madrid, Spain | 115A | AM282597 | |
| Soria, Spain | 133B | AM282598 | |
| Arkansas | AR4B | AM282599 | |
| Dictyostelium sphaerocephalum | Sevilla, Spain | 14A | AM282600 |
| Huelva, Spain | 20B | AM282601 | |
| Huesca, Spain | 88A | AM282602 | |
| Huelva, Spain | 89B | AM282603 | |
| Huelva, Spain | 89G | AM282604 | |
| Madrid, Spain | 118B | AM282605 | |
| Polysphondylium candidum | Ciudad Real, Spain | 3A | AM282606 |
| Ciudad Real, Spain | 7A | AM282607 | |
| Sevilla, Spain | 11F | AM282608 | |
| Sevilla, Spain | 15 | AM282609 | |
| Huelva, Spain | 89A | AM282610 | |
| Huelva, Spain | 89C2 | AM282611 | |
| Huelva, Spain | 89D | AM282612 | |
| Tennessee | PH8B | AM282613 | |
| Polysphondylium pallidum | Huelva, Spain | 23D | AM282614 |
| Ohio | PP1 | AM282615 |
(i) Acytostelium leptosomum Raper.
Sorocarps of small size (750 to 1,500 μm in length), erect, with typical cluster growth. Numerous fructifications (20 in a cluster). Sorophore acellular, no pigmentation, with piliform and very thin tip. Spores globose with polar granules distributed irregularly around them. Aggregations radiate with streams thin and long. Migratory slugs. There is considerable difference between the sorocarps of the first isolates in HI agar and those of pure cultures (smaller size in the latter).
(ii) Acytostelium sp.
Sorocarps of very small size (300 to 600 μm in length), solitary and very delicate. Sorophore acellular and slender without pigmentation. Spores globose with polar granules. Aggregation is mound type. Long and narrow slugs that do not migrate. Grows more slowly than genus Dictyostelium. Need charcoal to get good fructifications. Presence of microcysts in these isolates.
(iii) Dictyostelium giganteum B. N. Singh.
Sorocarps of big size (4 to 10 mm in length), usually prostrate. Sorophores very long and wavy with tip capitata, no branches and no pigmentation. Sori elipsoid. Spores elipsoid, 5 to 7 by 3 to 4 μm, without polar granule. Aggregation radiate. Slugs that migrate long distances producing stalk.
(iv) Dictyostelium minutum Raper.
Sorocarps erect, of small size (0.5 to 0.85 mm in length), with cluster growth. Sorophores colorless. Spores ellipsoid and small, 5 to 6 by 3 to 3.5 μm, without polar granules. Typical aggregation of the species, mound like. Slugs do not migrate.
(v) Dictyostelium mucoroides Bref.
Sorocarps of medium size (2 to 5 mm in length), erect or semiprostrate with solitary growth. Sorophore with no pigmentation and round base. Spores ellipsoid, 4.5 to 6 by 2.5 to 3.5 μm, without polar granules. Aggregations radiate and small, characteristic of this species.
(vi) Dictyostelium sphaerocephalum (Oud.) Sacc. et March.
Sorocarps of medium size (2 to 4 mm in length) in relation with others species of cellular slime molds, erect or semierect with solitary growth and no pigmentation. Sorophore hyaline, sometimes irregular and sparse branching, with round base and acuminate tip with many tiers of cells. Presence of collar near the tip. Sori white, big in relation with the whole sorocarp. Spores ellipsoid and wide, 5.5 to 7 by 3 to 3.5 μm, with vacuolated cells without polar granules. Aggregations radiate and small. Some slugs migrate but not long distances.
(vii) Polysphondylium candidum. H. Hagiw.
Sorocarps of small size and solitary growth. Sorophores delicate and long with one tier of cells. Presence of supporters. Sorophores with no pigmentation and 2 to 8 whorls of 2 to 7 branches. These branches are symmetric but with different sizes. Sometimes seems to be irregular branching more than whorls. Also there are some sorocarps with no branches and no whorls. Some sorophores finish in terminal sori but the majority finish in terminal elongation. Spores ellipsoid and wide, 7 to 8 by 4 to 5 μm, with unconsolidated, but clearly evident, polar granules. Aggregates very big, radiate. Slugs that migrate large distances.
(viii) Polysphondylium pallidum Olive.
Sorocarps solitary with 1 to 11 whorls of 2 to 6 branches, not phototropic and colorless. Sorophores without lengthened terminal segments. Spores 5 to 7 by 2.5 to 3 μm, with unconsolidated polar granules. Aggregation radiate smaller than that of P. candidum. Slugs that migrate.
Amoebae were grown in association with Klebsiella aerogenes, and their DNA was obtained using MasterAmp DNA extraction solution (Epicentre Technologies, Madison, WI). The rDNA locus coding for the 5.8S rRNA and the two flanking internal transcribed spacers, ITS1 and ITS2, was amplified by PCR using the oligonucleotides 5′-GAGGAAGGAGAAGTCGTAACAAGGTATC-3′ and 5′-GCTTACTGATATGCTTAAGTTCAGCGGG-3′. Amplified DNAs were sequenced in both strands by use of flanking and internal primers.
