Abstract
In Kluyveromyces lactis, the pentose phosphate pathway is an alternative route for the dissimilation of glucose. The first enzyme of the pathway is the glucose-6-phosphate dehydrogenase (G6PDH), encoded by KlZWF1. We isolated this gene and examined its role. Like ZWF1 of Saccharomyces cerevisiae, KlZWF1 was constitutively expressed, and its deletion led to increased sensitivity to hydrogen peroxide on glucose, but unlike the case for S. cerevisiae, the Klzwf1Δ strain had a reduced biomass yield on fermentative carbon sources as well as on lactate and glycerol. In addition, the reduced yield on glucose was associated with low ethanol production and decreased oxygen consumption, indicating that this gene is required for both fermentation and respiration. On ethanol, however, the mutant showed an increased biomass yield. Moreover, on this substrate, wild-type cells showed an additional band of activity that might correspond to a dimeric form of G6PDH. The partial dimerization of the G6PDH tetramer on ethanol suggested the production of an NADPH excess that was negative for biomass yield.
The pentose phosphate pathway (PPP) constitutes the major source of NADPH required for the neutralization of reactive oxygen species (ROS), for reductive biosynthetic reactions, and for the production of metabolic intermediates. Glucose-6-phosphate dehydrogenase (G6PDH) is the first enzyme of the oxidative part of the PPP, is highly conserved through evolution (2, 23), and catalyzes the rate-limiting NADPH-producing step of this metabolic pathway (18). G6PDH deficiency in humans is one of the most common enzymopathies; clinical symptoms associated with reduced activity are hemolytic anemia, favism, and other pathologies caused by enhanced sensitivity of erythrocytes to oxidants (6). In Saccharomyces cerevisiae, this protein, which shows 48% identity to the human one, is encoded by ZWF1, and the deletion of this gene resulted in increased sensitivity to hydrogen peroxide (H2O2) and in the requirement for organic sources of sulfur, such as methionine (15, 17, 24, 37). In this yeast, however, G6PDH does not seem to play a relevant metabolic role in that deletion of the gene has no evident effects during growth with fermentative and respiratory carbon sources (24). In contrast, the deletion of PGI1, the gene coding for the glycolytic phosphoglucose isomerase, impeded growth of S. cerevisiae in glucose due to the inability to regenerate NADP from the NADPH produced in the PPP (1, 7). Differently from S. cerevisiae, it has been reported that Kluyveromyces lactis pgi1 (rag2) mutants are able to grow in glucose (12). Similarly, a null mutant with deletion of the PFK genes, encoding subunits of the phosphofructokinase, was still able to grow in glucose (16), suggesting that the glycolytic block in K. lactis can be overcome by the PPP (12, 16). To study in more detail the metabolic importance of G6PDH in K. lactis, we disrupted the KlZWF1 gene in this yeast and studied the effects of the G6PDH deficiency on the utilization of carbon sources, on the expression of genes involved in the reoxidation of reducing equivalents, and in the response to oxidative stress.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The following K. lactis strains were used in this work: MW179-1D (MATα metA1 ade leu2 trip ura3) (31), CBS2359/3 (MATa hisA1) (provided by M. Wésolowsky), GG1996 (MATa rag2::LoxP) (34), and MS16-21B (MATa metA1 ade leu2 Klsdh1::kanMX4URA3 (this work).
The following S. cerevisiae strains were used in this work: BY4742 (MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) (Euroscarf) and BY4742/zwf1Δ (zwf1::kanMX4) (Euroscarf).
Cultures were grown with shaking at 28°C in YP (1% Difco yeast extract, 2% Bacto peptone [Difco]) or in minimal medium (6.7 g liter−1 Difco yeast nitrogen base) supplemented with different carbon sources at the concentration specified in the text. Geneticin (G418) was used on YPD (YP plus 2% glucose) agar medium at a concentration of 100 μg ml−1. Minimal medium was supplemented with auxotrophic requirements at a final concentration of 10 μg ml−1. Hydrogen peroxide was added to plates at concentrations of 3 and 5 mM (15).
Escherichia coli strain DH5α was used for the propagation of plasmid DNA. Cultures were grown at 37°C on LB medium (0.5% yeast extract, 1% Difco tryptone, 0.5% NaCl) supplemented with 100 μg ml−1 ampicillin.
ADH and G6PDH assay.
Cell extracts were prepared from 5 ml of rich medium or 20 ml of minimal medium cultures grown overnight or for 3 days, respectively. Preparation of extracts, native polyacrylamide gels, and electrophoresis conditions was performed according to the procedures described previously for alcohol dehydrogenase (ADH) activity (20, 30). G6PDH activity was visualized by incubating gels for 10 to 15 min in 5 ml of the following solution: 10 μl of 1 M MgCl2, 50 μl of NADP (100 mg ml−1 in 100 mM Tris-HCl, pH 8.0), 15 μl of phenazine methosulfate (40 mg ml−1 in H2O) (Sigma), 30 μl of nitroblue tetrazolium salt (50 mg ml−1 in H2O) (Sigma), 25 μl of glucose-6-phosphate (100 mg ml−1 in H2O), and H2O to a final volume of 5 ml.
Glucose and ethanol concentrations in culture supernatants were measured using commercial kits from R-Biopharma (Darmstadt, Germany) according to the manufacturer's instructions.
Intracellular ROS production was detected by staining growing cells with dihydrorhodamine (DHR) 123, at a concentration of 5 μg ml−1, according to the method of Madeo et al. (21) and visualizing them with a DHR optical filter in a Zeiss microscope.
Cell respiration.
The respiration rate was measured at 30°C, using a Clark-type electrode according to the method of Ferrero et al. (10) under the conditions described previously (31).
Quantitative determination of cytochromes was performed according to the method of Lodi and Ferrero (19).
Construction and amplification of KlZWF1 deletion cassette.
The KlZWF1 gene was amplified as a SalI fragment from the K. lactis genome with the following primers: forward, 5′-agggtcGACACTGTATTCCTCTCGTTACC-3′ (lowercase letters indicate the inserted half SalI site); and reverse, 5′-CTCATGTCGACGGTCATAGCG-3′.