The sizes of the 5.8S rDNA varied between 161 and 166 nucleotides (nt) (data not shown). Multiple alignment of the sequences, made using the ClustalW program (12), allowed us to determine the percentage of nucleotide identity between samples (Table 2; summarized in Table 3). Samples classified in the same species ranged between 97 and 100% identity, indicating a low level of intraspecific variability. Identities between species from the same genera varied between 92 and 98%, the exception being Dictyostelium minutum, which showed an identity of around 70% (69 to 73%) with other Dictyostelium species, very similar to the identities of D. minutum with Polysphondylium and Acytostelium species (62 to 71%). The identity between Acytostelium species (82%) was lower than that for Dictyostelium and Polysphondylium species. The identities between samples from different genera ranged from 69.3 to 88.3%.
TABLE 2.
Percentage of identity of the 5.8S rDNA regions between the different isolates
| Genus | Species | Isolate | % Identity with:
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 212 | GCE9 | DCB6B | 1880 | 1889 | 11C | L4B | AR4B | 1C | 133B | 115A | 89C | 20B | 88A | 89B | 89G | 14A | 118B | PH8B | 3A | 7A | 11F | 15 | 89A | 89C2 | 89D | 23D | Pp1 | |||
| Acytostelium | leptosomum | 212 | 100 | 82.0 | 82.0 | 73.2 | 74.3 | 61.8 | 62.2 | 73.2 | 73.2 | 73.2 | 73.8 | 73.8 | 73.0 | 73.8 | 73.8 | 72.6 | 73.8 | 73.8 | 79.5 | 79.5 | 77.5 | 79.5 | 79.5 | 79.5 | 79.5 | 79.5 | 78.8 | 78.8 |
| sp. | GCE9 | 82.0 | 100 | 100 | 81.9 | 83.1 | 66.9 | 67.3 | 83.1 | 83.1 | 83.1 | 82.5 | 82.5 | 83.3 | 84.4 | 84.4 | 82.8 | 84.4 | 84.4 | 88.3 | 88.3 | 86.0 | 88.3 | 88.3 | 88.3 | 88.3 | 88.3 | 88.8 | 88.8 | |
| sp. | DCB6B | 82.0 | 100 | 100 | 81.9 | 83.1 | 66.9 | 67.3 | 83.1 | 83.1 | 83.1 | 82.5 | 82.5 | 83.3 | 84.4 | 84.4 | 82.8 | 84.4 | 84.4 | 88.3 | 88.3 | 86.0 | 88.3 | 88.3 | 88.3 | 88.3 | 88.3 | 88.8 | 88.8 | |
| Dictyostelium | giganteum | 1880 | 73.2 | 81.9 | 81.9 | 100 | 98.7 | 69.9 | 70.4 | 97.5 | 97.5 | 97.5 | 98.1 | 98.1 | 95.7 | 96.9 | 96.9 | 95.1 | 96.9 | 96.9 | 85.8 | 85.8 | 83.5 | 85.8 | 85.8 | 85.8 | 85.8 | 85.8 | 84.5 | 84.5 |
| giganteum | 1889 | 74.3 | 83.1 | 83.1 | 98.7 | 100 | 71.2 | 71.6 | 98.7 | 98.7 | 98.7 | 99.4 | 99.4 | 96.9 | 98.1 | 98.1 | 96.3 | 98.1 | 98.1 | 87.0 | 87.0 | 84.8 | 87.0 | 87.0 | 87.0 | 87.0 | 87.0 | 85.7 | 85.7 | |
| minutum | 11C | 61.8 | 66.9 | 66.9 | 69.9 | 71.2 | 100 | 98.1 | 72.4 | 72.4 | 72.4 | 71.8 | 71.8 | 70.1 | 70.6 | 70.6 | 69.3 | 70.6 | 70.6 | 69.9 | 69.9 | 68.5 | 69.9 | 69.9 | 69.9 | 69.9 | 69.9 | 70.6 | 70.6 | |
| minutum | L4B | 62.2 | 67.3 | 67.3 | 70.4 | 71.6 | 98.1 | 100 | 72.8 | 72.8 | 72.8 | 72.2 | 72.2 | 70.6 | 71.0 | 71.0 | 69.7 | 71.0 | 71.0 | 70.4 | 70.4 | 68.9 | 70.4 | 70.4 | 70.4 | 70.4 | 70.4 | 71.0 | 71.0 | |
| mucoroides | AR4B | 73.2 | 83.1 | 83.1 | 97.5 | 98.7 | 72.4 | 72.8 | 100 | 100 | 100 | 99.4 | 99.4 | 96.9 | 98.1 | 98.1 | 96.3 | 98.1 | 98.1 | 85.8 | 85.8 | 83.5 | 85.8 | 85.8 | 85.8 | 85.8 | 85.8 | 85.7 | 85.7 | |
| mucoroides | 1C | 73.2 | 83.1 | 83.1 | 97.5 | 98.7 | 72.4 | 72.8 | 100 | 100 | 100 | 99.4 | 99.4 | 96.9 | 98.1 | 98.1 | 96.3 | 98.1 | 98.1 | 85.8 | 85.8 | 83.5 | 85.8 | 85.8 | 85.8 | 85.8 | 85.8 | 85.7 | 85.7 | |