The primer sequences were located 980 bp upstream of the ATG codon and 640 bp downstream of the stop codon. The amplified DNA was cloned into the SalI site of vector pTZ19 (Pharmacia) (pTZ19/KlZWF1). The 5′-terminal part of the gene was isolated by digestion of pTZ19/KlZWF1 with HpaI-PvuII, and the purified fragment was cloned into the PvuII site of the pFA6aKanMX4 vector (38) (pFA6a/5′Klzwf1). The 3′ end of the gene was similarly obtained as an EcoRI fragment and cloned into the EcoRI site of pFA6a/5′Klzwf1. The final product containing the deletion cassette was called pFA6/Klzwf1Δ.This plasmid was linearized by NotI digestion and transformed into the MW179-1D strain for gene replacement. Positive clones selected on G418 plates were then analyzed for growth on YPD plates containing 5 mM hydrogen peroxide. Of 150 clones analyzed, we isolated three H2O2-sensitive clones that were further characterized by Southern blotting. The genomic DNAs of these clones and of the wild-type strain were digested with SalI and XbaI, separated by gel electrophoresis, transferred to a nylon filter, and probed with the amplified fragment labeled with 32P. The autoradiograph showed one signal, of 4.3 kbp, for the wild type and two signals (2.1 and 2.3 kbp) for the disruptants, as expected from the integration of the cassette into the KlZWF1 locus (not shown). To exclude the presence of additional mutations, one of these disruptants, called MW179-1D/Klzwf1Δ, was complemented with the centromeric KCplac13 (LEU2) vector (provided by M. Wésolowski-Louvel) containing the KlZWF1 gene (HindIII-XbaI fragment from pTZ19/KlZWF1) and analyzed for the presence of G6PDH activity and for growth on different carbon sources.
It has been reported that S. cerevisiae zwf1Δ mutants are also methionine auxotrophs (37). Since the parental K. lactis strain from which we deleted KlZWF1 had methionine requirements, we crossed the Klzwf1Δ strain with CBS2359/3, a Met+ strain of the opposite mating type. The presence of many spores showing both G418+ and Met+ phenotypes among the meiotic segregants indicated that for K. lactis, the deletion of Klzwf1 did not lead to methionine auxotrophy (not shown). The introduction of the KlZWF1 gene into the S. cerevisiae zwf1Δ strain nevertheless fully recovered methionine auxotrophy, as well as resistance to H2O2 (not shown).
General methods.
DNA manipulation, plasmid engineering, and other techniques were performed according to standard procedures. Yeast transformation was performed by electroporation with a Bio-Rad Gene-Pulser apparatus following the method described by Becker and Guarente (5). The multicopy KEp6 (URA3) and centromeric KCplac13 (LEU2) vectors were used to introduce the KlZWF1 gene into the MW179-1D/Klzwf1Δ strain. pRS416 was the vector used to introduce the KlZWF1 gene into the S. cerevisiae BY4742 and BY4742/zwf1Δ strains. Preparation of total RNA has been described previously (30). Protein concentrations were determined by the method of Bradford (8).
RESULTS
Amplification and deletion of the KlZWF1 gene.
Analysis of the entire K. lactis genome sequence revealed the presence of a gene encoding a protein that showed nearly 70% identity to G6PDH of S. cerevisiae. The KlZWF1 gene and its flanking regions were amplified from the genomic DNA by using two primers, located 1 kbp upstream and 0.6 kbp downstream of the coding region. The amplified gene was then used for the construction in plasmid FA6a-KanMX4 (38) of a disruption cassette with a deletion of 75% of the coding region (see Materials and Methods). The linear DNA fragment containing the deleted gene was transformed into the MW179-1D strain, and transformants were selected for G418 resistance (28). Since it has been reported that S. cerevisiae strains lacking ZWF1 are more sensitive to H2O2 (17, 23, 24, 37), G418-resistant clones were further analyzed for their growth in the presence of hydrogen peroxide. Following this approach, we selected putative deletion mutants that were sensitive to H2O2. Southern analysis confirmed the correct integration of the disruption cassette into the chromosomal locus (not shown), and one of these strains, named MW179-1D/Klzwf1Δ, was used in the following experiments.
Carbon source utilization in the Klzwf1Δ mutant.