| mucoroides | 133B | 73.2 | 83.1 | 83.1 | 97.5 | 98.7 | 72.4 | 72.8 | 100 | 100 | 100 | 99.4 | 99.4 | 96.9 | 98.1 | 98.1 | 96.3 | 98.1 | 98.1 | 85.8 | 85.8 | 83.5 | 85.8 | 85.8 | 85.8 | 85.8 | 85.8 | 85.7 | 85.7 | |
| mucoroides | 115A | 73.8 | 82.5 | 82.5 | 98.1 | 99.4 | 71.8 | 72.2 | 99.4 | 99.4 | 99.4 | 100 | 100 | 96.3 | 97.5 | 97.5 | 95.7 | 97.5 | 97.5 | 86.4 | 86.4 | 84.1 | 86.4 | 86.4 | 86.4 | 86.4 | 86.4 | 85.1 | 85.1 | |
| mucoroides | 89C | 73.8 | 82.5 | 82.5 | 98.1 | 99.4 | 71.8 | 72.2 | 99.4 | 99.4 | 99.4 | 100 | 100 | 96.3 | 97.5 | 97.5 | 95.7 | 97.5 | 97.5 | 86.4 | 86.4 | 84.1 | 86.4 | 86.4 | 86.4 | 86.4 | 86.4 | 85.1 | 85.1 | |
| sphaerocephalum | 20B | 73.0 | 83.3 | 83.3 | 95.7 | 96.9 | 70.1 | 70.6 | 96.9 | 96.9 | 96.9 | 96.3 | 96.3 | 100 | 98.8 | 98.8 | 97.0 | 98.8 | 98.8 | 87.1 | 87.1 | 84.8 | 87.1 | 87.1 | 87.1 | 87.1 | 87.1 | 87.0 | 87.0 | |
| sphaerocephalum | 88A | 73.8 | 84.4 | 84.4 | 96.9 | 98.1 | 70.6 | 71.0 | 98.1 | 98.1 | 98.1 | 97.5 | 97.5 | 98.8 | 100 | 100 | 98.1 | 100 | 100 | 87.7 | 87.7 | 84.8 | 87.7 | 87.7 | 87.7 | 87.7 | 87.7 | 87.6 | 87.6 | |
| sphaerocephalum | 89B | 73.8 | 84.4 | 84.4 | 96.9 | 98.1 | 70.6 | 71.0 | 98.1 | 98.1 | 98.1 | 97.5 | 97.5 | 98.8 | 100 | 100 | 98.1 | 100 | 100 | 87.7 | 87.7 | 85.4 | 87.7 | 87.7 | 87.7 | 87.7 | 87.7 | 87.6 | 87.6 | |
| sphaerocephalum | 89G | 72.6 | 82.8 | 82.8 | 95.1 | 96.3 | 69.3 | 69.7 | 96.3 | 96.3 | 96.3 | 95.7 | 95.7 | 97.0 | 98.1 | 98.1 | 100 | 98.1 | 98.1 | 86.1 | 86.1 | 83.8 | 86.1 | 86.1 | 86.1 | 86.1 | 86.1 | 86.0 | 86.0 | |
| sphaerocephalum | 14A | 73.8 | 84.4 | 84.4 | 96.9 | 98.1 | 70.6 | 71.0 | 98.1 | 98.1 | 98.1 | 97.5 | 97.5 | 98.8 | 100 | 100 | 98.1 | 100 | 100 | 87.7 | 87.7 | 85.4 | 87.7 | 87.7 | 87.7 | 87.7 | 87.7 | 87.6 | 87.6 | |
| sphaerocephalum | 118B | 73.8 | 84.4 | 84.4 | 96.9 | 98.1 | 70.6 | 71.0 | 98.1 | 98.1 | 98.1 | 97.5 | 97.5 | 98.8 | 100 | 100 | 98.1 | 100 | 100 | 87.7 | 87.7 | 85.4 | 87.7 | 87.7 | 87.7 | 87.7 | 87.7 | 87.6 | 87.6 | |
| Polysphondylium | candidum | PH8B | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 |
| candidum | 3A | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| candidum | 7A | 77.5 | 86.0 | 86.0 | 83.5 | 84.8 | 68.5 | 68.9 | 83.5 | 83.5 | 83.5 | 84.1 | 84.1 | 84.8 | 85.4 | 85.4 | 83.8 | 85.4 | 85.4 | 97.6 | 97.6 | 100 | 97.6 | 97.6 | 97.6 | 97.6 | 97.6 | 90.2 | 90.2 | |
| candidum | 11F | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| candidum | 15 | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| candidum | 89A | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.5 | 85.5 | 85.5 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| candidum | 89C2 | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| candidum | 89D | 79.5 | 88.3 | 88.3 | 85.8 | 87.0 | 69.9 | 70.4 | 85.8 | 85.8 | 85.8 | 86.4 | 86.4 | 87.1 | 87.7 | 87.7 | 86.1 | 87.7 | 87.7 | 100 | 100 | 97.6 | 100 | 100 | 100 | 100 | 100 | 92.6 | 92.6 | |
| pallidum | 23D | 78.8 | 88.8 | 88.8 | 84.5 | 85.7 | 70.6 | 71.0 | 85.7 | 85.7 | 85.7 | 85.1 | 85.1 | 87.0 | 87.6 | 87.6 | 86.0 | 87.6 | 87.6 | 92.6 | 92.6 | 90.2 | 92.6 | 92.6 | 92.6 | 92.6 | 92.6 | 100 | 100 | |
| pallidum | Pp1 | 78.8 | 88.8 | 88.8 | 84.5 | 85.7 | 70.6 | 71.0 | 85.7 | 85.7 | 85.7 | 85.1 | 85.1 | 87.0 | 87.6 | 87.6 | 86.0 | 87.6 | 87.6 | 92.6 | 92.6 | 90.2 | 92.6 | 92.6 | 92.6 | 92.6 | 92.6 | 100 | 100 | |
TABLE 3.