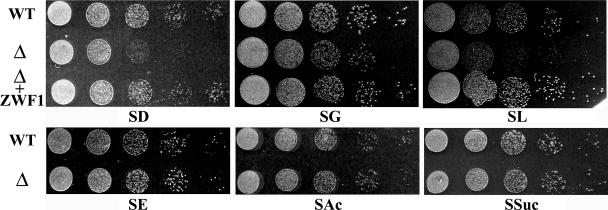
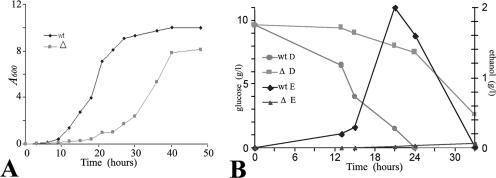
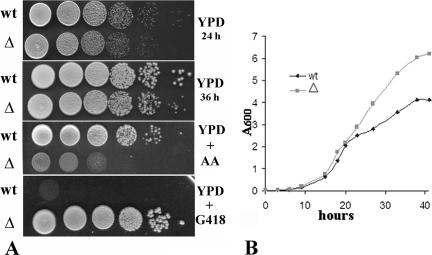
Both the wild type and the null strain were analyzed for the ability to grow in synthetic medium supplemented with carbon sources. In contrast to what has been reported for the corresponding mutant of S. cerevisiae (24), the Klzwf1Δ strain showed slower growth than the parental strain in the presence of fermentative carbon sources, such as glucose (Fig. 1) and fructose (not shown), and also in the presence of respiratory carbon sources, such as lactate and glycerol. To exclude the presence of additional mutations affecting the utilization of these substrates, we reintroduced the wild-type KlZWF1 gene into the Klzwf1Δ strain and, in all cases, observed the recovery of normal growth. No differences between the two strains were observed during growth on acetate and succinate, while on ethanol (SE) the mutant showed a slight growth enhancement that was more evident in rich medium (see below). We then analyzed in more detail the growth differences in glucose of the Klzwf1Δ strain and its parental strains. As shown in the growth curves obtained with rich glucose medium (Fig. 2A), the mutant showed a prolonged lag phase, reaching the stationary phase only after 40 h of cultivation, with a biomass yield that was only 80% that of the parental strain. We also determined the glucose consumption and ethanol production in culture supernatants. As shown in Fig. 2B, wild-type cultures completely exhausted the sugar after 24 h, while the mutant required more than 40 h to achieve the same result. In the latter strain, alcohol production reached a concentration of 0.025 g liter−1 after 33 h of growth, while the wild type produced about 2 g liter−1 after 24 h. These data strongly suggested that the KlZWF1 gene of K. lactis was also required for fermentation. To verify this hypothesis, we checked the ability of the mutant to grow in glucose in the presence of antimycin A, an inhibitor of the electron transport chain (11). As shown in Fig. 3A, the growth of the Klzwf1Δ strain compared to the parental strain was strongly inhibited in the presence of the mitochondrial drug, confirming that the absence of G6PDH impaired glucose fermentation. In agreement with this, it has been reported that the PPP in K. lactis is an important route for the dissimilation of glucose, since in Klpgi1Δ (rag2) mutants, which lack glycolytic phosphoglucose isomerase activity, glucose fermentation becomes respiration dependent (12, 16). When we crossed the Klzwf1Δ strain with the Klpgi1Δ mutant, no spores harboring both mutations were isolated among the meiotic segregants for 20 asci analyzed (not shown), confirming that the block of both glycolysis and PPP was lethal for the cell. The inability of the Klzwf1Δ strain to ferment suggested that, in this strain, glucose was essentially utilized through respiration. For this reason, we analyzed the respiratory capability of the Klzwf1Δ mutant. Cells grown in YPD until the late exponential phase were starved for 24 h, and oxygen consumption was measured in the presence of glucose, ethanol, and lactate. As reported in Table 1, the respiration rate of the mutant with glucose and ethanol was about 70% that of the wild type, while it was 37% with lactate. Consistent with these data, the cytochrome content of the Klzwf1Δ mutant grown in glucose was reduced to 50% to 66% of the wild-type level.
FIG. 1.
Growth of wild type (WT), Klzwf1Δ mutant (Δ), and Klzwf1Δ mutant complemented with the KlZWF1 gene (Δ + ZWF1) on different carbon sources. Plates contained synthetic medium with 2% glucose (SD), 2% glycerol (SG), 2% lactate (SL), 2% ethanol (SE), 1% acetate (SAc), or 1% succinate (SSuc). Each spot was 5 μl of a 10-fold cell suspension dilution series. The initial cell concentration was about 1 × 107 ml−1.
FIG. 2.
(A) Growth curves for wild-type and Klzwf1Δ mutant strains. Cultures were grown in YP medium containing 1% glucose, and cell density (A600) was determined at time intervals. (B) Glucose and ethanol determinations. The concentrations of glucose (D) and ethanol (E) in culture supernatants were determined at the indicated time intervals. wt, wild type; Δ, Klzwf1Δ strain. Each value in the figures represents the average of three independent determinations. In no case was the variation higher than 15%.
FIG. 3.
(A) Growth test of the wild-type and Klzwf1Δ strains on YP plates containing 2% glucose (YPD), 5% glucose plus 5 μM antimycin A (YPD + AA), and 2% glucose plus G418 (YPD + G418). Each spot was 5 μl of a 10-fold cell suspension dilution series. The initial cell concentration was about 1 × 108 ml−1. (B) Growth curves for wild type (wt) and Klzwf1Δ mutant (▵) in YP medium containing 2% ethanol.
TABLE 1.
Respiration rates and cytochrome contentsa
| Strain | O2 consumption (nmol O2 min−1 mg dry mass−1) in presence of:
|
Cytochrome content (μmol g dry mass−1)
|
||||
|---|---|---|---|---|---|---|
| Glucose | Ethanol | Lactate | aa3 | b | c | |
| Wild type | 36 ± 5 | 34 ± 6 | 27 ± 4 | 36 ± 5 | 56 ± 7 | 90 ± 15 |
| Klzwf1Δ mutant | 26 ± 5 (72) | 23 ± 3 (68) | 10 ± 2 (37) | 18 ± 2 (50) | 37 ± 6 (66) | 55 ± 6 (61) |
Data are the averages of three independent determinations. Values in parentheses are percentages relative to the wild-type values.
Growth of the Klzwf1Δ strain on respiratory carbon sources.
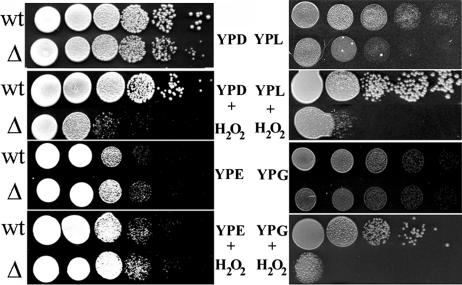
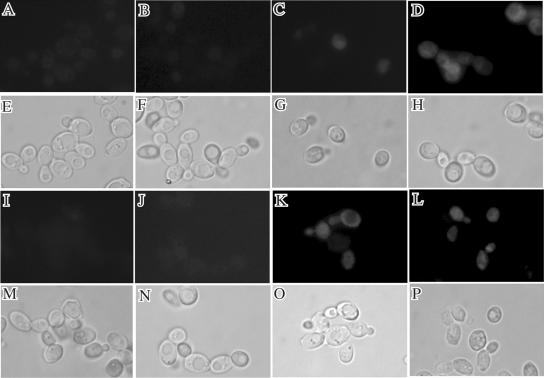
As shown in Fig. 1, the growth of the Klzwf1Δ mutant on ethanol was slightly increased compared to that of the wild type. To confirm this, the two strains were inoculated in YP medium containing 2% ethanol, with the growth monitored by cell density (A600) at time intervals. As shown in Fig. 3B, the Klzwf1Δ mutant showed prolonged exponential growth and increased biomass yield compared to the parent. This unexpected result suggested that KlZWF1 might have a negative effect during growth in ethanol. Since this gene is required for the production of NADPH to maintain the redox balance and for the neutralization of ROS, we tested the sensitivity of the Klzwf1Δ strain to hydrogen peroxide during growth on different carbon sources. As shown in Fig. 4, no differences were observed between the parental strain and the mutant grown on ethanol, while with all the other carbon sources the Klzwf1Δ strain showed increased sensitivity to hydrogen peroxide. These data indicated that, with the exception of growth on ethanol, the amount of NADPH produced in the mutant was not sufficient for cell needs. We wanted to verify if the reduced biomass yield of the Klzwf1Δ strain was caused by accumulation of ROS (32). Wild-type and mutant cells grown in glucose and ethanol until late exponential phase were incubated for 2 h with dihydrorhodamine according to the method of Madeo et al. (21). As shown in Fig. 5, no appreciable fluorescence was detected in the two strains growing exponentially with both carbon sources (A, B, I, and J), while in glucose, after H2O2 treatment, nearly 100% of Klzwf1Δ cells (D) and 50% of wild-type cells (C) produced ROS. The fractions of ROS-positive cells in wild-type and mutant cultures grown in ethanol after hydrogen peroxide addition were comparable (K and L), in agreement with their similar H2O2 sensitivities. These data showed that the reduced biomass yield of the Klzwf1Δ strain with glucose was not due to the accumulation of ROS.
FIG. 4.
Hydrogen peroxide sensitivity test. The wild-type (wt) and Klzwf1Δ (Δ) strains were grown on YP plates containing 2% glucose (YPD), 2% ethanol (YPE), 2% lactate (YPL), and 2% glycerol (YPG) and on the same plates with H2O2 at a concentration of 5 mM. Each spot was 5 μl of a 10-fold cell suspension dilution series. The initial cell concentration was about 1 × 108 ml−1.
FIG. 5.
Wild-type and Klzwf1Δ mutant cells stained with dihydrorhodamine 123. Cells are shown in fluorescence images (A to D and I to L) after 2 h of incubation with DHR and in phase-contrast images (E to H and M to P). The wild-type (A and C) and Klzwf1Δ (B and D) strains were grown in glucose without (A and B) and with (C and D) 5 mM H2O2, or the wild-type (I and K) and Klzwf1Δ (J and L) strains were grown in ethanol without (I and J) and with (K and L) 5 mM H2O2. Results are representative of three experiments.
Transcriptional analysis of genes involved in redox balance.
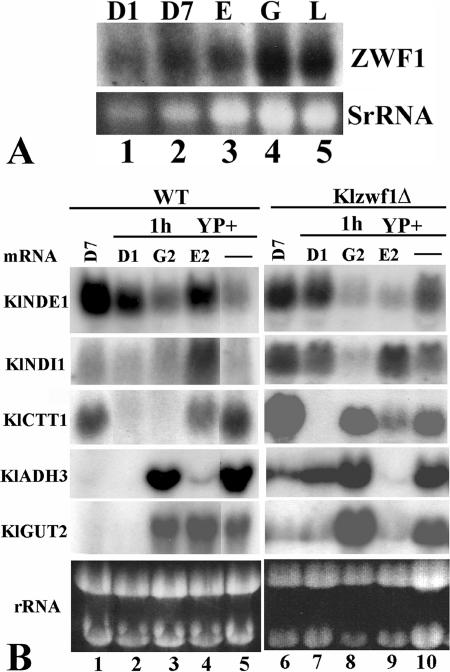
We wanted to verify if the expression of the KlZWF1 gene was under carbon source regulation. Northern analysis was performed with total RNAs prepared from late-exponential-phase cultures grown in YP medium containing different carbon sources. As shown in Fig. 6A, the transcript levels of KlZWF1 were similar in cells grown with low and high glucose concentrations as well as on respiratory carbon sources, indicating that the gene was constitutively transcribed, as reported for the S. cerevisiae ZWF1 gene (24).
FIG. 6.
(A) Transcriptional analysis of KlZWF1. The wild-type strain was grown in YP containing 1% (D1) or 7% (D7) glucose, 2% ethanol (E), 2% lactate (L), or 2% glycerol (G) to the late exponential phase. (B) Transcriptional analysis after substrate shift. The wild-type and Klzwf1Δ strains were grown to the late exponential phase in YP plus 7% glucose. Aliquots of cultures were collected, washed, and regrown for 1 hour in the same volume of YP (−), YP plus 1% glucose (D1), YP plus 2% glycerol (G2), or YP plus 2% ethanol (E2). Total RNA prepared from each culture was probed with the indicated genes. rRNAs are reported as an internal standard.
Since the deletion of KlZWF1 affected both respiration and fermentation and increased cell sensitivity to hydrogen peroxide, we wanted to analyze the expression of genes involved in the control of the cellular redox balance in the mutant. Among the genes analyzed in carbon source shift experiments, we report here those that responded to KlZWF1 deletion, including KlNDE1 and KlNDI1, coding for the external and internal mitochondrium-localized inner membrane trans-dehydrogenases, respectively, the cytoplasmic catalase gene (KlCTT1), the alcohol dehydrogenase gene (KlADH3), and the mitochondrial glycerol-3-phosphate dehydrogenase gene (KlGUT2). Wild-type and Klzwf1Δ cells pregrown in glucose were shifted for 1 h to different carbon sources, and their gene transcription was revealed by Northern analysis. As shown in Fig. 6B, we observed higher transcription levels of the KlNDI1, KlADH3, and KlGUT2 genes in the Klzwf1Δ strain grown on glucose (lanes 6 and 7) than in the wild type (lanes 1 and 2), while KlCTT1 was expressed more only in 7% glucose (lanes 1 and 6). Since KlNDI1, KlADH3, and KlGUT2 code for mitochondrium-localized activities devoted to the reoxidation of reducing equivalents, this suggested rerouting of the glucose intermediate flux into these organelles in the Klzwf1Δ strain. We also observed a strong induction of the KlCTT1 gene and an increased amount of the KlGUT2 transcript in the mutant grown with glycerol (lane 8). In contrast, in the presence of ethanol, both KlNDE1 and KlGUT2 transcripts were highly reduced in the mutant (lane 9) compared to those in the wild type (lane 4). It is noteworthy that with glucose and glycerol, in which the Klzwf1Δ strain showed reduced growth, we observed increased transcription of KlADH3, KlNDI1, KlCTT1, and KlGUT2, suggesting the activation of alternative routes to balance the absence of G6PDH. After the shift to ethanol, however, in which the Klzwf1Δ strain had a biomass yield higher than that of the wild type, we observed decreased transcription of KlNDE1 and KlGUT2.