Summary of identitiesa
| Type of comparison | Species or genus (genera)b | % Identity (mean ± SD)
|
||
|---|---|---|---|---|
| Complete region | 5.8S rDNA | ITS1 region | ||
| Species | D. mucoroides | 79.07 ± 12.34 | 99.64 ± 0.29 | 77.91 ± 12.26 |
| D. sphaerocephalum | 99.07 ± 0.89 | 98.97 ± 0.94 | 99.67 ± 0.47 | |
| P. candidum | 97.57 ± 2.64 | 99.40 ± 1.04 | 96.64 ± 3.97 | |
| Same genus | Dictyostelium | 56.78 ± 11.72 | 88.73 ± 12.39 | 49.61 ± 10.91 |
| Polysphondylium | 50.97 ± 0.99 | 92.3 ± 0.79 | 52.39 ± 0.62 | |
| Different genera | D/A | 39.79 ± 4.81 | 78.09 ± 6.67 | 39.32 ± 4.65 |
| P/D | 33.84 ± 3.21 | 84.20 ± 5.61 | 22.74 ± 4.43 | |
| P/A | 34.64 ± 5.71 | 85.17 ± 4.31 | 24.59 ± 4.56 | |
Identities were observed between different isolates from the same species and different species, from the same genus or different genera, and for the complete region (including the rDNA and ITS1 and ITS2 regions), the 5.8S rDNA, and the ITS1 region.
D, Dictyostelium; A, Acytostelium; P, Polysphondylium.
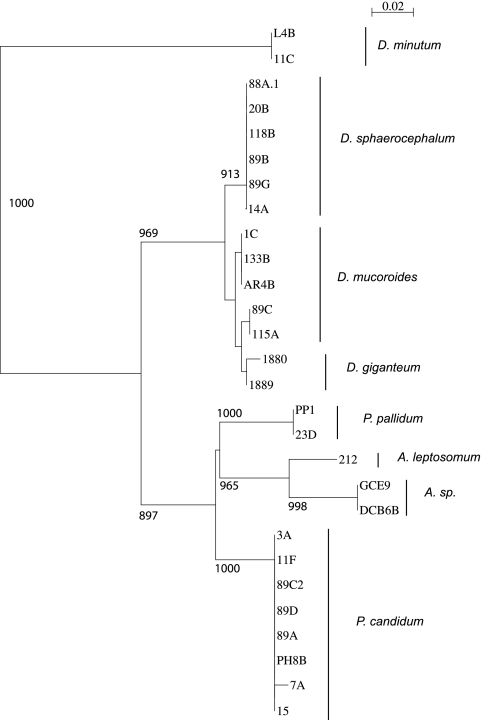
The 5.8S rDNA multiple alignment, excluding nonaligned positions, was used to construct a phylogenetic tree, shown in Fig. 1. Statistical significance, indicated by bootstrap values, is shown for the more consistent branches of the tree. As expected for the homology data, most of the samples for the same species branched closely together, except for the five samples of D. mucoroides, which associate in two groups: samples 1C, 133B, and AR4B and samples 89C and 115A. Samples from different species grouped in their specific genera, except for D. minutum, which segregated from the rest of the Dictyostelium samples. The closer association of Acytostelium species with Polysphondylium species than with Dictyostelium species is also noticeable and statistically significant.
FIG. 1.
Phylogenetic tree of dictyostelid species grouped according to their 5.8S rDNA sequences. The nucleotide sequences of the 5.8S rDNA from field and collection samples of different dictyostelid species, corresponding to the three genera described, Dictyostelium, Acytostelium, and Polysphondylium, were compared using the programs ClustalW, at the online Biology WorkBench facilities from the San Diego Supercomputer Center (http://workbench.sdsc.edu), and ClustalX (11). The 5.8S rDNA alignment was also visually optimized, and nonaligned positions were omitted. Nucleotide divergences between sequences were determined using the MVIEW program (5) at the Biology WorkBench facilities. Phylogenetic trees were determined using the neighbor-joining method (10) and the ClustalX program. A random generator seed of 111 and 1,000 bootstrap trials were calculated, and the number of times that each branch was obtained is indicated at left for values above 850. Trees were drawn using the njplot program. The evolutionary-distance scale, calculated as the fraction of nucleotide changes, is indicated in the upper right corner of the figure. The species in which the different isolates were classified are indicated at right.