G6PDH activity.
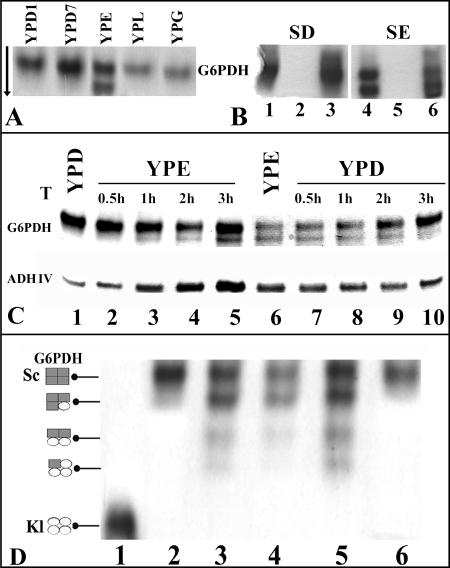
Cellular extracts prepared from the parental and mutant strains grown on fermentative and respiratory carbon sources were separated in native polyacrylamide gels. To detect the G6PDH activity, we developed a procedure modified from the ADH assay (see Materials and Methods) (20, 30). This assay revealed a single band of activity in wild-type extracts from glucose, glycerol, lactate (Fig. 7A), and acetate (not shown) cultures, while in the extracts from ethanol cultures, we unexpectedly found two bands of activity. The two bands were also present when ethanol was used together with other carbon sources, indicating a dominant effect of ethanol (not shown). No activity could be detected in the extracts from Klzwf1Δ mutant cultures grown in glucose and ethanol (Fig. 7B, lanes 2 and 5) or with any other carbon source (not shown). When we reintroduced the KlZWF1 gene into the Klzwf1Δ mutant, we observed the same pattern of G6PDH activity as that shown by the wild type both in glucose (Fig. 7B, lanes 1 and 3) and in ethanol (lanes 4 and 6), confirming that the two bands of activity were the products of the KlZWF1 gene. In carbon source shift experiments, we found that the faster-migrating G6PDH form appeared in K. lactis cells about 1 hour after the shift from glucose to ethanol and increased in intensity during the following 3 hours (Fig. 7C, lanes 1 to 5). For the opposite shift, from ethanol to glucose, we observed a decrease in the intensity of the faster-migrating G6PDH band in the first 2 hours and its complete disappearance after 3 hours, with a concomitant increase of the upper band (Fig. 7C, lanes 6 to 10). Interestingly, the induction of the faster-migrating band on ethanol (Fig. 7C, lanes 1 to 5) paralleled that of KlAdh4p, the mitochondrium-localized alcohol dehydrogenase reported to be induced by ethanol (23, 30). Crystal structure and biochemical analyses have shown that the human native enzyme exists in a dimer-tetramer equilibrium (3, 29, 33). The presence of two G6PDH forms in ethanol-grown cells raised the question of whether the two bands detected in K. lactis corresponded to dimer and tetramer forms. Since K. lactis and S. cerevisiae G6PDH proteins have nearly 70% identity, we asked if the expression of the K. lactis gene in a wild-type S. cerevisiae strain, generating heteromultimers with new migrating properties, would help to identify the tetramer and dimer forms. Extracts prepared from three independent transformants of S. cerevisiae harboring the KlZWF1 gene showed the presence of at least four bands of G6PDH (Fig. 7D, lanes 3 to 5) that migrated between the positions of the K. lactis (lane 1) and S. cerevisiae (lane 2) activities. These bands should correspond to combinations of the proteins of the two yeast species according to the scheme of Fig. 7D, although the slower-migrating band that corresponded to the K. lactis tetramer could not be detected (lanes 3 to 5). The latter result could be due to reduced expression of the K. lactis KlZWF1 gene in S. cerevisiae. In extracts of untransformed cells of S. cerevisiae prepared from all tested carbon sources, only one band of activity was ever present (lanes 2 and 6), indicating that the formation of the hypothetical dimeric form of G6PDH in K. lactis did not occur in S. cerevisiae.
FIG. 7.
Native G6PDH activities separated in acrylamide gels. (A) Extracts were prepared from wild-type cultures grown on YP containing 1% (YPD1) or 7% (YPD7) glucose, 2% ethanol (YPE), 2% lactate (YPL), or 2% glycerol (YPG). (B) G6PDHs from the wild type (lanes 1 and 4), the Klzwf1Δ strain (lanes 2 and 5), and the Klzwf1Δ strain transformed with KlZWF1 (lanes 3 and 6). The cultures were grown for 3 days in synthetic medium containing glucose (SD) or ethanol (SE). The arrow indicates the direction of protein migration. (C) G6PDH and ADH isoforms during substrate shift. Thirty-milliliter samples of wild-type cultures in YPD and YPE were grown to the late exponential phase. Cells were collected, washed, and suspended in the same volume of YPE and YPD, respectively. Samples from the shifted cultures, incubated with the new carbon sources, were taken after 0.5, 1, 2, and 3 h of growth. Cell extracts were fractioned in native gels and stained for G6PDH and ADH. (D) G6PDH from the wild-type S. cerevisiae strain harboring KlZWF1. The gel shows native G6PDHs from K. lactis (lane 1) and S. cerevisiae (lanes 2 and 6) wild-type strains and three independent clones of S. cerevisiae transformed with the KlZWF1 gene (lanes 3, 4, and 5). Extracts were prepared from cultures grown for 3 days in synthetic medium containing glucose (SD) (lanes 1 to 5) or ethanol (SE) (lane 6). The scheme on the left indicates the putative compositions of tetramers. The enzyme activities were visualized as described in Materials and Methods. Each slot was loaded with 10 μg of protein.
DISCUSSION
The PPP has been described as an alternative and relevant route for the dissimilation of glucose in K. lactis (12, 13, 16), but despite these reports, the role of this pathway has not been analyzed in detail. For this reason, we focused our attention on KlZWF1, the gene encoding G6PDH, the first enzyme of the oxidative part of the PPP. Like ZWF1 of S. cerevisiae, KlZWF1 was constitutively expressed on fermentative and respiratory carbon sources, and the deletion of the gene led to increased sensitivity to hydrogen peroxide (24, 37). Differently from the case in S. cerevisiae, the deletion of KlZWF1 led to reduced biomass yields in glucose, lactate, and glycerol and, in contrast, to increased yields in ethanol, indicating different roles of this gene in the utilization of carbon sources in the two yeast species.
Glucose and ethanol metabolism in the Klzwf1Δ mutant.
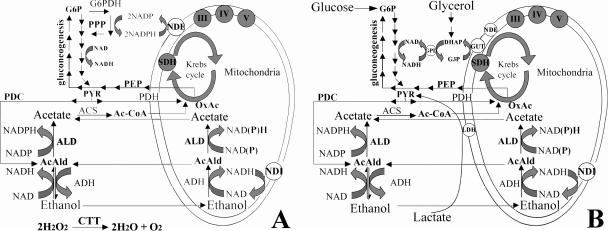
It has been reported that K. lactis mitochondria can directly oxidize NADPH (13, 26) due to a transdehydrogenase protein, encoded by KlNDE1, located on the external side of the inner mitochondrial membrane that, in S. cerevisiae, can accept only NADH as a cofactor (35, 36). This protein's activity then feeds the reduced equivalents to the electron transport chain. The smaller amounts of NADPH produced in glucose in the Klzwf1Δ mutant might explain the observed prolonged lag phase of growth and the reduced sugar consumption and respiration. In addition, the increased sensitivity to antimycin A and the very small amounts of ethanol produced indicated that the glycolytic flux in the mutant was reduced. These results suggested that other sources of NADPH present in the Klzwf1Δ mutant were not sufficient for cell growth and that only when enough intermediates were accumulated could alternative activities involved in the generation of NAD(P)H resume growth. The induction in Klzwf1Δ cells growing in glucose of the KlADH3, KlNDI1, and KlGUT2 genes, which are poorly expressed in the wild-type strain grown on this carbon source, point to their mitochondrial activities being among those that contribute to maintaining the redox balance (Fig. 8). It has been reported that the deletion of KlPGI1 (RAG2) led to the activation of the KlADH3-dependent ethanol/acetaldehyde shuttle during growth in glucose (26), and our results indicated that the block of PPP leads to the activation of the same shunt, suggesting that in K. lactis, glycolysis and PPP are both required for ethanol accumulation. In the scheme of Fig. 8, which summarizes our data, we report the role that G6PDH has in the wild-type strain. In lag and early log phases of growth, when the oxygen concentration in the culture is high, glucose is oxidized via pyruvate dehydrogenase into the Krebs cycle. The oxidative part of the PPP is the major source of NADPH for reductive biosynthesis, while the reoxidation of NADH, produced in the first part of glycolysis, occurs via the KlAdh3-dependent acetaldehyde/ethanol (26, 30) and/or dihydroxyacetone-P/glycerol-3P (27) shuttle. In stationary phase, when oxygen in the culture becomes limiting, glucose is oxidized through pyruvate decarboxylase, and the neutral redox balance is maintained by the accumulation of ethanol (Fig. 2B). Since K. lactis cultures did not have a diauxic curve, glucose and ethanol are both utilized at the same time. The ADH responsible for the oxidation of ethanol is most probably the mitochondrium-localized KlAdh4 protein (22, 30), as suggested by the induction of this protein's activity on this carbon source (Fig. 7C). Sequence analysis of the K. lactis genome has shown the presence of many hypothetical genes for acetaldehyde dehydrogenase (ALD) that, like ALD6 of S. cerevisiae (14, 23), might use NADP as a cofactor. Therefore, the acetaldehyde produced from ethanol in the mitochondria, which freely diffuses through mitochondrial membranes into the cytoplasm (26), can be converted to acetate, producing NADPH (Fig. 8A). We suggest that the production of excess NADPH by the contemporary expression of G6PDH and acetaldehyde activities leads to redox imbalance. In S. cerevisiae, which has a diauxic curve, the contemporary expression of these activities may be avoided (14, 23). Similarly, during growth of the wild type in ethanol (Fig. 8), in addition to the NADPH produced by ALD activity, part of the glucose-6-P produced during gluconeogenesis might be rerouted towards the G6PDH of the PPP, with the consequent production of an excess of NADPH that is partly reoxidized by KlNde1. In the mutant (Fig. 8B), the lack of G6PDH activity blocked the production of NADPH through the PPP but also blocked the transcription of KlNDE1 and KlGUT2 (Fig. 6), suggesting that the intracellular levels of reduced cofactors might also regulate the expression of genes/activities directly responsible for its reoxidation (27). It follows that KlNde1 and KlGut2 in the wild type participated in reoxidation of the NADPH excess and of gluconeogenesis intermediates, respectively. This indicates that all the glucose-6-P produced in the mutant was converted to biomass, explaining the higher cellular yield with a lower respiration rate obtained in ethanol. Since G6PDH activity is under very strong feedback inhibition by NADPH (33), we can imagine that in the presence of high levels of reduced cofactors, a fraction of the tetramers can be reduced and dissociate to form active dimers. It has been reported that G6PDH dimers and tetramers in human cells are functionally equivalent (3, 9), and at present, we have no evidence of whether the situation in K. lactis might be similar. In contrast to the G6PDH of S. cerevisiae, which has only one cysteine (Cys), the K. lactis protein has four Cys residues, and it has been reported that human G6PDH has a disulfide bond between Cys13 and Cys446 (3, 9). Moreover, reversible thiol-disulfide interchanges seem to regulate the activities of several chloroplast enzymes (39). We might suggest that, in K. lactis, the equilibrium between G6PDH tetramers/dimers and monomers, with the latter having no activity, might be due to cysteine modifications and that this mechanism might account for the quick response of cells to the NADP(H) and/or other reducing equivalent imbalance.