Analyses of these data indicate that the comparison of 5.8S rDNA sequences can be very informative to discriminate between samples from different genera. Interspecific comparisons within the same genera indicate that the 5.8S rDNA sequence is highly conserved, so that the divergence between 5.8S rDNA sequences of some species (for example, D. mucoroides and and D. giganteum) was similar to intraspecific variability. This similarity makes the 5.8S rDNA sequence comparison of restricted utility to discriminate between isolates of species from the same genera.
ITS1 regions are 278 to 312 nt long in Dictyostelium spp. and Acytostelium spp. and 192 to 253 nt long in Polysphondylium spp., with the exception of that of Acytostelium leptosomum, which is 245 nt long (data not shown). The differences are larger in the ITS2 region, where sizes vary between 311 and 383 nt in Acytostelium spp., 417 and 486 nt in Dictyostelium spp., and 667 and 856 nt in Polysphondylium spp. (data not shown).
Most species showed high similarity between samples in their ITS1 regions (Table 4; summarized in Table 3) (similar results were obtained for ITS2), which indicates low intraspecific variability. For example, eight samples from Dictyostelium sphaerocephalum and six from Polysphondylium candidum showed 98 to 100% identity between them. Acytostelium spp., D. minutum, and P. pallidum also showed over 95% (95 to 100%) identity between the two samples analyzed for each species. For some of these species, one of the samples was collected in the United States and the other in the south of Europe, two distant and different regions of the world. Only two species showed a larger divergence between samples (65 to 82%), Dictyostelium giganteum and D. mucoroides. The data on D. mucoroides are in concordance with those previously described for the 5.8S rDNA.
TABLE 4.
Percentage of identity of the ITS1 regions between different isolates
| Genus | Species | Isolate | % Identity with:
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 212 | GCE9 | DCB6B | 1880 | 1889 | 11C | L4B | AR4B | 1C | 133B | 115A | 89C | 20B | 88A | 89B | 89G | 14A | 118B | PH8B | 3A | 7A | 11F | 15 | 89A | 89C2 | 89D | 23D | Pp1 | |||
| Acytostelium | leptosomum | 212 | 100 | 36.0 | 36.0 | 38.1 | 26.7 | 35.2 | 35.2 | 29.4 | 36.7 | 36.7 | 33.9 | 33.9 | 35.2 | 33.2 | 35.2 | 35.2 | 35.2 | 35.2 | 28.7 | 31.5 | 32.2 | 31.8 | 31.8 | 31.8 | 31.8 | 31.8 | 27.7 | 27.7 |
| sp. | GCE9 | 36.0 | 100 | 100 | 43.9 | 40.5 | 35.1 | 36.5 | 38.2 | 42.2 | 42.3 | 41.3 | 41.3 | 45.1 | 40.4 | 45.1 | 45.1 | 45.1 | 45.1 | 18.8 | 21.0 | 21.6 | 21.3 | 21.3 | 21.3 | 21.3 | 21.3 | 23.8 | 23.8 | |
| sp. | DCB6B | 36.0 | 100 | 100 | 43.9 | 40.5 | 35.1 | 36.5 | 38.2 | 42.2 | 42.3 | 41.3 | 41.3 | 45.1 | 40.4 | 45.1 | 45.1 | 45.1 | 45.1 | 18.8 | 21.0 | 21.6 | 21.3 | 21.3 | 21.3 | 21.3 | 21.3 | 23.8 | 23.8 | |
| Dictyostelium | giganteum | 1880 | 38.1 | 43.9 | 43.9 | 100 | 74.1 | 36.1 | 37.0 | 49.7 | 52.2 | 52.4 | 52.0 | 52.0 | 57.2 | 53.4 | 57.2 | 57.2 | 57.2 | 57.2 | 20.5 | 21.7 | 22.0 | 22.0 | 22.0 | 21.7 | 21.7 | 21.7 | 24.7 | 24.7 |
| giganteum | 1889 | 26.7 | 40.5 | 40.5 | 74.1 | 100 | 30.5 | 31.0 | 49.3 | 49.6 | 49.6 | 51.8 | 51.8 | 54.6 | 49.6 | 54.6 | 54.6 | 54.6 | 54.6 | 14.5 | 16.7 | 16.9 | 16.9 | 16.9 | 16.7 | 16.7 | 16.7 | 19.7 | 19.7 | |