FIG. 8.
Schematic model of metabolism in the K. lactis wild-type (A) and Klzwf1Δ (B) strains. All reactions of glycolysis from glucose-6-phosphate (G6P) to pyruvate (PYR) and in gluconeogenesis from oxaloacetate (OxAc) and phosphoenolpyruvate (PEP) to G6P are indicated by series of arrows. Ac-CoA, acetyl-coenzyme A; AcAld, acetaldehyde; ACS, acetate synthase; ADH, alcohol dehydrogenase; ALD, aldehyde dehydrogenase; GPD, NAD-dependent glycerol-3-P dehydrogenase; GUT, FAD-dependent glycerol-3-P dehydrogenase; NDE and NDI, external and internal inner mitochondrial membrane transdehydrogenases, respectively; LDH, cytochrome c-dependent dl-lactate dehydrogenase; PDC and PDH, pyruvate decarboxylase and dehydrogenase, respectively; PPP, pentose phosphate pathway; SDH, succinate dehydrogenase complex; III, IV, and V, respiratory chain complexes. Glycerol and lactate entry into metabolism and the dihydroxyacetone-P (DHAP)/glycerol-3-P (G3P) shuttle are indicated only in panel B for simplicity.
Lactate and glycerol metabolism in the Klzwf1Δ mutant.
The reduced growth of the Klzwf1Δ mutant on lactate and glycerol, together with the increased sensitivity to hydrogen peroxide and the reduced respiration, indicated that, as observed in glucose, the NADPH levels produced by the oxidative part of the PPP were not sufficient for cell metabolism (Fig. 8A). Therefore, in wild-type cells, the glucose-6-phosphate produced by gluconeogenesis from lactate and glycerol, as described for ethanol, may be partially oxidized via the PPP to generate the NADPH required for assimilatory reactions, to feed the respiratory chain through KlNde1, and for ROS detoxification (35, 36). Indeed, Klzwf1Δ cells grown in glycerol showed high levels of the catalase (KlCTT1) transcript that were not observed in the wild-type strain. Moreover, we also found increased transcription of KlGUT2 (Fig. 6), one component of the dihydroxyacetone-P/glycerol-3P shuttle, that may intervene to compensate the redox imbalance that follows the deletion of KlZWF1 (Fig. 8A). We have previously shown that K. lactis Klsdh1Δ mutants devoid of succinate dehydrogenase activity were still able to grow in lactate (31). The requirement of G6PDH for growth on lactate is also supported by the observation that the capability of Klsdh1Δ mutants to grow on this substrate (31) was lost in Klzwf1Δ Klsdh1Δ double mutants (not shown).
Acknowledgments
We thank H. Y. Steensma (University of Leiden) for providing the GG1996 strain.
This work was funded by grant Ateneo 2005 from the University of Rome La Sapienza. We thank the Istituto Pasteur—Fondazione Cenci Bolognetti for providing Ilaria De Maria with a short fellowship.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Aguilera, A. 1986. Deletion of the phosphoglucose isomerase structural gene makes growth and sporulation glucose dependent in Saccharomyces cerevisiae. Mol. Gen. Genet. 204:310-316. [DOI] [PubMed] [Google Scholar]
- 2.Au, S. W., C. E. Naylor, S. Gover, L. Vandeputte-Rutten, D. A. Scopes, P. J. Mason, L. Luzzatto, V. M. Lam, and M. J. Adams. 1999. Solution of the structure of tetrameric human glucose 6-phosphate dehydrogenase by molecular replacement. Acta Crystallogr. D 55:826-834. [DOI] [PubMed] [Google Scholar]
- 3.Au, S. W., S. Gover, V. M. Lam, and M. J. Adams. 2000. Human glucose-6-phosphate dehydrogenase: the crystal structure reveals a structural NADP+ molecule and provides insights into enzyme deficiency. Structure 8:293-303. [DOI] [PubMed] [Google Scholar]
- 4.Bakker, B. M., K. M. Overkamp, A. J. van Maris, P. Kotter, M. A. Luttik, J. P. van Dijken, and J. T. Pronk. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:15-37. [DOI] [PubMed] [Google Scholar]
- 5.Becker, D. M., and L. Guarente. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194:182-187. [DOI] [PubMed] [Google Scholar]
- 6.Beutler, E. 1994. G6PD deficiency. Blood 84:3613-3636. [PubMed] [Google Scholar]
- 7.Boles, E., W. Lehnert, and F. K. Zimmermann. 1993. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 217:469-477. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, D. T. Y., J. M. Ou, S. W. Huang, and K. J. Tsai. 1996. The functional role of the eight cysteines in human glucose-6-phosphate dehydrogenase (G6PD). Blood 88:1213-1221. [Google Scholar]
- 10.Ferrero, I., A. M. Viola, and A. Goffeau. 1981. Induction by glucose of an antimycin-insensitive, azide-sensitive respiration in the yeast Kluyveromyces lactis. Antonie Leeuwenhoek 47:11-24. [DOI] [PubMed] [Google Scholar]
- 11.Goffrini, P., A. A. Algeri, C. Donnini, M. Wésolowski-Louvel, and I. Ferrero. 1989. RAG1 and RAG2: nuclear genes involved in the dependence/independence on mitochondrial respiratory function for growth on sugar. Yeast 5:99-105. [DOI] [PubMed] [Google Scholar]
- 12.Goffrini, P., M. Wésolowski-Louvel, and I. Ferrero. 1991. A phosphoglucose isomerase gene is involved in the Rag phenotype of the yeast Kluyveromyces lactis. Mol. Gen. Genet. 228:401-409. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez Siso, M. I., M. A. Freire Picos, and M. E. Cerdan. 1996. Reoxidation of the NADPH produced by the pentose phosphate pathway is necessary for the utilization of glucose by Kluyveromyces lactis rag2 mutants. FEBS Lett. 387:7-10. [DOI] [PubMed] [Google Scholar]