| minutum | 11C | 35.2 | 35.1 | 35.1 | 36.1 | 30.5 | 100 | 95.3 | 35.5 | 37.7 | 37.7 | 36.6 | 36.6 | 34.4 | 31.4 | 34.4 | 34.4 | 34.4 | 34.4 | 29.9 | 32.5 | 33.3 | 32.8 | 32.8 | 33.0 | 33.0 | 33.0 | 35.2 | 35.1 | |
| minutum | L4B | 35.2 | 36.5 | 36.5 | 37.0 | 31.0 | 95.3 | 100 | 36.3 | 38.5 | 38.5 | 36.5 | 36.5 | 35.2 | 32.3 | 35.2 | 35.2 | 35.2 | 35.2 | 30.6 | 32.8 | 33.3 | 33.0 | 33.0 | 33.0 | 33.0 | 33.0 | 34.2 | 34.2 | |
| mucoroides | AR4B | 29.4 | 38.2 | 38.2 | 49.7 | 49.3 | 35.5 | 36.3 | 100 | 82.3 | 82.5 | 65.7 | 65.7 | 58.0 | 54.3 | 58.0 | 58.0 | 58.0 | 58.0 | 20.8 | 22.7 | 23.2 | 22.9 | 22.9 | 22.9 | 22.9 | 22.9 | 22.8 | 22.8 | |
| mucoroides | 1C | 36.7 | 42.2 | 42.2 | 52.2 | 49.6 | 37.7 | 38.5 | 82.3 | 100 | 99.7 | 70.7 | 70.7 | 59.2 | 55.2 | 59.2 | 59.2 | 59.2 | 59.2 | 20.6 | 22.6 | 23.2 | 22.9 | 22.9 | 22.9 | 22.9 | 22.9 | 24.6 | 24.6 | |
| mucoroides | 133B | 36.7 | 42.3 | 42.3 | 52.4 | 49.6 | 37.7 | 38.5 | 82.5 | 99.7 | 100 | 70.9 | 70.9 | 59.4 | 55.4 | 59.4 | 59.4 | 59.4 | 59.4 | 20.6 | 22.7 | 23.2 | 23.0 | 23.0 | 23.0 | 23.0 | 23.0 | 24.6 | 24.6 | |
| mucoroides | 115A | 33.9 | 41.3 | 41.3 | 52.0 | 51.8 | 36.6 | 36.5 | 65.7 | 70.7 | 70.9 | 100 | 100 | 63.0 | 57.9 | 63.0 | 63.0 | 63.0 | 63.0 | 20.0 | 21.6 | 21.6 | 21.9 | 21.9 | 21.3 | 21.3 | 21.3 | 25.1 | 25.4 | |
| mucoroides | 89C | 33.9 | 41.3 | 41.3 | 52.0 | 51.8 | 36.6 | 36.5 | 65.7 | 70.7 | 70.9 | 100 | 100 | 63.0 | 57.9 | 63.0 | 63.0 | 63.0 | 63.0 | 20.0 | 21.6 | 21.6 | 21.9 | 21.9 | 21.3 | 21.3 | 21.3 | 25.1 | 25.4 | |
| sphaerocephalum | 20B | 35.2 | 45.1 | 45.1 | 57.2 | 54.6 | 34.4 | 35.2 | 58.0 | 59.2 | 59.4 | 63.0 | 63.0 | 100 | 99 | 100 | 100 | 100 | 100 | 18.7 | 20.0 | 20.9 | 20.6 | 20.6 | 20.6 | 20.6 | 20.6 | 21.0 | 21.3 | |
| sphaerocephalum | 88A | 33.2 | 40.4 | 40.4 | 53.4 | 49.6 | 31.4 | 32.3 | 54.3 | 55.2 | 55.4 | 57.9 | 57.9 | 99.0 | 100 | 99.0 | 99.0 | 99.0 | 99.0 | 19.3 | 20.4 | 21.3 | 21.0 | 21.0 | 21.0 | 21.0 | 21.0 | 17.7 | 18.0 | |
| sphaerocephalum | 89B | 35.2 | 45.1 | 45.1 | 57.2 | 54.6 | 34.4 | 35.2 | 58.0 | 59.2 | 59.4 | 63.0 | 63.0 | 100 | 99.0 | 100 | 100 | 100 | 100 | 18.7 | 20.0 | 20.9 | 20.6 | 20.6 | 20.6 | 20.6 | 20.6 | 21.0 | 21.3 | |
| sphaerocephalum | 89G | 35.2 | 45.1 | 45.1 | 57.2 | 54.6 | 34.4 | 35.2 | 58.0 | 59.2 | 59.4 | 63.0 | 63.0 | 100 | 99.0 | 100 | 100 | 100 | 100 | 18.7 | 20.0 | 20.9 | 20.6 | 20.6 | 20.6 | 20.6 | 20.6 | 21.0 | 21.3 | |
| sphaerocephalum | 14A | 35.2 | 45.1 | 45.1 | 57.2 | 54.6 | 34.4 | 35.2 | 58.0 | 59.2 | 59.4 | 63.0 | 63.0 | 100 | 99.0 | 100 | 100 | 100 | 100 | 18.7 | 20.0 | 20.9 | 20.6 | 20.6 | 20.6 | 20.6 | 20.6 | 21.0 | 21.3 | |
| sphaerocephalum | 118B | 35.2 | 45.1 | 45.1 | 57.2 | 54.6 | 34.4 | 35.2 | 58.0 | 59.2 | 59.4 | 63.0 | 63.0 | 100 | 99.0 | 100 | 100 | 100 | 100 | 18.7 | 20.0 | 20.9 | 20.6 | 20.6 | 20.6 | 20.6 | 20.6 | 21.0 | 21.3 | |
| Polysphondylium | candidum | PH8B | 28.7 | 18.8 | 18.8 | 20.5 | 14.5 | 29.9 | 30.6 | 20.8 | 20.6 | 20.6 | 20.0 | 20.0 | 18.7 | 19.3 | 18.7 | 18.7 | 18.7 | 18.7 | 100 | 89.4 | 89.8 | 90.2 | 90.2 | 89.8 | 89.8 | 89.8 | 51.0 | 51.4 |
| candidum | 3A | 31.5 | 21.0 | 21.0 | 21.7 | 16.7 | 32.5 | 32.8 | 22.7 | 22.6 | 22.7 | 21.6 | 21.6 | 20.0 | 20.4 | 20.0 | 20.0 | 20.0 | 20.0 | 89.4 | 100 | 97.4 | 99.2 | 99.2 | 98.1 | 98.1 | 98.1 | 51.9 | 52.3 | |
| candidum | 7A | 32.2 | 21.6 | 21.6 | 22.0 | 16.9 | 33.3 | 33.3 | 23.2 | 23.2 | 23.2 | 21.6 | 21.6 | 20.9 | 21.3 | 20.9 | 20.9 | 20.9 | 20.9 | 89.8 | 97.4 | 100 | 98.0 | 98.1 | 99.2 | 99.2 | 99.2 | 52.7 | 53.1 | |
| candidum | 11F | 31.8 | 21.3 | 21.3 | 22.0 | 16.9 | 32.8 | 33.0 | 22.9 | 22.9 | 23.0 | 21.9 | 21.9 | 20.6 | 21.0 | 20.6 | 20.6 | 20.6 | 20.6 | 90.2 | 99.2 | 98.0 | 100 | 100 | 98.8 | 98.8 | 98.8 | 51.9 | 52.3 | |
| candidum | 15 | 31.8 | 21.3 | 21.3 | 22.0 | 16.9 | 32.8 | 33.0 | 22.9 | 22.9 | 23.0 | 21.9 | 21.9 | 20.6 | 21.0 | 20.6 | 20.6 | 20.6 | 20.6 | 90.2 | 99.2 | 98.1 | 100 | 100 | 98.9 | 98.9 | 98.9 | 51.9 | 52.3 | |
| candidum | 89A | 31.8 | 21.3 | 21.3 | 21.7 | 16.7 | 33.0 | 33.0 | 22.9 | 22.9 | 23.0 | 21.3 | 21.3 | 20.6 | 21.0 | 20.6 | 20.6 | 20.6 | 20.6 | 89.8 | 98.1 | 99.2 | 98.8 | 98.9 | 100 | 100 | 100 | 52.7 | 53.1 | |
| candidum | 89C2 | 31.8 | 21.3 | 21.3 | 21.7 | 16.7 | 33.0 | 33.0 | 22.9 | 22.9 | 23.0 | 21.3 | 21.3 | 20.6 | 21.0 | 20.6 | 20.6 | 20.6 | 20.6 | 89.8 | 98.1 | 99.2 | 98.8 | 98.9 | 100 | 100 | 100 | 52.7 | 53.1 | |
| candidum | 89D | 31.8 | 21.3 | 21.3 | 21.7 | 16.7 | 33.0 | 33.0 | 22.9 | 22.9 | 23.0 | 21.3 | 21.3 | 20.6 | 21.0 | 20.6 | 20.6 | 20.6 | 20.6 | 89.8 | 98.1 | 99.2 | 98.8 | 98.9 | 100 | 100 | 100 | 52.7 | 53.1 | |
| pallidum | 23D | 27.7 | 23.8 | 23.8 | 24.7 | 19.7 | 35.2 | 34.2 | 22.8 | 24.6 | 24.6 | 25.1 | 25.1 | 21.0 | 17.7 | 21.0 | 21.0 | 21.0 | 21.0 | 51.0 | 51.9 | 52.7 | 51.9 | 51.9 | 52.7 | 52.7 | 52.7 | 100 | 99.6 | |
| pallidum | Pp1 | 27.7 | 23.8 | 23.8 | 24.7 | 19.7 | 35.1 | 34.2 | 22.8 | 24.6 | 24.6 | 25.4 | 25.4 | 21.3 | 18.0 | 21.3 | 21.3 | 21.3 | 21.3 | 51.4 | 52.3 | 53.1 | 52.3 | 52.3 | 53.1 | 53.1 | 53.1 | 99.6 | 100 | |
Identities between species from the same genera were on the order of 50 to 60% (Tables 3 and 4). The exception is D. minutum, which showed an identity of 30 to 38% with other Dictyostelium species, and A. leptosomum, which was 36% identical to other Acytostelium spp. Samples of different genera are less than 45% identical, which is too low for phylogenetic analyses.
The data on ITS1 alignments were used to generate the phylogenetic tree shown in Fig. 2. All of the samples from D. sphaerocephalum and P. candidum grouped closely together between them. However, for D. mucoroides, samples 133/1C and 115A/89C grouped together but segregated significantly between them and with sample AR4B. The two samples of D. giganteum also differed significantly. These data were confirmed by analyses of the 5.8S rDNA and ITS2 sequences. The larger differences found in these species could indicate higher intraspecific variability. Alternatively, these samples could have been grouped erroneously as belonging to the same species due to their similar morphological characteristics. Nucleotide sequence data would be in agreement with the division of D. mucoroides into, at least, two species, one represented by samples 1C, 133B, and AR4B and the other represented by samples 115A and 89C. Some morphological data also support this proposal; in particular, spores had a size of 7 by 4 μm in samples 115A and 89C and 6 by 3 μm in samples 1C, 133B, and AR4B.