- 14.Grabowska, D., and A. Chelstowska. 2003. The ALD6 gene product is indispensable for providing NADPH in yeast cells lacking glucose-6-phosphate dehydrogenase activity. J. Biol. Chem. 278:13984-13988. [DOI] [PubMed] [Google Scholar]
- 15.Izawa, S., K. Maeda, T. Miki, Y. Inoue, and A. Kimura. 1998. Importance of glucose-6-phosphate dehydrogenase in the adaptive response to hydrogen peroxide in Saccharomyces cerevisiae. Biochem. J. 330:811-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby, J., C. P. Hollenberg, and J. J. Heinisch. 1993. Transaldolase mutants in the yeast Kluyveromyces lactis provide evidence that glucose can be metabolized through the pentose phosphate pathway. Mol. Microbiol. 10:867-876. [DOI] [PubMed] [Google Scholar]
- 17.Juhnke, H., B. Krems, P. Kotter, and K. D. Entian. 1996. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. 252:456-464. [DOI] [PubMed] [Google Scholar]
- 18.Kletzien, R. F., P. K. W. Harris, and L. A. Foellmi. 1994. Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB J. 8:174-181. [DOI] [PubMed] [Google Scholar]
- 19.Lodi, T., and I. Ferrero. 1993. Isolation of the DLD1 gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme d-lactate ferricytochrome c oxidoreductase. Mol. Gen. Genet. 238:315-324. [DOI] [PubMed] [Google Scholar]
- 20.Lutstorf, U., and R. Megnet. 1968. Multiple forms of alcohol dehydrogenase in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 126:933-944. [DOI] [PubMed] [Google Scholar]
- 21.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. Sigrist, D. H. Wolf, and K.-U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazzoni, C., M. Saliola, and C. Falcone. 1992. Ethanol-induced and glucose-insensitive alcohol dehydrogenase in the yeast Kluyveromyces lactis. Mol. Microbiol. 6:2279-2286. [DOI] [PubMed] [Google Scholar]
- 23.Minard, K. I., and L. McAlister-Henn. 2005. Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 280:39890-39896. [DOI] [PubMed] [Google Scholar]
- 24.Nogae, I., and M. Johnston. 1990. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene 96:161-169. [DOI] [PubMed] [Google Scholar]
- 25.Notaro, R., A. Afolayan, and L. Luzzatto. 2000. Human mutations in glucose-6-phosphate dehydrogenase reflect evolutionary history. FASEB J. 14:485-494. [DOI] [PubMed] [Google Scholar]
- 26.Overkamp, K. M., B. M. Bakker, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 2002. Two mechanisms for oxidation of cytosolic NADPH by Kluyveromyces lactis mitochondria. Yeast 19:813-824. [DOI] [PubMed] [Google Scholar]
- 27.Rigoulet, M., H. Aguilaniu, N. Avéret, O. Bunoust, N. Camougrand, X. Grandier-Vazeille, C. Larsson, I. L. Pahlman, S. Manon, and L. Gustafsson. 2004. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biochem. 256/257:73-81. [DOI] [PubMed] [Google Scholar]
- 28.Rothstein, R. 1983. One step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 29.Rowland, P., A. K. Basak, S. Gover, H. R. Levy, and M. J. Adams. 1994. The three dimensional structure of glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides refined at 2.0 A. Structure 2:1073-1087. [DOI] [PubMed] [Google Scholar]
- 30.Saliola, M., and C. Falcone. 1995. Two mitochondrial alcohol dehydrogenase activities of Kluyveromyces lactis are differentially expressed during respiration and fermentation. Mol. Gen. Genet. 249:665-672. [DOI] [PubMed] [Google Scholar]
- 31.Saliola, M., P. C. Bartoccioni, I. De Maria, T. Lodi, and C. Falcone. 2004. The deletion of the succinate dehydrogenase gene KlSDH1, in Kluyveromyces lactis, does not lead to respiratory deficiency. Eukaryot. Cell 3:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz, J. B., M. Weller, and T. Klockgether. 1996. Potassium deprivation-induced apoptosis of cerebellar granule neurons: a sequential requirement for new mRNA and protein synthesis, ICE-like protease activity, and reactive oxygen species. J. Neurosci. 16:4696-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scopes, D., J. M. Bautista, C. E. Naylor, M. J. Adams, and P. J. Mason. 1998. Amino acid substitutions at the dimer interphase of human glucose-6-phosphate dehydrogenase that increase thermostability and reduce the stabilising effect of NADP. Eur. J. Biochem. 251:382-388. [DOI] [PubMed] [Google Scholar]
- 34.Steensma, H. Y., and J. J. M. Ter Linde. 2001. Plasmids with the Cre recombinase and the dominant nat marker, suitable for use in prototrophic strains of Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast 18:469-472. [DOI] [PubMed] [Google Scholar]
- 35.Tarrio, N., S. Diaz Prado, M. E. Cerdan, and M. I. Gonzalez Siso. 2005. The nuclear genes encoding the internal (KlNDI1) and external (KlNDE1) alternative NAD(P)H:ubiquinone oxidoreductases of mitochondria from Kluyveromyces lactis. Biochim. Biophys. Acta 1707:199-210. [DOI] [PubMed] [Google Scholar]
- 36.Tarrio, N., M. Becerra, M. E. Cerdan, and M. I. Gonzalez Siso. 2006. Reoxidation of cytosolic NADPH in Kluyveromyces lactis. FEMS Yeast Res. 6:371-380. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, D., H. Cherest, and Y. Surdin-Kerian. 1991. Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 10:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 39.Wenderoth, I., R. Scheibe, and A. von Schaewen. 1997. Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. J. Biol. Chem. 272:26985-26990. [DOI] [PubMed] [Google Scholar]