FIG. 2.
Phylogenetic tree of dictyostelid species grouped according to their ITS1 nucleotide sequences. The nucleotide sequences of the ITS1 region were determined from field and collection samples of eight different species from the three dictyostelid genera: Dictyostelium, Acytostelium, and Polysphondylium. Sequences were aligned using the ClustalW and ClustalX programs, and the multiple alignment was used to construct a phylogenetic tree using the neighbor-joining method, as described in the legend for Fig. 1. The evolutionary-distance scale, calculated as the fraction of nucleotide changes, is shown in the upper right corner of the figure. One thousand bootstrap trials were calculated, and the values higher than 850 are shown at left for the corresponding branches. The species in which the different isolates were classified are shown at right.
These data indicate that, despite the large differences between the ITS regions of dictyostelid genera, the smaller differences between isolates from the same genera and species make these regions of utility to study their phylogenetic relationships. Actually, comparison between ITS1 sequences can be very useful to discriminate between samples from close species whose 5.8S rDNA sequences are almost identical.
The phylogenetic analyses of the 5.8S rDNA and ITS sequences have shown a good general agreement with the current systematics of the phyla, with some exceptions. The more remarkable is D. minutum, which showed low similarity with other Dictyostelium species, in either the 5.8S rDNA (70%) or the ITS1 (35%) region, compared to the average similarity that exists between Dictyostelium species (88.73 or 49.61%, respectively). Analyses of the small subunit rRNA sequences also indicated that D. minutum diverged from Dictyostelium rosarium and Dictyostelium discoideum, which were closely related between them (2). In the same previous study, D. minutum was closer to these two Dictyostelium species than to P. pallidum, although it was even more distant from Dictyostelium fasciculatum. The data presented in the present study showed significant bootstrap values for the divergence between D. minutum and the other Dictyostelium and Polysphondylium species but did not allow us to establish their evolutionary relationship.
The larger divergence of D. minutum with respect to the other Dictyostelium species and also to Polysphondylium and Acytostelium species could be due to a faster molecular evolution in this species. Alternatively, this divergence could indicate that D. minutum is distant from the three existing genera and might be the founder species of a new genus yet to be defined. Recent data obtained from the analysis of protein-coding genes support this hypothesis (10a).
Acknowledgments
We thank J. C. Cavender (Ohio University) and J. Landolt (Shepherd University) for kindly providing several samples.
This work was supported by grants CGL2005-00320/BOS, BMC02-01501, and BFU2005-00138 from the Dirección General de Investigación, Ministerio de Educación y Ciencia, Spain, and an I3P grant to M.R.
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Álvarez, I., and J. Wendel. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29:417-434. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Curto, E., D. Rozen, A. Ritchie, C. Fouquet, S. L. Baldauf, and P. Schaap. 2005. Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc. Natl. Acad. Sci. USA 102:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldauf, S. L., and W. F. Doolittle. 1997. Origin and evolution of the slime molds (Mycetozoa). Proc. Natl. Acad. Sci. USA 94:12007-12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brefeld, O. 1869. Dictyostelium mucoroides. Ein neuer Organismus aus der Verwandtschaft der Myxomyceten. Abh. Senckenb. Naturforsch. Ges. 7:85-107. [Google Scholar]
- 5.Brown, N., C. Leroy, and C. Sandaer. 1998. MView: a web compatible database search or multiple alignment viewer. Bioinformatics 14:380-381. [DOI] [PubMed] [Google Scholar]
- 6.Cavender, J. C. 1990. Phylum Dictyostelida, p. 88-101. In L. Margulis, J. O. Corliss, M. Melkonian, and D. J. Chapman (ed.), Handbook of Protoctista. Jones and Bartlett, Boston, Mass.
- 7.Hagiwara, H. 1989. The taxonomic study of Japanese dictyostelid cellular slime molds. National Science Museum, Tokyo, Japan.
- 8.Loomis, W. F., and D. W. Smith. 1995. Consensus phylogeny of Dictyostelium. Experientia 51:1110-1115. [DOI] [PubMed] [Google Scholar]
- 9.Raper, K. B. 1984. The Dictyostelids. Princeton University Press, Princeton, N.J.
- 10.Saitou, N., and M. Nei. 1987. The neighbor-joining method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 10a.Schaap, P., T. Winckler, M. Nelson, E. Alvarez-Curto, B. Elgie, H. Hagiwara, J. Cavender, A. Milano-Curto, D. E. Rozen, T. Dingermann, R. Mutzel, and S. L. Baldauf. 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science 314:661-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougins, and D. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uilenberg, G., F. Thiaucourt, and F. Jongejan. 2004. On molecular taxonomy: what is in a name? Exp. Appl. Acarol. 32:301-312. [DOI] [PubMed] [Google Scholar]